Abstract
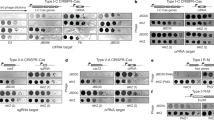
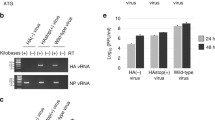
The coliphage mEp021 belongs to a phage group with a unique immunity repressor, and its life cycle requires the host factor Nus. mEp021 has been classified as non-lambdoid based on its specific characteristics. The mEp021 genome carries a gene encoding an Nλ-like antiterminator protein, termed Gp17, and three nut sites (nutL, nutR1, and nutR2). Analysis of plasmid constructs containing these nut sites, a transcription terminator, and a GFP reporter gene showed high levels of fluorescence when Gp17 was expressed, but not in its absence. Like lambdoid N proteins, Gp17 has an arginine-rich motif (ARM), and mutations in its arginine codons inhibit its function. In infection assays using the mutant phage mEp021ΔGp17::Kan (where gp17 has been deleted), gene transcripts located downstream of transcription terminators were obtained only when Gp17 was expressed. In contrast to phage lambda, mEp021 virus particle production was partially restored (>1/3 relative to wild type) when nus mutants (nusA1, nusB5, nusC60, and nusE71) were infected with mEp021 and Gp17 was overexpressed. Our results suggest that RNA polymerase reads through the third nut site (nutR2), which is more than 7.9 kbp downstream of nutR1.





Similar content being viewed by others
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request.
References
Das A et al (2003) Genetic and biochemical strategies to elucidate the architecture and targets of a processive transcription antiterminator from bacteriophage lambda. Methods Enzymol 371:438–459
Lazinski D, Grzadzielska E, Das A (1989) Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell 59(1):207–218
Said N et al (2017) Structural basis for lambdaN-dependent processive transcription antitermination. Nat Microbiol 2:17062
Stagno JR et al (2011) Structural basis for RNA recognition by NusB and NusE in the initiation of transcription antitermination. Nucleic Acids Res 39(17):7803–7815
Li J, Mason SW, Greenblatt J (1993) Elongation factor NusG interacts with termination factor rho to regulate termination and antitermination of transcription. Genes Dev 7(1):161–172
Campbell A (1994) Comparative molecular biology of lambdoid phages. Annu Rev Microbiol 48:193–222
Banik-Maiti S, King RA, Weisberg RA (1997) The antiterminator RNA of phage HK022. J Mol Biol 272(5):677–687
Hernandez-Sanchez J et al (2008) Analysis of some phenotypic traits of feces-borne temperate lambdoid bacteriophages from different immunity groups: a high incidence of cor+ FhuA-dependent phages. Arch Virol 153(7):1271–1280
Neely MN, Friedman DI (1998) Functional and genetic analysis of regulatory regions of coliphage H-19B: location of shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol Microbiol 28(6):1255–1267
Neely MN, Friedman DI (2000) N-mediated transcription antitermination in lambdoid phage H-19B is characterized by alternative NUT RNA structures and a reduced requirement for host factors. Mol Microbiol 38(5):1074–1085
Ravin NV, Rech J, Lane D (2008) Extended function of plasmid partition genes: the Sop system of linear phage-plasmid N15 facilitates late gene expression. J Bacteriol 190(10):3538–3545
Hershko-Shalev T et al (2016) Gifsy-1 prophage IsrK with dual function as small and messenger RNA modulates vital bacterial machineries. PLoS Genet 12(4):e1005975
Kameyama L et al (1999) Characterization of wild lambdoid bacteriophages: detection of a wide distribution of phage immunity groups and identification of a nus-dependent, nonlambdoid phage group. Virology 263(1):100–111
Kameyama L, Fernandez L, Bermudez RM, Garcia-Mena J, Ishida C, Guarneros G (2001) Properties of a new coliphage group from human intestinal flora. Recent Res Dev Virol 3:297–303
Madeira F et al (2022) Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res
Friedman DI et al (1990) Transcription-dependent competition for a host factor: the function and optimal sequence of the phage lambda boxA transcription antitermination signal. Genes Dev 4(12A):2210–2222
Su L et al (1997) RNA recognition by a bent alpha-helix regulates transcriptional antitermination in phage lambda. Biochemistry 36(42):12722–12732
Guzman LM et al (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177(14):4121–4130
Scharpf M et al (2000) Antitermination in bacteriophage lambda. The structure of the N36 peptide-boxB RNA complex. Eur J Biochem 267(8):2397–2408
DeVito J, Das A (1994) Control of transcription processivity in phage lambda: Nus factors strengthen the termination-resistant state of RNA polymerase induced by N antiterminator. Proc Natl Acad Sci U S A 91(18):8660–8664
Roberts JW (1969) Termination factor for RNA synthesis. Nature 224(5225):1168–1174
Rees WA et al (1996) Bacteriophage lambda N protein alone can induce transcription antitermination in vitro. Proc Natl Acad Sci U S A 93(1):342–346
Cocozaki AI, Ghattas IR, Smith CA (2008) Bacteriophage P22 antitermination boxB sequence requirements are complex and overlap with those of lambda. J Bacteriol 190(12):4263–4271
Guarneros G et al (1982) Posttranscriptional control of bacteriophage lambda gene expression from a site distal to the gene. Proc Natl Acad Sci U S A 79(2):238–242
Kameyama L et al (1991) RNaselll activation of bacteriophage lambda N synthesis. Mol Microbiol 5(12):2953–2963
Legault P et al (1998) NMR structure of the bacteriophage lambda N peptide/boxB RNA complex: recognition of a GNRA fold by an arginine-rich motif. Cell 93(2):289–299
Furusawa H, Fukusho S, Okahata Y (2014) Arginine arrangement of bacteriophage lambda N-peptide plays a role as a core motif in GNRA tetraloop RNA binding. ChemBioChem 15(6):865–871
Cocozaki AI, Ghattas IR, Smith CA (2008) The RNA-binding domain of bacteriophage P22 N protein is highly mutable, and a single mutation relaxes specificity toward lambda. J Bacteriol 190(23):7699–7708
Cai Z et al (1998) Solution structure of P22 transcriptional antitermination N peptide-boxB RNA complex. Nat Struct Biol 5(3):203–212
Franklin NC (1993) Clustered arginine residues of bacteriophage lambda N protein are essential to antitermination of transcription, but their locale cannot compensate for boxB loop defects. J Mol Biol 231(2):343–360
Cilley CD, Williamson JR (2003) Structural mimicry in the phage phi21 N peptide-boxB RNA complex. RNA 9(6):663–676
Kroger M, Hobom G (1982) A chain of interlinked genes in the ninR region of bacteriophage lambda. Gene 20(1):25–38
Court D, Sato K (1969) Studies of novel transducing variants of lambda: dispensability of genes N and Q. Virology 39(2):348–352
Silhavy T, Enquist L (1984) Experiments with gene fusions. Cold Spring Harbor Laboratory Press, New York
Altschul SF et al (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Altschul SF et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402
Gish W, States DJ (1993) Identification of protein coding regions by database similarity search. Nat Genet 3(3):266–272
Madden TL, Tatusov RL, Zhang J (1996) Applications of network BLAST server. Methods Enzymol 266:131–141
Quevillon E et al (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33:W116–W120
Klucar L, Stano M, Hajduk M (2010) phiSITE: database of gene regulation in bacteriophages. Nucleic Acids Res 38:366–370
Gautheret D, Lambert A (2001) Direct RNA motif definition and identification from multiple sequence alignments using secondary structure profiles. J Mol Biol 313(5):1003–1011
Hofacker IL et al (1994) Fast folding and comparison of rna secondary structures. Monatshefte Fur Chemie 125(2):167–188
Lesnik EA et al (2001) Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res 29(17):3583–3594
Macke TJ et al (2001) RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res 29(22):4724–4735
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31(13):3406–3415
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9(7):671–675
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, New York
Strauch MA et al (1989) The transition state transcription regulator abrB of Bacillus subtilis is a DNA binding protein. EMBO J 8(5):1615–1621
Friedman DI et al (1981) Evidence that ribosomal protein S10 participates in control of transcription termination. Proc Natl Acad Sci U S A 78(2):1115–1118
Ward DF, DeLong A, Gottesman ME (1983) Escherichia coli nusB mutations that suppress nusA1 exhibit lambda N specificity. J Mol Biol 168(1):73–85
Patterson TA et al (1994) Bacteriophage lambda N-dependent transcription antitermination. Competition for an RNA site may regulate antitermination. J Mol Biol 236(1):217–228
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97(12):6640–6645
Mogridge J, Mah TF, Greenblatt J (1995) A protein-RNA interaction network facilitates the template-independent cooperative assembly on RNA polymerase of a stable antitermination complex containing the lambda N protein. Genes Dev 9(22):2831–2845
Chattopadhyay S et al (1995) Bipartite function of a small RNA hairpin in transcription antitermination in bacteriophage lambda. Proc Natl Acad Sci U S A 92(9):4061–4065
Deatherage CL, Hadziselimovic A, Sanders CR (2012) Purification and characterization of the human gamma-secretase activating protein. Biochemistry 51(25):5153–5159
Greiner S, Lehwark P, Bock R (2019) OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res 47(W1):W59–W64
Acknowledgments
We appreciate the valuable, sharp, careful comments and observations by the anonymous reviewers. Also, we thank Dr. Jaime Ortega and Dr. Norma Oviedo for their invaluable comments throughout this study. We are grateful to M.Sc. Ma. Guadalupe Aguilar-Gonzalez and Dr. Dulce Maria del Carmen Delgadillo Alvarez from LaNse-Cinvestav for technical assistance in DNA sequencing. We also thank M.Sc. Jose Bueno-Martinez and M.Sc. Carlos Osorio for helpful technical assistance, and M.Sc. Jairo Hurtado-Cortes and Dr. Victor Flores for bioinformatic assistance. During this work, G.V.T. and E.P.B.T. were granted doctoral fellowships from CONACyT, Mexico (No. 627423) and (No. 535549), respectively.
Funding
CINVESTAV supported this work.
Author information
Authors and Affiliations
Contributions
GV-T did experimental and analysis work, wrote the first manuscript draft, and conceptualized the work. Supporting experimental work was performed by EPB-T. EM-P did genome sequence assembly. Manuscript editing, corrections, suggestions, and comments were made by RMB-C. Supervision, suggestions, comments, and advice were given by GG. LK contributed to conceptualization, management, direction, and manuscript writing. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no financial interests to disclose.
Ethical approval
This article does not include any studies with animals or human participants performed by any of the authors.
Additional information
Handling Editor: Johannes Wittmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Valencia-Toxqui, G., Ballinas-Turrén, E.P., Bermúdez-Cruz, R.M. et al. Early antitermination in the atypical coliphage mEp021 mediated by the Gp17 protein. Arch Virol 168, 92 (2023). https://doi.org/10.1007/s00705-023-05721-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00705-023-05721-w