More Information
Submitted: November 08, 2022 | Approved: December 28, 2022 | Published: December 29, 2022
How to cite this article: Zehra M, Parveen R, Irfan M, Nasir M, Bashir S, Forensic analysis of raw meat adulteration using mtDNA. J Forensic Sci Res. 2022; 6: 088-092.
DOI: 10.29328/journal.jfsr.1001041
Copyright License: © 2022 Zehra M, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Forensic analysis of raw meat adulteration using mtDNA
Marium Zehra1*, Rukhsana Parveen2, Muhammad Irfan1, Mahrukh Nasir1 and Sidra Bashir2
1Jamil-ur-Rahman Center for Genome Research, International Center for Chemical and Biological Sciences (ICCBS), University of Karachi, Karachi, Pakistan
2Center for Applied Molecular Biology (CAMB), University of Punjab, Lahore, Pakistan
*Address for Correspondence: Marium Zehra, Jamil-ur-Rahman Center for Genome Research, International Center for Chemical and Biological Sciences (ICCBS), University of Karachi, Karachi, Pakistan, E-mail: mariumzehra999@gmail.com
Meat species identification has become essential with the increasing events of frauds like the illegal slaughter of cows, meat adulteration, and substitution. Food scam directly influences public well-being, trade, and wildlife. In Pakistan, donkey meat is used as adulterants for cow meat and is considered Haram in Islamic concepts. In this study PCR, based detection methods are used for identification purposes. The mitochondrial gene cytochrome b has been used in this study to identify the origin of meat specie. Specie-specific primers of cyt b of cow and donkey were used for identification. DNA from different binary ratios of cow and donkey meat was extracted by the phenol-chloroform method. Ratios were made from 1-10 and extracted DNA was subjected to PCR to amplify the target fragment of the cyt b gene. Primers were sensitive to identifying species origin in all meat ratios. Multiplex PCR was designed to identify both species and the results were analyzed by gel electrophoresis. Fragment size of 309bp for cow and 475bp for donkey was observed. Results of the current study conclude that PCR assays, including multiplex PCR, is efficient and has a high sensitivity for even small amount of meat. It is concluded that multiplex PCR is useful and reliable for adulterated meat detection.
Food fraud is a hot topic nowadays and is growing with each passing day. Developing countries highly suffer from the problem. Different types of food are adulterated across the world e.g. spices, cereals, fruits, milk, oil, ghee, bakery products, vegetables, honey, fish, honey, rice, and especially meat [1-3]. Meat is a vital part of human nutrition as it is rich in protein and other important components needed for the human body. Due to ever increasing demand for meat and decreasing meat availability rate of meat adulteration and substitution has been raised. Adulteration of meat has serious issues related to health, religion, ethics, and [4-6]. Pakistan is a Muslim-majority country that follows dietary rules according to Islam. Meat adulteration is a serious issue majorly affecting Muslims. Muslims eat halal meat only and meat from haram organisms e.g. pig, dog, donkey, cat, etc. is prohibited in Islam [7]. These organisms also are potential carriers of different zoonotic diseases [8]. This issue has created a red alert for Muslims and authenticity and identification of food ingredients become necessary before food consumption [9].
Meat when sold fresh can be identified and differentiated easily, however, cooked, processed, and minced meat cannot be differentiated in the final product so the rate of adulteration is also high [10]. Cases of meat adulteration are increasing with time and it is now necessary to solve this problem and protect consumers and producers from falling into this trap of meat adulteration. As Pakistan is a Muslim country, its citizens also need certification of the product they consume. The components utilized in food cannot be distinguished & discriminated by simple visual inspection, hence sophisticated molecular procedures are needed [11,12].
Different methods and analytical research have been done to identify meat origin and to investigate food adulteration. Methods to detect meat adulteration are often based on physical, chemical, biological, and other factors. Often for food detection, two types of methods are used (a) protein-based and (b) DNA-based [13,14].
Protein-based methods include electrophoretic assay, sodium lauryl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) [14,15], immunological techniques [16], immunosorbent assay [17,18], chromatographic techniques [19] and mass spectrometry techniques. While DNA-based techniques include polymerase chain reaction (PCR) and real-time PCR (RT-PCR) [16].
Despite various advantages, protein analysis has certain limitations. Although protein-based methods are effective in fresh meat analysis protein denatures easily by salts, heat, and pressure and thus has low effectiveness in highly processed food [10]. Thermally treated products have denatured protein which causes hindrances in protein analysis. Immunoassay analysis is based on antibodies against different proteins while cross-reaction occurs among different species [9]. These limitations lead us toward DNA analysis. DNA is a stable molecule and is abundantly present as compared to proteins. DNA is identically present in each cell of the organism, so any part of the organism can be used as a source. These properties make DNA a good source for different identification processes. The degeneracy of DNA is a plus point for the identification and discrimination of species by DNA analysis [12].
DNA-based techniques include polymerase chain reaction (PCR) and DNA hybridization. DNA hybridization is a process of hybridization of labeled DNA probes with DNA of the source linked covalently to a membrane. This is identified by different detection techniques such as DNA microarray and real-time PCR [20,21].
PCR is preferable over other techniques because of its certain properties i.e. specific, sensitive, less time-consuming, and easily employed compared to other techniques [22]. There are 2 types of DNA targets used in PCR chromosomal DNA and mitochondrial DNA. These both are present inside the cell. Mitochondrial DNA (mt-DNA) is in majority as compared to chromosomal DNA and mostly specie specific identification methods are concerned with it as it contains marker genes that help in the identification of organisms [23]. Mt DNA is abundant and has a high copy number in contrast to nuclear DNA because each mitochondrion has 2-10 Mt DNA present in it. Mitochondrion presence in each cell is ample almost 1000 mitochondrion per cell therefore it is very useful for processed meat analysis and short sample size [23,24]. Its evolution rate is also high which helps in distinguishing closely related organisms also [24,25]. Cytochrome b gene is a mitochondrial gene used as a marker for identification purposes [26]. Its interspecific and intraspecific variations help in the identification of the organism and also discriminate closely related organisms [27].
Our study has been undertaken for the identification of raw meat using the mitochondrial gene i.e. cyt b gene. This is due to the increase in meat fraud in Pakistan.
In the current study, we collected the Cow meat sample from an authentic slaughterhouse while donkey meat was collected from the University of Veterinary & Animal sciences UVAS Lahore. Meat samples were stored at -20 °C till further processing.
DNA extraction
Genomic including mitochondrial was isolated from meat samples by using the CTAB method with slight modification. Meat samples from both species with various binary ratios (1:0, 1:1,1:2, 1:4, 1:6, 1:8,1:10, 1:50, and 1:100) were minced and used as starting material for DNA isolation. Approximately 250 mg of minced meat was transferred to the micro-centrifuge tube. The lysis buffer (400 μl) lysis buffer and protein K (10 μl) was added to the tube and incubated at 56 °C overnight. Phenol Chloroform and isoamyl alcohol (PCI) with a 25:24:1 ratio were added to the tube and briefly vortexed followed by centrifugation at 1000rpm for five min. The upper layer containing the aqueous phase was transferred to the new tube. Pre-chilled ethanol was added and the tube was kept at -20 °C for 15 minutes. The tube was centrifuged and the DNA pellet was washed with 70% ethanol two times. The DNA pellet was dissolved in IX TE buffer. The quality of DNA was evaluated by 1% Agarose gel electrophoresis and the concentration of DA was estimated photometrically. DNA samples were stored at -20oC till further processed.
Specie-specific primers of the mitochondrial cyt b gene were designed for DNA amplification by using Primer3. Two different specific primer pairs were designed (Table 1). Features and specificity of designed primers were checked by Primer-BLAST NCBI. The synthesized primers were lyophilized to 100 pmole in IX TE buffer.
| Table 1: Primer sequences of Cytochrome b gene. | ||||
| Organism Names | Primers | Sequences | No of Bases. | Product length |
| COW | F1 | TACTATTCGCACCCGACCTC | 20 | 309bp |
| R1 | GGTGTTCGACTGGTTGTCCT | 20 | ||
| DONKEY | F2 | ATCAGCAATCCCCTACATCG | 20 | 475bp |
| R2 | GTGTAGGGTGGGGATGAGT | 19 | ||
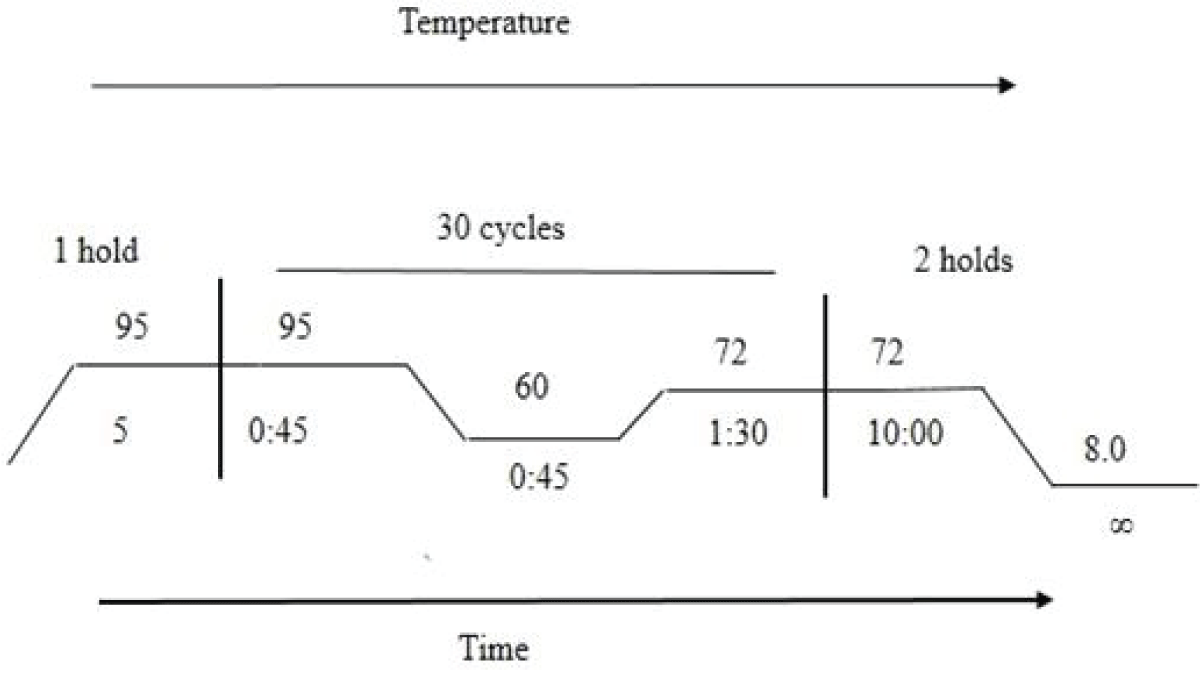
The targeted region of both species was amplified by PCR. The primer’s optimum annealing temperature was done by using gradient PCR and it was noted that both sets of primers were best annealed at 60 °C. For amplification, 07 μL PCR Master mix (2X) (Thermo Fisher Scientific) containing Taq DNA polymerase (0.05 U/μL), 4 mM MgCl2, 0.4 mM of each dNTP and reaction buffer, 1.0 μL each of forward and reverse primer, forward and reverse primers with 10 pmole, genomic DNA (50 ng) were into the PCR tube and total volume was adjusted to 15 μL with nuclease-free water. The temperature conditions; initial denaturation at 95 °C for 5min followed by 30 cycles of 95 °C for 0:45s, 60 °C for 0:45s, 72 °C for 1 min, and final extension for 10 min at 72 °C and infinite at 8 °C (Figure 1).
Figure 1: Graphical representation of PCR cycling condition.
The amplified products were evaluated by using 2% agarose gel electrophoreses.
Multiplex PCR was designed for both species cow and donkey in a single tube containing the accurate amount of primers and DNA. Multiplex PCR was applied on different binary ratios to check the sensitivity level land detection limit. The aim was to identify both organisms in a single reaction. This is helpful for the detection of meat adulteration in the meat mixture.
Genomic DNA extraction
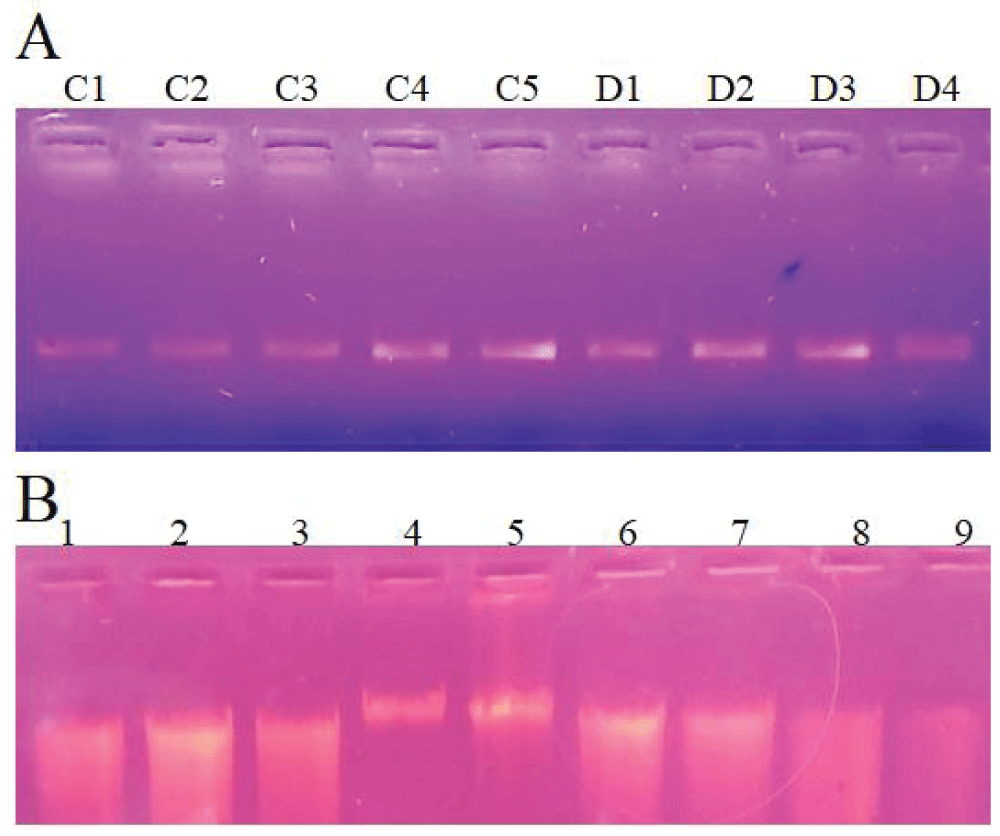
DNA was extracted from both cow and donkey with meat ratios including; 1:0, 1:1,1:2, 1:4, 1:6, 1:8,1:10, 1:50, and 1:100. DNA was also extracted separately from both species for the evaluation of the sensitivity level. The isolated DNA from both species was evaluated by Agarose gel electrophoresis. DNA isolated from the species-specific meat sample is visualized as intact DNA bands (Figure 2A) while DNA extracted from the mixed meat samples of both species with the binary ratio is shown in the smear as well as the intact band (Figure 2B).
Figure 2: Isolated DNA visualization on Agarose gel. Figure A illustrate the DNA extracted from both species. Sample C1-C5 represents the DNA isolated from cow meat while D1-D4 represents the sample from donkey meat. Figure B represents the DNA isolated from minced meat samples with binary ratios of both species. Lane 1-4 represents Cow: Donkey mixed DNA sample with 1:5, 1:10, 1:10, and 1:0 ratio. Lane 5-9 represents Donkey: Cow mixed DNA sample with 1:0, 1:5, 1:10, 1:50, and 1:100 ratio.
Amplified product gel estimation
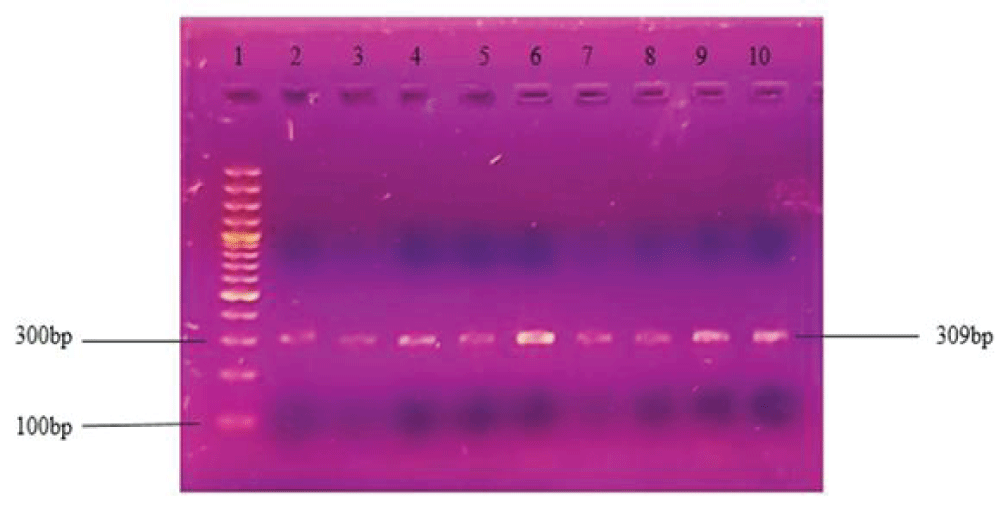
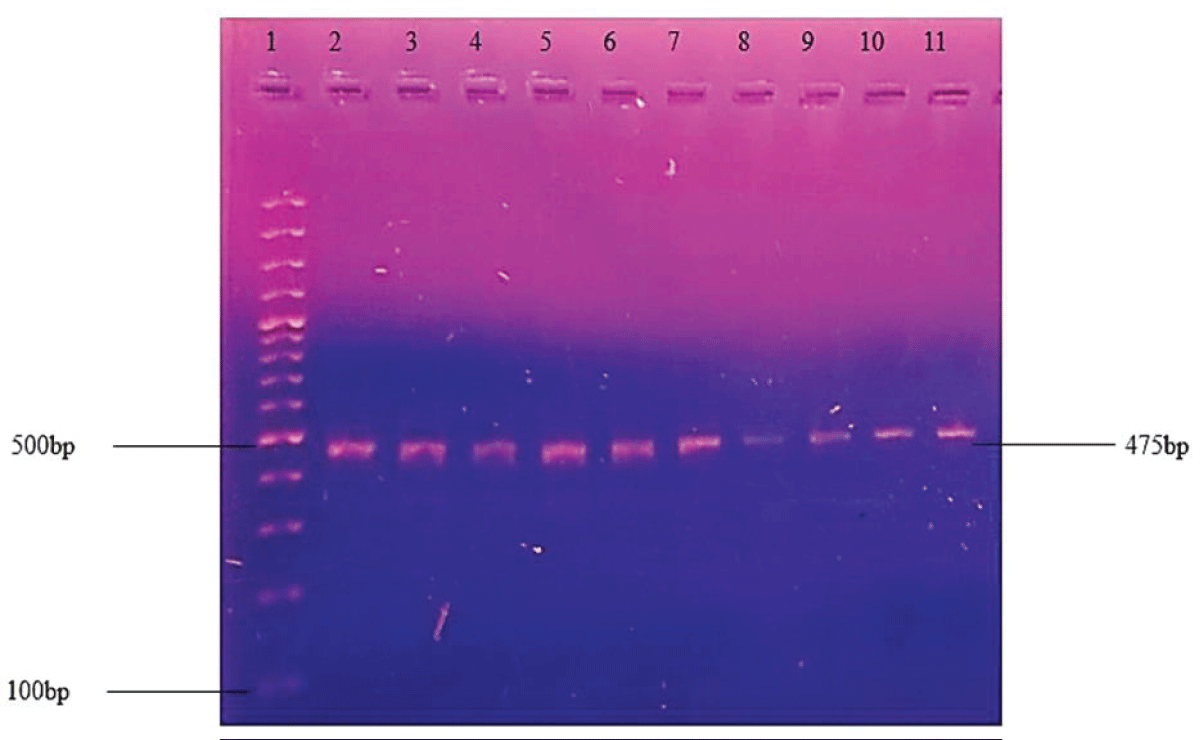
DNA amplification through PCR was optimized to get the best bands of the desired products. Different conditions were changed i.e. annealing temperature, the concentration of Taq polymerase, primers concentration, and the master mixture was changed. The best results were obtained at 60 °C along with the Hot-start master mixture buffer and Taq polymerase of ThermoFisher USA. Donkey and cow ratios were amplified separately with individual primers respectively. The amplification of the targeted mitochondrial cyt b gene from the cow meat sample represents the PCR band of 309 bp (Figures 3,4).
Figure 3: PCR product of mitochondrial cyt b gene of cow meat ratios. Checking sensitivity of cow primers in the binary ratio of cow: donkey. Lane 1 contains a 100bp standard ladder and lane 2-10 represents PCR products of samples having the ratio 1:0, 1:1,1:2, 1:4, 1:6, 1:8,1:10, 1:50, and 1:100 of Cow: Donkey respectively.
Figure 4: PCR amplified product of cyt b gene of the donkey and different donkey: cow ratios. Lane 1 has a 100bp standard ladder, lane 2 has donkey lanes 3-11 represent DNA bands of a mixed donkey: cow samples with 1:0, 1:1,1:2, 1:4, 1:6, 1:8,1:10, 1:50, and 1:100 ratio respectively.
Multiplex PCR
Multiplex PCR for the targeted cyt b gene was subjected to different binary ratios of cow and donkey to detect the sensitivity level of PCR. Multiplex PCR successfully detected both species in the meat mixture. In all 1-10 ratios of both species, results were positive.
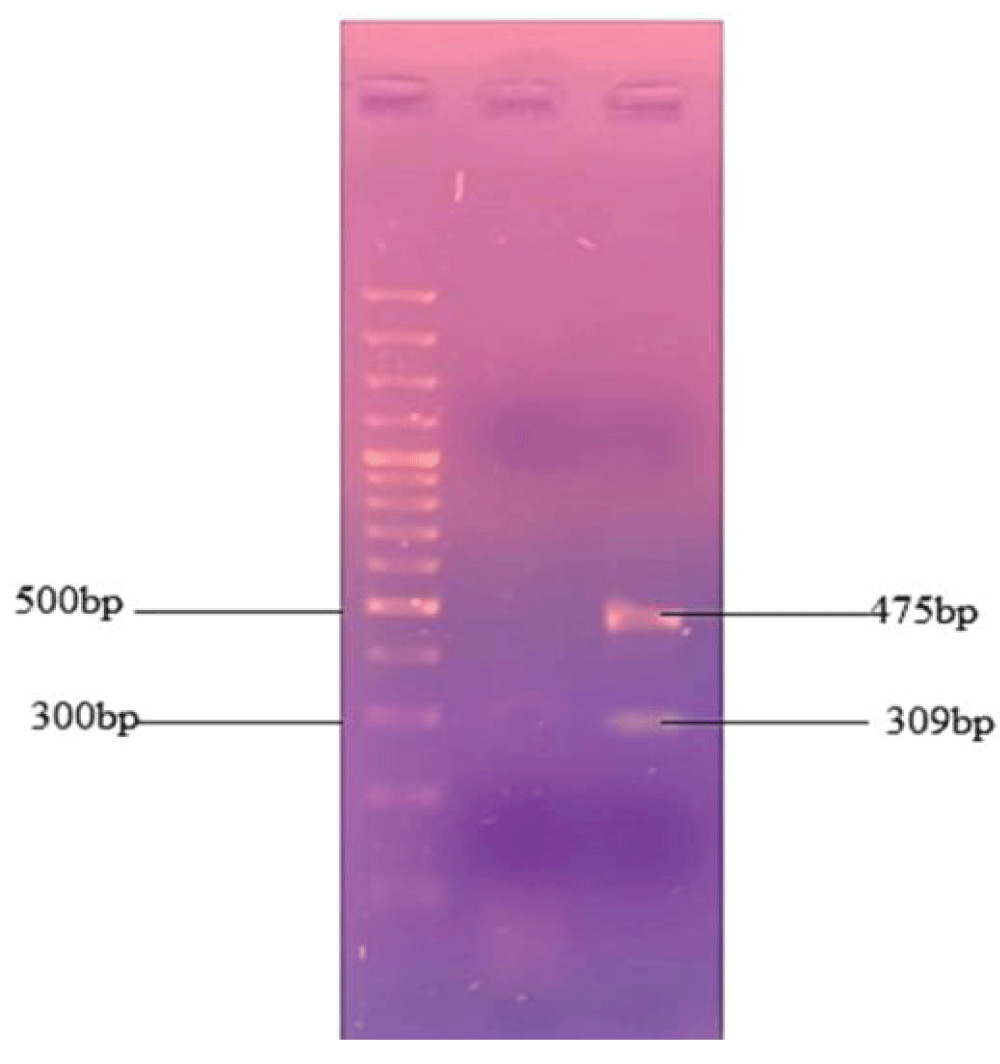
The temperature of all primers was 60 °C. PCR thermal cycling conditions were the same as those of simple PCR. The PCR product was evaluated by 2% gel electrophoresis and the band pattern was visualized with a UV transilluminator. The multiplex PCR amplify the targeted region of the Cow with a PCR band of 309 bp and 475 bp of the Donkey DNA Figure 5.
Figure 5: Multiplex PCR for the targeted cyt b amplification of cow and donkey. DNA bands of 309 and 475 were obtained from the cow and donkey samples respectively.
Meat adulteration is an arising issue nowadays and it is a big challenge to develop certain authentication techniques for detecting species origin [28].
Protein analysis and molecular-based techniques have been used successfully for the detection of meat adulteration. DNA-based approaches i.e. PCR methods are preferred to identify specie origin in raw meat substitution and adulteration [10] due to the facts that DNA is stable, temperature resistant, more informative than protein and present in ample amounts [16,29].
In this study mitochondrial DNA is used for specie detection and identification from raw meat [26]. Mitochondrial DNA is preferred because its presence in high copy number, more thermostable and this increases the chances of positive results even in samples having degraded DNA [30]. Research conducted b [25] showed results in favor of using mitochondrial genes for identification purposes. Cyt b gene is used due to its intra and interspecific sequences which play a promising role in the phylogenetic analysis of closely related and distant species [28].
Simplex PCR was designed to identify single species and check the sensitivity of designed primers as described by [10]. PCR is efficient in the identification of species origin in raw meat DNA from different binary meat ratios were extracted and PCR was used to identify the specie from a 1-10 ratio in a mixture.
Multiplex PCR designed to identify both species of cow and donkey in a mixture showed successful results and this is helpful in the identification of meat adulteration in meat mixtures [8]. The specificity of multiplex PCR and simple PCR was checked by changing the quantity of DNA in ratios and checking the sensitivity and detection level of primers and PCR reactions.
This designed study was used to check the adulteration of donkey meat in cow meat and find the sensitivity level of primers and how much quantity of adulterant can be detected in the meat mixture. The results obtained were positive in correlation with the results of [8,28] also detected five meat species origin by using multiplex PCR and showed that multiplex PCR can detect even a small amount of DNA present [31], conduct research and detected the origin of meat species using multiplex PCR of the cyt b gene.
It is concluded that this study was helpful in the identification of meat adulteration in meat mixtures by simple and multiplex PCR. Even a small amount of DNA was detected by PCR and it takes less time and shows authentic results. The results of this study are positive.
This was a small-scale study based on a small amount of sample. Results can fluctuate based on a commercial level, so this should also be checked on a large scale with a large number of samples.
- Abidfaheem, T., Nayak, B. S., & Andrade, M. (2013). Food adulteration and family's knowledge on food adulteration in selected village of Udupi Taluk, Karnataka. Nitte University Journal of Health Science. 3(2): 33.
- Akhtar S, Randhawa M, Riaz M, Hameed A, Ismail T, Ismail A, Ali Z. Food safety conundrum: a Pakistan's scenario. Quality Assurance and Safety of Crops & Foods. 2014; 7(4): 559-567.
- Kartheek M, Smith AA, Muthu AK, Manavalan R. Determination of adulterants in food: a review. Journal of Chemical and Pharmaceutical Research. 2011; 3(2): 629-636.
- Hossain MAM, Uddin SMK, Sultana S, Bonny SQ, Khan MF, Chowdhury ZZ, Johan MR, Ali ME. Heptaplex Polymerase Chain Reaction Assay for the Simultaneous Detection of Beef, Buffalo, Chicken, Cat, Dog, Pork, and Fish in Raw and Heat-Treated Food Products. J Agric Food Chem. 2019 Jul 24;67(29):8268-8278. doi: 10.1021/acs.jafc.9b02518. Epub 2019 Jul 16. PMID: 31283221.
- Stanciu S. Horse meat consumption− between scandal and reality. Procedia Economics and Finance. 23: 697-703.
- Trivedi DK, Hollywood KA, Rattray NJ, Ward H, Trivedi DK, Greenwood J, Ellis DI, Goodacre R. Meat, the metabolites: an integrated metabolite profiling and lipidomics approach for the detection of the adulteration of beef with pork. Analyst. 2016 Apr 7;141(7):2155-64. doi: 10.1039/c6an00108d. PMID: 26911805; PMCID: PMC4819684.
- Doosti A, Ghasemi Dehkordi P, Rahimi E. Molecular assay to fraud identification of meat products. J Food Sci Technol. 2014 Jan;51(1):148-52. doi: 10.1007/s13197-011-0456-3. Epub 2011 Jul 16. PMID: 24426061; PMCID: PMC3857419.
- Ali ME, Razzak MA, Hamid SB, Rahman MM, Amin MA, Rashid NR, Asing. Multiplex PCR assay for the detection of five meat species forbidden in Islamic foods. Food Chem. 2015 Jun 15;177:214-24. doi: 10.1016/j.foodchem.2014.12.098. Epub 2015 Jan 2. PMID: 25660879.
- Nakyinsige K, Man YB, Sazili AQ. Halal authenticity issues in meat and meat products. Meat Sci. 2012 Jul;91(3):207-14. doi: 10.1016/j.meatsci.2012.02.015. Epub 2012 Feb 22. PMID: 22405913.
- Ashwani K, Singh Y, Prasad G. Cytochrome-b gene-based PCR for identification and differentiation of cooked meat of sheep, goat, cattle, pig, and poultry. Haryana Veterinarian. 2009; 48: 53-57.
- Lin CC, Fung LL, Chan PK, Lee CM, Chow KF, Cheng SH. A rapid low-cost high-density DNA-based multi-detection test for routine inspection of meat species. Meat Sci. 2014 Feb;96(2 Pt A):922-9. doi: 10.1016/j.meatsci.2013.09.001. Epub 2013 Sep 8. PMID: 24211550.
- Sentandreu MÁ, Sentandreu E. The authenticity of meat products: Tools against fraud. Food Research International. 2014; 60: 19-29.
- Ballin NZ. Authentication of meat and meat products. Meat Science. 2010; 86(3): 577-587.
- El Sheikha AF, Mokhtar NFK, Amie C, Lamasudin DU, Isa NM, Mustafa S. Authentication technologies using DNA-based approaches for meats and halal meats determination. Food Biotechnology. 2017; 31(4): 281-315.
- Mousavi SM, Khaniki GJ, Eskandari S, Rabiei M, Samiee SM, Mehdizadeh M.. Applicability of species-specific polymerase chain reaction for fraud identification in raw ground meat commercially sold in Iran. Journal of Food Composition and Analysis. 2015; 40: 47-51.
- Montowska M, Pospiech E. Authenticity determination of meat and meat products on the protein and DNA basis. Food Reviews International. 2010; 27(1): 84-100.
- Barai B, Nayak R, Singhal R, Kulkarni P. Approaches to the detection of meat adulteration. Trends in Food Science & Technology. 1992; 3: 69-72.
- Hurley IP, Elyse Ireland H, Coleman RC, Williams JH. Application of immunological methods for the detection of species adulteration in dairy products. International Journal of Food Science Technology. 2004; 39(8): 873-878.
- Chou CC, Lin SP, Lee KM, Hsu CT, Vickroy TW, Zen JM. Fast differentiation of meats from fifteen animal species by liquid chromatography with electrochemical detection using copper nanoparticle plated electrodes. J Chromatogr B Analyt Technol Biomed Life Sci. 2007 Feb 1;846(1-2):230-9. doi: 10.1016/j.jchromb.2006.09.006. Epub 2006 Sep 27. PMID: 17008137.
- Chikuni K, Ozutsumi K, Koishikawa T, Kato S. Species identification of cooked meats by DNA hybridization assay. Meat Sci. 1990;27(2):119-28. doi: 10.1016/0309-1740(90)90060-J. PMID: 22055225.
- Rahmati S, Julkapli NM, Yehye WA, Basirun WJ. Identification of meat origin in food products–A review. Food Control. 2016; 68: 379-390.
- Aida AA, Che Man YB, Raha AR, Son R. Detection of pig derivatives in food products for halal uthentication by polymerase-chain-reaction–restriction fragment length polymorphism. Journal of the Science of Food and Agriculture. 2007; 87(4): 569-572.
- Wolf C, Rentsch J, Hübner P. PCR-RFLP analysis of mitochondrial DNA: a reliable method for species identification. J Agric Food Chem. 1999 Apr;47(4):1350-5. doi: 10.1021/jf9808426. PMID: 10563979.
- Murugaiah C, Noor ZM, Mastakim M, Bilung LM, Selamat J, Radu S. Meat species identification and Halal authentication analysis using mitochondrial DNA. Meat Sci. 2009 Sep;83(1):57-61. doi: 10.1016/j.meatsci.2009.03.015. Epub 2009 Apr 18. PMID: 20416658.
- Rastogi G, Dharne MS, Walujkar S, Kumar A, Patole MS, Shouche YS. Species identification and authentication of tissues of animal origin using mitochondrial and nuclear markers. Meat Sci. 2007 Aug;76(4):666-74. doi: 10.1016/j.meatsci.2007.02.006. Epub 2007 Feb 22. PMID: 22061243.
- Gajbe R, Banubakode S, Dalvi R, Kurkure N, Charjan R, Nandeshwar N, Rana J. Differentiation of meat species using species-specific repeat and PCR-RFLP technique.
- Broll H. New approaches for verifying food species and variety. In New analytical approaches for verifying the origin of food. Elsevier 2013: 81-93.
- Tauma J, Abdul-Hassan I. Identification of Some Meat Species Using PCR and Multiplex PCR of Mitochondrial Cytochrome B Gene. Iraqi Poultry Sciences Journal. 2014; 8(1): 1-9.
- Abbas O, Zadravec M, Baeten V, Mikuš T, Lešić T, Vulić A, Prpić J, Jemeršić L, Pleadin J. Analytical methods used for the authentication of food of animal origin. Food Chem. 2018 Apr 25;246:6-17. doi: 10.1016/j.foodchem.2017.11.007. Epub 2017 Nov 3. PMID: 29291879.
- Mane B, Mendiratta S, Tiwari A. Polymerase chain reaction assay for identification of chicken in meat and meat products. Food Chemistry. 2009; 116(3): 806-810.
- Jain S, Brahmbhatt M, Rank D, Joshi C, Solanki J. Use of cytochrome b gene variability in detecting meat species by multiplex PCR assay. Indian Journal of Animal Sciences. 2007; 77(9): 880.