Abstract
Purpose
The potential toxic effects due to the application of sophorolipid and the novel saponin biosurfactant extracted from the leaves of Eucalyptus camaldulensis for the purpose of bioremediating PAH-contaminated soils and sediments were studied.
Methods
The phytotoxic effects of sophorolipid and eucalyptus saponin were determined based on the seed germination assays carried out using the seeds of Lactuca sativa L and compared with commonly used rhamnolipid and Tween 20 surfactants. Further, biosurfactant-induced changes in soil/sediment bacterial structure and diversity were investigated by conducting Miseq amplicon sequencing of the bacterial genes.
Results
Germination indices (GI) demonstrated the non-phytotoxic effects (GI > 80%) of saponin and sophorolipid biosurfactants (100–500 mg/L), while rhamnolipid demonstrated greater phytotoxicity than Tween 20 at high concentrations (500 mg/L). Saponin-amended soil resulted in greater bacterial diversity and richness compared to controls, while sophorolipid produced the opposite effect. These significant variations were not observed in sediment samples. Incubation of biosurfactants for 20 and 40 days did not result in significant changes in bacterial diversity and structure in any of the samples. Increased abundance of some of the PAH-degrading bacteria was noted at OTU level, in the presence of saponin and sophorolipid. Saponin had less impact on native soil/sediment bacteria relative to sophorolipid based on the prevalence of the significantly shifted OTUs.
Conclusion
As saponin and sophorolipid were shown to have no adverse impacts on the microbiome, and non-phytotoxic effects, their sustainable applications to remediate PAH-contaminated soils and sediments can be recommended.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Polycyclic aromatic hydrocarbons (PAHs) are toxic and carcinogenic organic pollutants that have accumulated in the natural environment mainly as a result of anthropogenic activities such as the combustion of fossil fuels and biomass (Kumar et al. 2021). Due to their hydrophobicity and low aqueous solubility, PAHs are primarily associated with soil and sediment particulate matter. Many PAH-contaminated soils and sediments host active populations of bacteria that can degrade PAHs. Bacterial species such as Pseudomonas, Bacillus, Mycobacterium, Rhodococcus and other Alpha-, Beta- and Gammaproteobacteria have been studied extensively for PAH degradation (Srivastava and Kumar 2019). However, the failure of soil/sediment bound PAHs to desorb back into the aqueous phase can severely limit microbial degradation due to low bioavailability (Li and Chen 2009). Desorption of soil/sediment bound PAHs through thermal or chemical methods enhances the bioavailability of PAHs, promoting microbial degradation (Kuppusamy et al. 2017; Li and Chen 2009). Nevertheless, the implementation of thermal methods and synthetic industrial solvents such as surfactants, to desorb PAHs, is offset by their energy intensiveness, treatment expense and environmental incompatibilities. Biosurfactants are promising substitutes for chemical surfactants and are used in various industries due to their surface-active properties (Jahan et al. 2020). When compared to artificially synthesised chemical surfactants derived from petroleum feedstock, biosurfactants produced by plants and microbes possess low toxicity, higher biodegradability and effectiveness under a wide range of environmental conditions (Lee et al. 2018).
Rhamnolipid, sophorolipid, saponin and lipopeptide are well-known biosurfactants used in the remediation of soils contaminated with PAHs and other hydrocarbons (Kariyawasam et al. 2022a, b). By reducing surface tension and interfacial tension, micelles formed by biosurfactants solubilise or emulsify and release of PAHs sorbed to soil organic matter. Consequently, micelle-bound PAHs and PAHs desorbed to the aqueous phase become available for microbial uptake (Li and Chen 2009). Despite the fact that biosurfactants are generally considered low or non-toxic biomolecules, potential toxic effects on soil/sediment microbiome and plants have received little attention, essentially at high concentrations. Moreover, it is vital to consider the impact of these biosurfactants on native PAH-degrading bacteria in soils and sediments.
Moderate to non-toxic effects of biosurfactants have been detected for plants, invertebrates and selected microbial species by employing bioluminescence assays, seed germination assays, determining biomass, microbial respiration and survival percentages (de Bezerra Souza et al. 2013; Edwards et al. 2003; Wolf and Gan 2018). Further, shifts in soil microbial communities in the presence of rhamnolipid were observed by conducting Miseq sequencing of bacterial genes (Lu et al. 2019; Wang et al. 2016). Considerable toxic effects and antimicrobial properties associated with high application rates of biosurfactants have also been reported for rhamnolipids (Ławniczak et al. 2013; Millioli et al. 2009; Vatsa et al. 2010). Although sophorolipid and saponin biosurfactants were effective in removing PAHs from soil and sediment (Kobayashi et al. 2012; Schippers et al. 2000), the individual toxic effects of these surfactants on seed germination and soil/sediment microbes have not been studied.
This study aims to reveal the potential toxic effects of sophorolipid from Candida bombicola and saponin biosurfactant extracted from the leaves of Eucalyptus camaldulensis on lettuce seed germination and compare those with a commonly used biosurfactant, rhamnolipid and synthetic industrial surfactant Tween 20. Further, the response of soil and sediment microbial communities to sophorolipid and saponin was investigated.
2 Materials and methods
2.1 Chemicals and other materials
Tween 20 and rhamnolipid biosurfactant (90% purity) extracted from Pseudomonas aeruginosa were purchased from Sigma-Aldrich, Australia. Sophorolipid biosurfactant (> 80% purity) from the yeast Candida bombicola produced by Cayman Chemical, USA, was purchased from Sapphire Biosciences, Australia. Saponin biosurfactant was extracted from the leaves of Eucalyptus camaldulensis (Hajimohammadi et al. 2016).
The soil was collected from an agricultural field in Yanco, NSW, Australia (34°360ʹ S, 146°240ʹ E), and was characterised as a brown Chromosols. Sediment was collected from Bullenbung Creek, Galore (35°182ʹ S, 146°944ʹ E), NSW, Australia. While PAHs were detected in the sediment, their concentrations were below limit of quantification for sediments. Any PAHs in the soil were below limit of detection for soil. Characteristics of the soil and sediment have been previously reported (Kariyawasam et al. 2022c). Lettuce seeds (Lactuca sativa L.) were purchased from a local market in Wagga Wagga, NSW, Australia.
2.2 Inhibition of seed germination assay
An experiment determining inhibition of seed germination by biosurfactants was performed in Petri dishes (9 cm in diameter) containing one layer of filter paper and 3 mL of sterile distilled water (control) or aqueous surfactant solutions prepared in sterile distilled water. Each dish (n = 4) contained a single surfactant at a concentration of 100 mg/L, 200 mg/L or 500 mg/L. Ten seeds were placed in each dish, and covered dishes were incubated at 22 °C in continuous white light in a growth chamber for 4 days. The viability of seeds was pre-determined following the above procedure in the absence of surfactants and was shown to be > 90%. Roots longer than 3 mm were only considered, and root lengths of emerged plantlets were measured after 4 days, and the percentage germination, root elongation and germination index (GI) were calculated according to the following formulae (Agrawal and Shahi 2017):
2.3 Incubation of the surfactant-amended soil and sediment
Surfactant solutions (500 mg/L) prepared in deionised water were sprayed (5 mL) into soil/sediment samples (20 g) in 1 L glass jars (n = 8), and the moisture contents were adjusted to 70% field capacity. Jars with surfactant-free soil/sediment were used as the controls. Sealed jars were aerobically incubated at 22 °C, maintaining the moisture contents. Four replicate jars were removed for each treatment after 20 and 40 days of incubation periods, and samples were collected for DNA extraction.
2.4 DNA extraction
Total bacterial DNA was extracted from 250 mg soil/sediment (n = 4) with the DNeasy PowerSoil Kit (QIAGEN, Chadstone, Victoria, Australia) according to the manufacturer’s instructions. The concentration and purity of extracted DNA samples was determined both via gel electrophoresis and a NanoDrop Spectrophotometer (Thermo Fisher Scientific, Scoresby, Victoria, Australia), and the concentration of each was adjusted to approximately 10 ng/μL prior to the polymerase chain reactions (PCR) and gene sequencing.
2.5 PCR amplification of the 16S rRNA gene and MiSeq amplicon sequencing
PCR amplification and sequencing were performed at the Australian Genome Research Facility (AGRF, Sydney, NSW, Australia). The V3–V4 hypervariable region of the bacterial 16S rRNA gene was amplified with the forward primer (341F: 5ʹ-CCTAYGGGRBGCASCAG-3ʹ) and the reverse primer (806R: 5ʹ-GGACTACNNGGGTATCTAAT-3ʹ) according to the protocol of Yu et al. (2005). Thermocycling was performed on an Applied Biosystem 384 Veriti and using AmpliTaq Gold 360 Master Mix (Life Technologies, Australia) as follows: initial denaturation at 95 °C for 7 min, followed by 29 cycles of 94 °C for 30 s, 50 °C for 60 s and 70 °C for 60 s and a final extension at 72 °C for 7 min.
Illumina indexing of the amplicons was carried out in a second PCR utilising TaKaRa Taq DNA Polymerase (Clontech). Quantification of the indexed amplicon libraries was achieved by fluorometry (Promega Quantifluor) and normalised. An equimolar pool was created and adjusted to 5 nM for sequencing on an Illumina MiSeq (San Diego, CA, USA) with a V3, 600 cycle kit (2 × 300 base pairs paired end).
2.6 Data analysis
Sequences were clustered into operational taxonomic units (OTUs) following the default Quantitative Insights into Microbial Ecology (QIIME 1.8) pipeline with reference to 97% sequence similarity against the SILVA database. Microbial diversity profiling data generated from soil and sediment samples treated with biosurfactants or deionised water (controls) and sampled at 20 and 40 days were analysed using the Marker Data Profiling module of MicrobiomeAnalyst (Dhariwal et al. 2017), which implements R version 3.6.1. The OTU and Simpson rarefaction curve were used to evaluate whether the sequencing data amount was enough to cover all the sampled species and to reflect the species richness in samples. The alpha diversity was evaluated by Chao1 and Shannon indices. Beta diversity was assessed with Bray–Curtis distance between two samples and visualised by principal coordinate analysis (PCoA). Core microbiome analysis was conducted by considering the sample prevalence and the relative abundance (fractions) of taxa or features in order to consider them as a part of the core member.
Statistical analysis was conducted using one-way ANOVA and Tukey’s post hoc test (Wawra et al. 2018). A two‑tailed P < 0.05 was considered statistically significant.
3 Results
3.1 Seed germination study
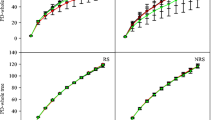
Phytotoxicities of the saponin biosurfactant extracted from eucalyptus leaves and sophorolipid were assessed based on their GI and compared with those of rhamnolipid and the industrial chemical surfactant Tween 20. The percentage seed germination for all the biosurfactants was above 80% at concentrations of 100, 200 and 500 mg/L (Fig. 1a). The results suggested no significant difference (P > 0.05) in lettuce seed germination with the addition of any of the surfactants. In contrast, root elongation was markedly varied among the surfactants and with the doses of surfactants applied (Fig. 1b). Root elongation was over 80% for saponin and sophorolipid biosurfactants over the 100–500 mg/L range, while the rhamnolipid biosurfactant reduced root length to 33% at the 500 mg/L treatment. Tween 20 was the only chemical surfactant tested and consistently reduced root growth to 50–67% across the entire concentration range tested. Among the three biosurfactants, sophorolipid and saponin exhibited the least impact on root elongation at all the concentrations tested. Rhamnolipid resulted in the retardation of root growth at 500 mg/L.
GIs calculated based on the percentages of seed germination and root elongation were all greater than 80% for all four of the surfactants, with the exception of rhamnolipid at 500 mg/L (Fig. 1c). GI values < 50%, 50–80% and > 80% represent high toxicity, low and non-toxicity, respectively (Agrawal and Shahi 2017). This defines the non-toxicity of biosurfactants towards the lettuce plant. However, rhamnolipid at 500 mg/L concentration demonstrated high toxicity, whereas Tween 20 showed low toxicity under the three concentrations tested. Based on our study on the optimisation of the experimental conditions for the surfactant mediated desorption of PAHs (Kariyawasam et al. 2022b), optimal concentrations of the biosurfactants were within 60–100 mg/L. It is likely that surfactants will not be uniformly distributed throughout the soil/sediment when applied, and discrete regions of high surfactant concentration will result. Hence, toxicity studies on the concentrations above the applied rates are necessary.
3.2 Response of soil and sediment microbial communities to surfactant addition
3.2.1 Biosurfactant-induced changes in soil and sediment bacterial structure and diversity
According to the previous section, rhamnolipid and Tween 20 toxicity was manifested as suppression of root growth in lettuce plants, while sophorolipid and saponin showed much lower impacts, particularly at higher concentrations. A previous study also indicated that rhamnolipid exhibited toxicity directly towards soil microbes (Ławniczak et al. 2013; Millioli et al. 2009; Vatsa et al. 2010). Consequently, the impact of only sophorolipid and saponin biosurfactant application on soil and sediment microbial communities was investigated using amplicon sequencing. Rarefaction analysis of identified OTUs indicated that the sequencing depths of these samples were well represented (Fig. S1). Beta diversity profiling based on Bray–Curtis distance between soil and sediment samples depicted the presence of significantly (P < 0.05) different microbial communities in the two matrices (Fig. S2).
Bacteria in non-treated soil- and biosurfactant-amended soil showed distinct community structure (P < 0.05, F = 9.37) as was revealed by PCoA based on Bray–Curtis distance (Fig. 2a). Two alpha diversity indices were calculated to compare the richness and the diversity of the bacterial communities in soil with the treatments. According to Chao 1 index (Fig. 2b), a significant difference (P < 0.05, F = 5.74) in the species richness among the two treatments and control was observed. Furthermore, diversity in terms of species richness was greater for saponin applied soil compared to the non-treated soil, and it was the lowest in sophorolipid applied soil. Shannon index indicated that the species evenness in control soil samples was significantly (P < 0.05, F = 8.09) higher than in the treated samples (Fig. 2c). Alpha and beta diversity profiling at phylum and feature levels for sediment samples treated with two biosurfactants showed no significant difference in diversity and richness. Further, soil and sediment samples treated with saponin and sophorolipid did not show a significant difference in bacterial communities in 20- and 40-day incubated samples, as revealed by alpha diversity indices and PCoA based on Bray–Curtis distance.
Shifts in soil microbial communities due to surfactant amendment (E, eucalyptus saponin-amended soil; S, sophorolipid-amended soil; none, non-treated soil). a PCoA analysis of soil bacterial communities based on Bray–Curtis distance at the phylum level. Different colours of dots represent different treatments. Alpha diversity including Chao1 (b) and Shannon (c) indices of bacterial communities
According to the univariate analysis conducted at the feature level, there were 11 OTUs that reported significantly (P < 0.05) different abundances in biosurfactant-treated and control soils. OTUs and the corresponding bacterial genera are listed in Table S1. Higher abundances of bacterial OTU50 (Methylobacterium), OTU109 (Bradyrhizobium), OTU401 (Caulobacteraceae) and OTU637 (Xanthobacteraceae), which represent the bacterial classes Alphaproteobacteria and Gammaproteobacteria were detected in the presence of saponin compared to the control soils (Fig. S3). A significantly (P < 0.05) lower overall abundance of bacteria was noted in sophorolipid-added soil samples compared to the controls and saponin-treated samples. Notably, OTU1288 (Gemmatimonadetes) found in non-treated samples was absent in the presence of biosurfactants. The univariate analysis reported significant changes (P < 0.05) in the abundance of six bacterial OTUs (Table S1), which represent the classes Alphaproteobacteria and Actinobacteria due to the biosurfactant treatment in sediment. Enrichment of bacteria representing OTUs 50 (Methylobacterium), 57 (Phenylobacterium), 1058 (Microbacteriaceae) and 2114 (Solirubrobacter) was observed in the presence of sophorolipid, whereas OTU50 was predominately found in saponin applied sediment samples (Fig. S4).
The identification of core bacterial communities in biosurfactant-added samples may help to predict the community responses to the addition of biosurfactants. The core microbiome of biosurfactant-added soil samples consisted of Firmicutes, Proteobacteria, Actinobacteria and Bacteroidetes (Fig. 3a). The prevalence of nearly 50% soil microbial phyla was observed in both treated and non-treated samples. At the feature level (Fig. S3), Bacillus (OTU2), Nocardioides (OTU5), Massilia (OTU10) and Phenylobacterium (OTU17) were the most prevalent members, which represented the above phyla. In biosurfactant-supplemented sediment, Proteobacteria, Actinobacteria and Bacteroidetes were the prominent phyla in the core microbiome (Fig. 3b). Rhodanobacter (OTU1), Streptomyces (OTU3) and Streptacidiphilus (OTU8) were the most prevalent OTUs found in the core microbiome of all the sediment samples (Fig. S4).
3.2.2 Relative abundance of soil and sediment bacteria in the presence of biosurfactants
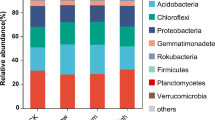
Taxonomic analysis based on representative sequences of OTUs showed (Fig. 4) that Bacilli was the most dominant bacterial class in soils with sophorolipid, saponin and non-treated soil accounting for 47%, 37% and 31% abundance, respectively. This was followed by Actinobacteria, Alphaproteobacteria and Gammaproteobacteria in sophorolipid-added soil and controls. In saponin-amended soil, the bacterial class Alphaproteobacteria was more abundant than Actinobacteria. Although there were no significant differences in the abundance of the four most dominant classes among treatments, a shift of bacterial community composition was observed mainly in sophorolipid-amended soil compared to the other samples. Most prominently, the relative abundances of Bacilli and Alphaproteobacteria were influenced by the sophorolipid biosurfactant, resulting in a slight increase in Bacilli and a reduction in Alphaproteobacteria compared to the control.
Actinobacteria, Gammaproteobacteria and Alphaproteobacteria were the most dominant classes of bacteria both in treated and non-treated sediment samples (Fig. 5). The abundance of Actinobacteria was higher in treated samples (44–49%) compared to non-treated sediment (33–37%). Members of the class Gammaproteobacteria showed a slight reduction in the presence of surfactants. Alphaproteobacteria and Bacteroides also accounted for around 6 to 10% of abundance and had not been changed by the addition of the biosurfactant. Notably, Bacilli the most predominant bacterial class in soil was found only less than 2% in sediment samples.
Taxonomic analysis based on representative sequences of OTUs showed a shift in microbial communities in treated soil and sediments compared to the controls. All the sediment samples after day 20 and 40 incubations demonstrated an increase in Actinobacteria and a decrease in Gammaproteobacteria abundance. Although the abundance of bacterial genera in sophorolipid applied sediments had not changed during the 20 to 40 days of incubation, the increase in Gammaproteobacteria composition was detected with time in saponin-amended sediment samples.
4 Discussion
4.1 Potential phytotoxic effects of biosurfactants
This study evaluated the phytotoxic potential of sophorolipid and eucalyptus saponin based on lettuce seed germination and root elongation. Although the impact of sophorolipid on seed germination in the presence of soil has been explored, this is the first study that investigated the individual effect of sophorolipid and saponin in the absence of soil. The lack of toxic effects of the studied biosurfactants, compared to Tween 20, during the early stages of lettuce growth, suggests their applicability to remediate contaminated soils.
When compared to sophorolipid and saponin, rhamnolipid exhibited higher toxicity at high concentrations, which demonstrates the suitability and more environmental compatibility of sophorolipid and saponin. Millioli et al. (2009) investigated the impact of the rhamnolipid application on lettuce seed germination and reported a decreased seed germination from 120 to < 40% when the surfactant concentration in soil was increased from 2 to 4 mg/g. Mekwichai et al. (2020) did not observe any toxic effects of rhamnolipid on the growth of corn plants when low concentrations (up to 3.2 mg/g) were applied. However, in the above studies, observed phytotoxic/non-phytotoxic effects of rhamnolipid may not only be due to the influence of rhamnolipid. Rather, interactions of soil constituents and microbes with rhamnolipid, as well as the interactions of soil microbes with seed constituents, may have affected the final outcome. In the presence of soil, the phytotoxic effects of surfactants can be affected by the soil composition and types of microbes present. Priji et al. (2017) conducted a phytotoxicity study in the absence of soil and observed a non-inhibitory effect of rhamnolipid at low concentrations (50 and 100 mg/L) on rice seed germination. While individual phytotoxic effects of sophorolipid have not been previously explored, Vaughn et al. (2014) observed no impact on corn seed biomass when plants were grown in sophorolipid-amended potting substrate. Moreover, Shah and Daverey (2021) observed stimulation of shoot and root growth of Medicago sativa and Bidens pilosa in sophorolipid augmented soil (100 mg/kg) in the presence of cadmium.
Based on the GIs, rhamnolipid at low concentration (< 500 mg/L) and sophorolipid and saponin were found to be less inhibiting of root growth when applied to soil than Tween 20. According to Wen et al. (2009), 20% of rhamnolipid applied at 1.3 g/kg to soil was degraded after 3–7 days in three different soil types, while rhamnolipid applied at 6.6 g/kg took 5–11 days. As the current study was undertaken in an aqueous solution, the toxic effect of rhamnolipid on lettuce root growth may have been more exaggerated than if it was performed in soil. Moreover, it is important to note that soil constituents also play a significant role in rhamnolipid dissipation.
4.2 Shifts in soil and sediment microbial community structure in response to biosurfactants
The use of two matrices having significantly (P < 0.05) different microbial diversities (Fig. S1) helped to demonstrate the responses of a wide range of soil and sediment microbes to the addition of biosurfactants. Although Actinobacteria, Alphaproteobacteria and Gammaproteobacteria were the most abundant classes in both soil and sediment, the abundance of Bacilli was markedly higher in soil compared to sediments with varied treatments (Figs. 4 and 5). Despite the insignificant shifts in soil and sediment bacterial classes due to treatments and incubation times (20–40 days), no bacterial class seems to have been totally inhibited by biosurfactant application.
Shifts in bacterial abundance in the soil due to the addition of surfactants could be attributed to the ability of microbes to utilise the surfactants as carbon sources. Surfactant addition could also wipe out bacterial communities due to inhibitory effects and favour the more surfactant tolerant bacterial communities to thrive under reduced competition. Lu et al. (2019) and Singleton et al. (2016) also observed shifts in different classes of microbes due to rhamnolipid supplementation. Feng et al. (2021) reported stimulation of cell growth and microbial activity in sophorolipid-amended contaminated soil. Similar to the present study (Fig. 5), the enrichment of Actinomycetes in the presence of surfactants has also been noted (Kappeler and Gujer 1994). However, in the presence of contaminants in soils and sediments, toxic effects can be expected owing to the synergistic effects of the surfactants with the contaminants. Lu et al. (2019) revealed a marked reduction in the soil microbial classes Alphaproteobacteria, Gammaproteobacteria and Bacilli in the presence of PAHs and rhamnolipid when compared to the PAH-free rhamnolipid applied soil. This underscored the influence of PAHs in the presence of surfactants in driving the variations in soil microbial communities.
Sophorolipid and eucalyptus biosurfactants are reported to have a potential application in soil/sediment bioremediation of PAH-contaminated sites (Kariyawasam et al. 2022b). Hence, it is vital to consider the impact of these biosurfactants on PAH-degrading microbes in soils and sediments. An increased abundance of known PAH-degrading bacteria would be beneficial to utilise targeted assays to determine if this increase results in increased gene activity. Gaiella, Massilia, Lysobacter, Tumebacillus, Geobacillus, Solirubrobacter, Nocardioides, Phenylobacterium, Novosphingobium, Methylobacterium and Arthrobacter belonging to phyla Alphaproteobacteria, Gammaproteobacteria and Bacilli are known PAH degraders in soil (Lu et al. 2019; Singleton et al. 2016; Wang et al. 2016), and the most prevalent OTUs in the core microbiome of the treated and non-treated soil in the present study were potential PAH degraders. Despite the bacteria belonging to phyla Alpha and Gammaproteobacteria being found in the core microbiome according to the univariate analysis, increases in the abundances of some of the OTUs (Figs. S5 and S6) affiliated with the above phyla in biosurfactant-treated soil have shown to be beneficial, as these OTUs are relevant for PAH degradation. Biosurfactant application negatively impacted the existence of OTU1288 (Fig. 6), which belongs to the bacterial class Gemmatimonadetes, although it is not a prominent class of bacteria associated with PAH degradation. Complete inhibition of some of the OTUs (Fig. 6) that demonstrated significant shifts in abundance based on univariate analysis was observed in the presence of sophorolipid. As presented in Fig. 6, most of the shifted OTUs can still be found in saponin-amended samples. Thus, saponin biosurfactant resulted in a lesser impact on the soil and sediment microbial communities than sophorolipid, as most of the OTUs shown in Fig. 6 were prevalent both in saponin-amended and control samples.
5 Conclusion
Eucalyptus saponin and sophorolipid biosurfactants are more environmentally compatible than the chemical surfactant Tween 20 due to the lack of phytotoxic effects based on lettuce root elongation and seed germination studies. In addition, Miseq sequencing indicated that the relative abundance, diversity and structure of soil and sediment microbial communities were not significantly affected by the saponin and sophorolipid amendment after 20 and 40 days of incubation. However, at the OTU level, stimulation of the growth of potential PAH degraders Methylobacterium and Phenylobacterium was observed mainly in the presence of saponin biosurfactant, while the growth of Methylobacterium, Solirubrobacter and Phenylobacterium was stimulated by sophorolipid. Thus, the application of eucalyptus saponin and sophorolipid to remediate PAH-contaminated soils and sediments may not cause adverse effects on the native microbiome. In terms of the prevalence of the significantly shifted OTUs, saponin is preferred by soil/sediment microbes over sophorolipid. Future work should focus on the impact of different doses of biosurfactants under varying environmental conditions (pH, salinity, temperature) on soil/sediment microbial communities. Further, the effect of residual concentrations of the biosurfactants on microbial populations over the incubation period needs to be considered in future studies.
Data availability
The datasets generated and/or analysed during the current study are not publicly available but are available from the corresponding author on reasonable request.
References
Agrawal N, Shahi SK (2017) Degradation of polycyclic aromatic hydrocarbon (pyrene) using novel fungal strain Coriolopsis byrsina strain APC5. Int Biodeterior Biodegrad 122:69–81. https://doi.org/10.1016/j.ibiod.2017.04.024
de Bezerra Souza SH, de Luna JM, Rufino RD, Figueiredo Porto AL, Sarubbo LA (2013) Assessment of toxicity of a biosurfactant from Candida sphaerica UCP 0995 cultivated with industrial residues in a bioreactor. Electron J Biotechnol 16:4–4. https://doi.org/10.2225/vol16-issue4-fulltext-4
Dhariwal A, Chong J, Habib S, King IL, Agellon LB, Xia J (2017) MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res 45:W180–W188. https://doi.org/10.1093/nar/gkx295
Edwards KR, Lepo JE, Lewis MA (2003) Toxicity comparison of biosurfactants and synthetic surfactants used in oil spill remediation to two estuarine species. Mar Pollut Bull 46:1309–1316. https://doi.org/10.1016/S0025-326X(03)00238-8
Feng L, Jiang X, Huang Y, Wen D, Fu T, Fu R (2021) Petroleum hydrocarbon-contaminated soil bioremediation assisted by isolated bacterial consortium and sophorolipid. Environ Pollut 273:116476. https://doi.org/10.1016/j.envpol.2021.116476
Hajimohammadi R, Hosseini M, Amani H, Najafpour GD (2016) Production of saponin biosurfactant from Glycyrrhiza glabra as an agent for upgrading heavy crude oil. J Surfactants Deterg 19:1251–1261. https://doi.org/10.1007/s11743-016-1871-2
Jahan R, Bodratti AM, Tsianou M, Alexandridis P (2020) Biosurfactants, natural alternatives to synthetic surfactants: physicochemical properties and applications. Adv. Colloid Interface Sci 275:102061. https://doi.org/10.1016/j.cis.2019.102061
Kappeler J, Gujer W (1994) Scumming due to actinomycetes: towards a better understanding by modelling. Water Res 28:763–779. https://doi.org/10.1016/0043-1354(94)90084-1
Kariyawasam T, Doran GS, Howitt JA, Prenzler PD (2022a) Polycyclic aromatic hydrocarbon contamination in soils and sediments: sustainable approaches for extraction and remediation. Chemosphere 291:132981. https://doi.org/10.1016/j.chemosphere.2021.132981
Kariyawasam T, Prenzler PD, Howitt JA, Doran GS (2022b) Eucalyptus saponin- and sophorolipid-mediated desorption of polycyclic aromatic hydrocarbons from contaminated soil and sediment. Environ Sci Pollut R 30:21638–21653. https://doi.org/10.1007/s11356-022-23562-z
Kariyawasam T, Prenzler PD, Howitt JA, Doran GS (2022c) Greener extraction of polycyclic aromatic hydrocarbons from soil and sediment using eucalyptus oil. Environ Chem Lett 20:2757–2764. https://doi.org/10.1007/s10311-022-01467-0
Kobayashi T, Kaminaga H, Navarro RR, Iimura Y (2012) Application of aqueous saponin on the remediation of polycyclic aromatic hydrocarbons-contaminated soil. J Environ Sci Health A Tox Hazard Subst Environ Eng 47:1138–1145. https://doi.org/10.1080/10934529.2012.668106
Kumar M, Bolan NS, Hoang SA, Sawarkar AD, Jasemizad T, Gao B, Keerthanan S, Padhye LP et al (2021) Remediation of soils and sediments polluted with polycyclic aromatic hydrocarbons: to immobilize, mobilize, or degrade? J Hazard Mater 420:126534. https://doi.org/10.1016/j.jhazmat.2021.126534
Kuppusamy S, Thavamani P, Venkateswarlu K, Lee YB, Naidu R, Megharaj M (2017) Remediation approaches for polycyclic aromatic hydrocarbons (PAHs) contaminated soils: technological constraints, emerging trends and future directions. Chemosphere 168:944–968. https://doi.org/10.1016/j.chemosphere.2016.10.115
Ławniczak Ł, Marecik R, Chrzanowski Ł (2013) Contributions of biosurfactants to natural or induced bioremediation. Appl Microbiol Biotechnol 97:2327–2339. https://doi.org/10.1007/s00253-013-4740-1
Lee DW, Lee H, Kwon BO, Khim JS, Yim UH, Kim BS, Kim JJ (2018) Biosurfactant-assisted bioremediation of crude oil by indigenous bacteria isolated from Taean beach sediment. Environ Pollut 241:254–264. https://doi.org/10.1016/j.envpol.2018.05.070
Li J-L, Chen B-H (2009) Surfactant-mediated biodegradation of polycyclic aromatic hydrocarbons. Materials 2:76–94. https://doi.org/10.3390/ma2010076
Lu L, Zhang J, Peng C (2019) Shift of soil polycyclic aromatic hydrocarbons (PAHs) Dissipation pattern and microbial community composition due to rhamnolipid supplementation. Water Air Soil Pollut 230:107. https://doi.org/10.1007/s11270-019-4118-9
Mekwichai P, Tongcumpou C, Kittipongvises S, Tuntiwiwattanapun N (2020) Simultaneous biosurfactant-assisted remediation and corn cultivation on cadmium-contaminated soil. Ecotoxicol Environ Saf 192:110298. https://doi.org/10.1016/j.ecoenv.2020.110298
Millioli V, Servulo E, Sobral L (2009) Bioremediation of crude oil-bearing soil: evaluating the effect of rhamnolipid addition to soil toxicity and to crude oil biodegradation efficiency. Global NEST J 11:181–188
Priji P, Sajith S, Unni KN, Anderson RC, Benjamin S (2017) Pseudomonas sp. BUP6, a novel isolate from Malabari goat produces an efficient rhamnolipid type biosurfactant. J Basic Microbiol 57:21–33. https://doi.org/10.1002/jobm.201600158
Schippers C, Geßner K, Müller T, Scheper T (2000) Microbial degradation of phenanthrene by addition of a sophorolipid mixture. J Biotechnol 83:189–198. https://doi.org/10.1016/S0168-1656(00)00304-7
Shah V, Daverey A (2021) Effects of sophorolipids augmentation on the plant growth and phytoremediation of heavy metal contaminated soil. J Clean Prod 280:124406. https://doi.org/10.1016/j.jclepro.2020.124406
Singleton DR, Adrion AC, Aitken MD (2016) Surfactant-induced bacterial community changes correlated with increased polycyclic aromatic hydrocarbon degradation in contaminated soil. Appl Microbiol Biotechnol 100:10165–10177. https://doi.org/10.1007/s00253-016-7867-z
Srivastava S, Kumar M (2019) Biodegradation of polycyclic aromatic hydrocarbons (PAHs): a sustainable approach. In S Shah, V Venkatramanan and R Prasad (Eds.), Sustainable green technologies for environmental management (pp. 111–139). Singapore: Springer Singapore
Vatsa P, Sanchez L, Clement C, Baillieul F, Dorey S (2010) Rhamnolipid biosurfactants as new players in animal and plant defense against microbes. Int J Mol Sci 11:5095–5108. https://doi.org/10.3390/ijms11125095
Vaughn SF, Behle RW, Skory CD, Kurtzman CP, Price NPJ (2014) Utilization of sophorolipids as biosurfactants for postemergence herbicides. Crop Protect 59:29–34. https://doi.org/10.1016/j.cropro.2014.01.014
Wang L, Li F, Zhan Y, Zhu L (2016) Shifts in microbial community structure during in situ surfactant-enhanced bioremediation of polycyclic aromatic hydrocarbon-contaminated soil. Environ Sci Pollut R 23:14451–14461. https://doi.org/10.1007/s11356-016-6630-4
Wawra A, Friesl-Hanl W, Puschenreiter M, Soja G, Reichenauer T, Roithner C, Watzinger A (2018) Degradation of polycyclic aromatic hydrocarbons in a mixed contaminated soil supported by phytostabilisation, organic and inorganic soil additives. Sci Total Environ 628–629:1287–1295. https://doi.org/10.1016/j.scitotenv.2018.02.156
Wen J, Stacey SP, McLaughlin MJ, Kirby JK (2009) Biodegradation of rhamnolipid, EDTA and citric acid in cadmium and zinc contaminated soils. Soil Biol Biochem 41:2214–2221. https://doi.org/10.1016/j.soilbio.2009.08.006
Wolf DC, Gan J (2018) Influence of rhamnolipid biosurfactant and Brij-35 synthetic surfactant on 14C-pyrene mineralization in soil. Environ Pollut 243:1846–1853. https://doi.org/10.1016/j.envpol.2018.10.031
Yu Y, Lee C, Kim J, Hwang S (2005) Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol Bioeng 89:670–679. https://doi.org/10.1002/bit.20347
Acknowledgements
Thiloka Kariyawasam acknowledges Charles Sturt University for the provision of the Australian Government Research Training Program Scholarship (AGRTP) for her PhD study. The authors also wish to acknowledge the School of Agricultural, Environmental and Veterinary Sciences, Charles Sturt University, Australia, for their financial support to complete the work. Julia Howitt, who contributed greatly to this paper, unfortunately passed away before this paper was submitted. She will be deeply missed by her loved ones.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. This work was supported by the Australian Government Research Training Program Scholarship (AGRTP).
Author information
Authors and Affiliations
Contributions
Thiloka Kariyawasam: conceptualisation, methodology, validation, formal analysis, investigation and writing — original draft. Gregory S. Doran: conceptualisation, methodology, formal analysis, supervision and writing — review and editing. Paul D. Prenzler: conceptualisation, methodology, formal analysis, supervision and writing — review and editing. Julia A. Howitt: conceptualisation and supervision. Benjamin Stodart: conceptualisation, methodology, formal analysis, investigation, supervision and writing — review and editing.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
The authors have read and approved the final manuscript.
Consent for publication
The authors agree to publish the paper upon acceptance.
Competing interests
The authors declare no competing interests.
Additional information
Responsible editor: Yuan Ge
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kariyawasam, T., Doran, G.S., Prenzler, P.D. et al. Effect of eucalyptus saponin and sophorolipid amendment on soil and sediment microbial communities and seed germination: potential application for PAH bioremediation. J Soils Sediments 23, 2544–2555 (2023). https://doi.org/10.1007/s11368-023-03499-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11368-023-03499-7