Abstract
Biotreatment of oily sludge and the involved microbial communities, particularly in saline environments, have been rarely investigated. We enriched a halophilic bacterial consortium (OS-100) from petroleum refining oily sludge, which degraded almost 86% of the aliphatic hydrocarbon (C10-C30) fraction of the oily sludge within 7 days in the presence of 100 g/L NaCl. Two halophilic hydrocarbon-degrading bacteria related to the genera Chromohalobacter and Halomonas were isolated from the OS-100 consortium. Hydrocarbon degradation by the OS-100 consortium was relatively higher compared to the isolated bacteria, indicating potential synergistic interactions among the OS-100 community members. Exclusion of FeCl2, MgCl2, CaCl2, trace elements, and vitamins from the culture medium did not significantly affect the hydrocarbon degradation efficiency of the OS-100 consortium. To the contrary, hydrocarbon biodegradation dropped from 94.1 to 54.4% and 5% when the OS-100 consortium was deprived from phosphate and nitrogen sources in the culture medium, respectively. Quantitative PCR revealed that alkB gene expression increased up to the 3rd day of incubation with 11.277-fold, consistent with the observed increments in hydrocarbon degradation. Illumina-MiSeq sequencing of 16 S rRNA gene fragments revealed that the OS-100 consortium was mainly composed of the genera Halomonas, Idiomarina, Alcanivorax and Chromohalobacter. This community structure changed depending on the culturing conditions. However, remarkable changes in the community structure were not always associated with remarkable shifts in the hydrocarbonoclastic activity and vice versa. The results show that probably synergistic interactions between community members and different subpopulations of the OS-100 consortium contributed to salinity tolerance and hydrocarbon degradation.
Key points
• A halophilic bacterial consortium efficiently degrades oily sludge hydrocarbons.
• Nitrogen starvation and high salinity have the greatest impact on hydrocarbon loss.
• Change in the consortium structure is not a prerequisite to shift biodegradation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The petroleum industry generates huge amounts of highly toxic waste oily sludge, which is produced during both upstream and downstream operations (Kondaveeti et al. 2023; Murungi and Sulaimon 2022; Wang et al. 2021; Yin et al. 2023). It is estimated that about 0.3–0.5% of waste oily sludge is produced from 1 ton of crude oil processed in petroleum refineries (Jerez et al. 2021; Muneeswari et al. 2023). Waste oily sludge is characterized by complex composition including water, hydrocarbons, heavy metals, sediments, and other chemicals, and its accumulation leads to devastating damage to natural habitats (Hu et al. 2013; Kondaveeti et al. 2023; Murungi and Sulaimon 2022; Sarkar et al. 2020; Wang et al. 2021; Chand et al. 2022). As a consequence of its serious ecological and health concerns, the oily sludge was listed as a priority hazardous pollutant by many organizations including the United States Environmental Protection Agency (USEPA) (Kondaveeti et al. 2023; US EPA 2016). Microbial biodegradation has been largely recognized as the most appropriate approach toward remediation of hydrocarbon- and oil-polluted environments (Ławniczak et al. 2020). However, despite significant advances in the understanding of the process, several misconceptions on hydrocarbon microbiology still prevail, which should be rectified through further investigations of the fundamental aspects (Ławniczak et al. 2020). Although biological treatment or bioremediation can be potentially applied on oily sludge-polluted environments (Ławniczak et al. 2020; Murungi and Sulaimon 2022; Sarkar et al. 2020), its success is often hampered by the characteristics of the impacted site, including high salinity (Akbari et al. 2021; Jimoh et al. 2022; Li et al. 2022; Murungi and Sulaimon 2022; Peng et al. 2020).
Saline and hypersaline environments are particularly susceptible to petroleum hydrocarbon contamination including oily sludge due to their close association to the oil industry operations and facilities (Paniagua-Michel and Fathepure 2018). Among these, natural gas and oil production sites worldwide, including hundreds of kilometers of coastlines in the arid regions of the Arabian Gulf countries, are of serious concern because of the extent and magnitude of contamination (Fathepure 2014; Jimoh et al. 2022). The pollution problem in those locations is much more pronounced due to the high salinity, which makes the removal of petroleum toxic components more challenging (Jimoh et al. 2022). In fact, the shallowness of the Arabian Gulf water together with the arid climate lead to a strong evaporation that significantly exceeds freshwater input from river runoff and precipitation, making the waters at this region to be hypersaline (Ibrahim et al. 2020; Paparella et al. 2022). The surface salinity in the Arabian Gulf is about 37–40‰ in the central part and can reach 60–70‰ in remote bays and lagoons (Simmonds and Lamboeuf 1981; Taher et al. 2012). Moreover, several studies reported that the desalination of seawater in the Arabian Gulf region, which is crucial to provide freshwater supply, might strongly contribute to the increase of salinity (Ibrahim et al. 2020; Paparella et al. 2022).
It is known that high salt levels impose a natural barrier for the degradation of hydrocarbon pollutants (Truskewycz et al. 2019). High salinity not only inhibits the metabolic functions of microorganisms, but also decrease oxygen levels and aqueous solubility of petroleum hydrocarbons (Fathepure 2014; Imron et al. 2020). Nonetheless, hydrocarbon-degrading halophilic microorganisms could be appropriate for the bioremediation of saline environments (Akbari et al. 2021; Jimoh et al. 2022; Saeedi et al. 2022). Halophilic and halotolerant microorganisms developed specific strategies to cope with the osmotic stress caused by the excessive salt concentrations (Atashgahi et al. 2018; Neifar et al. 2019; Srivastava et al. 2022). This could be achieved through the accumulation of a variety of small molecules in the cytoplasm known as compatible solutes (sugars, amino acids, sugars polyols and ectoine, etc.), or inorganic salt ions (K+ and Cl−), in order to maintain their intracellular and extracellular osmotic equilibrium (Akbari et al. 2021; Li et al. 2022; Neifar et al. 2019; Peng et al. 2020). Therefore, application of halophilic microorganisms in bioremediation has received intense attention in recent years (Jimoh et al. 2022; Li et al. 2022).
In general, genes and enzymes of hydrocarbon biodegradation in the presence of high salt levels have not been sufficiently studied (Cao et al. 2020; Imron et al. 2020; Li et al. 2022). In addition, compared to hydrocarbon- and oil-polluted environments, biotreatment of oily sludge has been rarely investigated (Ke et al. 2021), particularly in saline environments. A recent research described the efficiency of a salt-tolerant bacterium Pseudomonas aeruginosa SD to remediate oily sludge with salt tolerance up to 8% (Sun et al. 2023).
In this study, we isolated an autochthonous halophilic bacterial consortium from waste oil sludge and studied its hydrocarbon biodegradation capability under various bioprocess conditions of incubation time, salinity, temperature, and nutrient amendments. Furthermore, we characterized the consortium in terms of salinity tolerance, bacterial population dynamics, and hydrocarbon degradation genes.
Materials and methods
Oily sludge source and characterization
Two types of waste oily sludge (aged and fresh) were collected from the weathering pit of a petroleum plant in Bahrain on march 2020. The aged sludge was used as an inoculum to isolate oil-degrading bacteria and the fresh sludge served as the carbon, sulfur, and energy source for all microbial cultures used in the biodegradation experiments. Physicochemical characteristics of the fresh oily sludge were previously described (Hentati et al. 2022).
Culture media
Luria-Bertani (LB) broth and agar media were prepared according to the supplier’s instructions. Chemically defined medium (CDM) is composed of basal medium (phosphate buffer: KH2PO4 + K2HPO4, ammonium chloride: NH4Cl, and deionized water) and supplemented with FeCl2, MgCl2, CaCl2, trace elements and vitamins (these components are designated hereafter as the “complement”), as described in Table S1.
Enrichment of a halophilic bacterial consortium from of oily sludge
Five grams of aged oily sludge were inoculated into 100 mL of CDM containing 2% (w/v) of fresh oily sludge as the carbon and sulfur source in the presence of 100 g/L NaCl. The enrichment cultures were incubated at 30 °C under shaking at 180 rpm for 14 days. Subsequently, 10% (v/v) from this original enrichment was transferred to a fresh medium with the same composition and further incubated under the same conditions for 14 days. After three times of subculturing, a bacterial consortium (designated as OS-100) that maintained growth on 2% (w/v) oily sludge was obtained and served to isolate pure bacteria. The OS-100 bacterial consortium is available in the laboratory of the corresponding author.
Isolation of halophilic bacteria from the OS-100 consortium
Aliquots (100 µL) of 10− 1 to 10− 5 dilutions from the third enrichment culture of OS-100 were spread onto CDM agar plates supplemented with 2% oily sludge (w/v) in the presence of 100 g/L NaCl. The plates were incubated at 30 °C for 4 to 5 days. Two single colonies that were surrounded by a clear zone were picked and transferred into fresh CDM containing 2% (w/v) of oily sludge and incubated for 14 days to confirm hydrocarbon degradation phenotype.
Preparation of precultures
Considering that the culture medium type could have an effect on the initial composition of the community structure, which may affect phenotype under investigation, the preculture of the OS-100 bacterial consortium was prepared in two different media, namely; LB broth and CDM + oily sludge. An aliquot of the OS-100 consortium (1 mL) was retrieved from a frozen stock preserved at -80 °C, inoculated in 20 mL LB broth, and incubated for 24 h at 30 ºC and 180 rpm. Then, 10 mL from this preculture were transferred into a 1-L Erlenmeyer flask containing 400 mL of LB broth and incubated under the same conditions for 24 h. The cells were harvested by centrifugation (6000 rpm, 10 min, 4 ºC) and washed twice with phosphate buffer (0.1 M, pH 7). The washed cell pellet was resuspended in 50 mL of phosphate buffer and this cell suspension (9.61 ± 1.23 g dry cell mass/L, OD600 ≈ 5) was used as the inoculum. Alternatively, other precultures of the OS-100 consortium were prepared in 50 mL CDM containing 2% (w/v) oily sludge and incubated at 30 ºC under shaking (180 rpm) for 4 days. The cells were harvested, washed twice, and resuspended in 25 mL of phosphate buffer (0.1 M, pH 7) to reach OD600 ≈ 2 (4.72 ± 0.83 g dry cell mass/L) to be used as the inoculum. The precultures of the two isolated bacteria (designated OS100-3 and OS100-4) were prepared in LB broth as described above for the OS-100 consortium where the final cell suspensions in phosphate buffer had OD600 ≈ 3 (5.01 ± 0.6 g dry cell mass/L). All precultures contained NaCl at a concentration of 100 g/L.
Salt tolerance of the OS-100 bacterial consortium and the isolated bacteria
The salinity tolerance profile of the consortium OS-100 and bacteria isolated therefrom was tested at different NaCl concentrations. To study the effect of the culture medium composition, salinity tolerance was tested in LB broth (rich medium) and CDM + oily sludge cultures inoculated with 5% from the respective precultures described above. Accordingly, LB cultures were inoculated with cell suspensions prepared from LB cultures, and CDM cultures were inoculated with cell suspensions prepared from CDM + oily sludge cultures. The LB cultures (50 mL) contained different NaCl concentrations (10, 30, 50, 100, 150, 200, 250, and 300 g/L) in addition to a culture without addition of NaCl. The same was done in 50 mL of CDM containing 2% (w/v) of oily sludge as the sole carbon and sulfur source in the presence of 0, 50, 100, 150, and 200 g/L NaCl. All cultures (in biological triplicates) were incubated at 30 °C under shaking (180 rpm). In LB cultures, the growth was monitored by measuring the optical density at 600 nm (OD600), while growth in CDM cultures was followed by measuring the biomass dry weight. The latter was measured in 2 mL of the cultures collected every day and centrifuged at 14.000 rpm for 20 min to separate the cells from the culture broth. The cell pellets were washed twice by phosphate buffer (0.1 M, pH 7), resuspended in 1 mL, and poured into an Eppendorf tube which was incubated in an oven at 100 ºC until a constant weight is attained. Then, the cell dry mass was calculated gravimetrically as g/L.
The specific growth rate of each culture was calculated by determining the slope of lnX vs. t in the exponential phase of growth using the following equation:
where, µ = specific growth rate (h− 1); Xt = biomass concentration (OD600) at time t; X0 = biomass concentration (OD600) at time 0, and t = operational time (Zahari et al. 2022).
Biotreatment of the oily sludge with the OS100-3 and OS100-4 isolates
Precultures of the isolates OS100-3 and OS100-4 were prepared as described above and used to inoculate 50 mL of CDM containing 2% (w/v) of oily sludge as the sole carbon and sulfur source in the presence of 0, 50, 100, 150 and 200 g/L NaCl. Uninoculated medium was prepared under the same conditions and used as an abiotic control. After 7 days of incubation (30 °C and 180 rpm), all cultures and abiotic controls were extracted by an equal volume of dichloromethane (50 mL). Then, the organic phase was evaporated and the residual oil was analyzed by Gas Chromatography-Mass Spectrometry (GC-MS) according to Hentati et al. (2022). All cultures were in biological triplicates.
Biotreatment of the oily sludge with the consortium OS-100
The biodegradation experiments were carried out in 50 mL of CDM batch cultures (in 250 mL Erlenmeyer flasks) containing 2% (w/v) of oily sludge as the sole carbon and sulfur source, in the presence of 0, 50, 100, 150, and 200 g/L NaCl. Two different inocula (5%) were used; one inoculum prepared in LB broth and the other in CDM + oily sludge as described above. All cultures were incubated at 30 °C and 180 rpm for 7 days. Uninoculated flasks were incubated under the same conditions and used as abiotic controls. All experiments were performed in biological triplicates. At the end of the incubation period, all cultures and abiotic controls were extracted by an equal volume of dichloromethane. Then, the organic phase was evaporated and the residual oil was analyzed by Gas Chromatography-Mass Spectrometry (GC-MS) according to Hentati et al. (2022).
Growth profile of the OS-100 consortium
To assess growth of the bacterial consortium OS-100 during treatment of oily sludge, the biomass dry weight was monitored every day during 10 days of incubation. The OS-100 consortium was inoculated (5%) into 2-L Erlenmeyer flasks (triplicates) containing 400 mL of CDM in the presence of 2% (w/v) of oily sludge and 100 g/L of NaCl. The cultures were incubated in an orbital shaker at 30 ºC and 180 rpm. Ten mL of the cultures were collected every day and centrifuged at 14.000 rpm for 20 min to harvest the cells and measure the biomass dry weight.
Biotreatment of the oily sludge under different bioprocess conditions
All the subsequent experiments were conducted using precultures of the OS-100 consortium prepared in LB medium. The effect of culturing conditions on oily sludge hydrocarbon biodegradability by the OS-100 consortium was investigated in batch cultures following a one variable-at-a time approach. In each experiment one condition was individually manipulated, while all other conditions were fixed. The studied factors included incubation time (0, 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10 days), temperature (30, 35, 40, and 45 °C), and culture medium composition [(complete CDM, CDM without complement (FeCl2, MgCl2, CaCl2, trace elements and vitamins), CDM without phosphate buffer, and CDM without NH4Cl)]. All experiments were conducted in biological triplicates, and the cultures were extracted and analyzed after 7 days of incubation as described earlier.
Analysis of bacterial community structure under different bioprocess conditions
Bacterial genomic DNA was extracted from the OS-100 cultures using QIAamp DNA Mini kit, (Qiagen, Germany). Quantification of DNA was carried out using a Qubit Fluorometer (Invitrogen, Thermo Fisher Scientific, Singapore), and the DNA purity was checked with a DS-11 FX+ Spectrophotometer/Fluorometer (DeNovix Inc, USA). Purified DNA extracts were submitted to Genomics BioSci & Tech Co., Ltd. (New Taipei City, Taiwan) for paired-end Illumina- MiSeq sequencing of the V3-V4 hypervariable region of the bacterial 16 S rRNA gene using the forward primer: CCTACGGGNGGCWGCAG and reverse primer: GACTACNVGGGTATCTAATCC (Klindworth et al. 2013) as described before (Hentati et al. 2022) (see Supplementary Material for details).
The sequences were generated on two separate MiSeq runs using a single indexing strategy. Samples were demultiplexed and sequencing adapters, barcodes, and primers were removed using the fastqprocessor developed by MRDNA (Shallowater, TX, USA), which further ensured the correct orientation of all reads as forward (R1) and reverse (R2). After demultiplexing, intact read pairs were extracted using pairfq_lite (https://github.com/sestaton/Pairfq). Further processing was conducted in R v3.5.2 (R Core Team 2018) using the R package dada2 v1.10.1 (Callahan et al. 2016) with default parameters if not otherwise indicated. Quality filtering was conducted at a maximum expected error rate of 3 for both forward and reverse reads after truncation to 230 bp. Error learning (based on at least 108 bases) and denoising (pooling all sequences per run) were executed separately for each MiSeq run. Then, forward and reverse reads of each sample were merged with a minimum overlap of 10 bp, and chimera detection was performed with the method ‘consensus’ on the whole dataset. Furthermore, only sequences between 400 and 430 bp as well as those occurring at least twice in the data set were retained. Taxonomic classification was conducted using the SILVA NGS web-service with the SILVA ribosomal database (version 132) (Quast et al. 2012). Only bacterial sequences classified on phylum level with a sequence similarity of at least 93% to the reference database and not affiliated with chloroplasts and mitochondria were used for further analysis. Operational taxonomic units (OTUs) were defined as unique amplicon sequence variants. Ordination of the Bray-Curtis dissimilarities between the different culturing conditions was performed using nonmetric MDS, with 100 random restarts and the results were plotted in two dimensions. The Illumina-MiSeq data set was submitted to the Sequence Read Archives under the accession number PRJNA921925.
Molecular identification of the isolated strains
Genomic DNA of pure strains OS3-100 and OS4-100 isolated from the mixed culture OS-100 was extracted using BioVision kit (Biovision Inc, CA, USA). Amplification of the 16 S rRNA gene was carried out in Applied Biosystems™ Veriti™ 96-well fast Themal Cycler (Singapore) using the universal primers 27 F and 1492R (Table S2). The 16 S rRNA gene sequencing and phylogenetic analysis were performed as described (Hentati et al. 2016).
Detection of functional genes involved in hydrocarbon biodegradation and in the synthesis of osmoprotectants
Primers listed in Table S2 were adopted for the detection of genes involved in hydrocarbon degradation including alkanes hydroxylating monooxygenases (alkB), PAH (polycyclic aromatic hydrocarbon)-ring hydroxylating dioxygenases (PAH-RHDα), and naphthalene dioxygenase (nahAc), as well as the biosynthesis of the osmoprotectants betaine and ectoine. Genomic DNA was isolated from the OS-100 bacterial cultures grown on oily sludge (at 100 g/L NaCl, 30 °C and 180 rpm) using QIAamp DNA Mini kit, (Qiagen, Germany) and was used as a template. The 50 µL PCR assays contained 1 µL of template DNA (148 ng), 5 µL of each primer (2 pmol/µL), 25 µL Qiagen Taq PCR master mix, and 14 µL nuclease-free water. The PCR conditions were set according to the protocols provided in the respective reference (Table S2).
Quantification of the alkB gene using real-time qPCR (RT-qPCR)
The alkB gene encodes a membrane-bound non-heme di-iron monooxygenase which catalyzes n-alkane terminal hydroxylation, the first step of the most common alkane degradation pathway (Moreno and Rojo 2017). It is usually used as a proxy or functional marker for alkane degradation. Therefore, it was selected to monitor alkane degradation by the consortium OS-100 during biotreatment of the oily sludge. Two-L flasks (5 replicates) containing 400 mL of CDM, 2% (w/v) of oily sludge and 100 g/L NaCl were inoculated with the OS-100 consortium (5%, v/v) and incubated at 30 °C for 7 days. Ten mL of the cultures were collected every 24 h and centrifuged at 4800 x g for 20 min. The cell pellets were washed twice with phosphate buffer (0.1 M, pH 7) and used to isolate total RNA using RNeasy Protect kit (Qiagen, Germany). To remove the residual genomic DNA, the isolated RNA was treated with DNase I (Amplification Grade, Invitrogen, ThermoFisher Scientific, USA). The integrity of the RNA samples was checked on agarose gel electrophoresis (1%), while the purity and concentration were estimated with a DS-11 FXC Spectrophotometer/Fluorometer (DeNovix Inc., USA) (Table S3). The cDNA was synthesized from RNA using High Capacity cDNA Reverse Transcription kit (Applied Biosystems, ThermoFisher Scientific, USA) following the manufacturer’s instructions, after which the cDNA samples were checked for concentration and purity and stored at -20 °C to be used as a template in RT-qPCR (Table S4).
RT-qPCR was conducted according to the MIQE guidelines (Bustin et al. 2009) (Table S5) in 96-well plates on an Applied Biosystems 7500 Fast Real-Time PCR System using PowerUp™ SYBR™ Green Master Mix (Applied Biosystems, ThermoFisher Scientific, USA). Two primer sets were used for qPCR (Table S2). Primers alkB-1 F and alkB-1R amplified the alkane-hydroxylating monooxygenase gene, and the other primer set, 16 S-F/16S-R (selected as an internal standard), was used to amplify bacterial 16 S rRNA gene fragments. Amplification was performed in a total volume of 15 µL containing 7.5 µL of SYBR Green PCR Master Mix, 1 µL of each primer (10 µM), 2 µL of normalized cDNA (250 ng/µL) and the rest was completed with nuclease-free water. For each cDNA replicate, two RT-qPCR assays were carried out. To ensure the absence of nonspecific primer dimers, control reactions with no cDNA template were conducted. PCR cycling conditions were as follows: 10 min incubation at 95 °C followed by 40 cycles of 15 s at 95 °C, 30 s at 62 °C, and 45 s at 72 °C. Finally, melt curve was obtained from 60 to 95 °C at an increment of 0.2 °C/cycle. The relative changes in alkB gene expression were determined using the 2−∆∆CT method (Akbari et al. 2021).
Statistical analysis
All values represent the mean ± standard deviation. Data were statistically analyzed by one-way ANOVA using Tukey’s multiple comparisons test and applying a significance level of p < 0.05 using GraphPad Prism 6 (Trial version).
Results
Enrichment of a halophilic bacterial consortium from aged oily sludge
The aerobic enrichment procedures using aged oily sludge as an inoculum produced a mixed culture (OS-100) which sustained growth on 2% (w/v) oily sludge as the sole carbon, sulfur, and energy source in the presence of 100 g/L NaCl after repeated subculturing. Utilization of the oily sludge hydrocarbons by the OS-100 bacterial consortium was confirmed by GC-MS analysis which revealed 85.75 ± 7.5% loss of hydrocarbons (C10-C30 alkanes) in the dichloromethane-extractable fraction within 7 days of incubation (Fig. 1).
Total ion chromatograms showing degradation of oily sludge hydrocarbons with the OS-100 consortium after 7 days of incubation at 30 °C. The oily sludge was added to chemically defined medium (2%, w/v) as the sole carbon and sulfur source in the presence of 100 g/L NaCl. The oily sludge hydrocarbons were extracted from whole cultures in dichloromethane
Isolation of two bacterial strains from the consortium OS-100
The detection of several morphologically distinct colonies on CDM-oily sludge agar plates indicated that OS-100 is a bacterial consortium. Two pure strains, designated OS100-3 and OS100-4, were isolated. Molecular analysis revealed that strain OS100-3 was closely related to Chromohalobacter salexigens (Type strain AJ295146) (Arahal et al. 2001), with 99.5% 16 S rRNA gene sequence identity, whereas strain OS100-4 shared 98.17% sequence identity with Halomonas halophila (Type strain FN257740) (Dobson and Mcmeekin 1993) and Halomonas salina (Type strain AJ295145) (Arahal et al. 2001) (Fig. S1). The 16 S rRNA gene sequences of strains OS100-3 (composed of 1440 nucleotides) and OS100-4 (composed of 1437 nucleotides) were deposited in the GenBank under accession numbers OQ283852 and OQ283853, respectively.
Salinity tolerance profile of the two bacteria isolated from the consortium OS-100
The salinity tolerance of both isolates was tested in two different media, LB broth and CDM + oily sludge (Figs. S2 and S3). The growth profile of the two isolates was different depending on the type of the culture medium and salt concentration. In LB broth, both isolates grew in the presence of 50 up to 250 g/L NaCl. Absence of NaCl or 300 g/L NaCl did not allow growth. Strain OS100-3 attained the highest OD600 at a NaCl concentration 100 and 150 g/L, while strain OS100-4 reached the highest OD600 at 50 and 100 g/L NaCl. In both cultures, the lowest OD600 was reached in the presence of 250 g/L NaCl. However, the OD600 values in the OS100-4 cultures were always higher compared to the OS100-3 cultures. This was also the case for the maximum specific growth rates. In the OS100-3 culture, the maximum specific growth rate was about 0.2 ± 0.01 h− 1 at 50 and 100 g/L NaCl. On the contrary, the OS100-4 culture exhibited the highest specific growth rate (0.47 ± 0.02 h− 1) at 50 g/L NaCl. Generally, in CDM + oily sludge cultures, specific growth rates of both isolates were lower than those of the corresponding LB cultures. Moreover, and similar to the LB cultures, the biomass yield of the OS100-4 cultures in CDM + oily sludge was relatively higher than the corresponding values attained in the OS100-3 cultures. Growth of the OS100-3 isolate was higher in the presence of 100 and 150 g/L NaCl, while the OS100-4 isolate grew better within the range 50–150 g/L NaCl.
Biotreatment of the oily sludge with the two bacteria isolated from the consortium OS-100
GC-MS analysis revealed biodegradation of oily sludge hydrocarbons by the two isolated bacteria OS100-3 and OS100-4 (Figs. S4 and S5). In both cultures, hydrocarbon degradation was the lowest when NaCl was excluded. In the OS100-3 culture, as the NaCl concentration increased to 100 g/L, hydrocarbon degradation increased then declined at 150 and 200 g/L. In the OS100-4 culture, the highest degradation was at NaCl concentration of 50 g/L, followed by a drop as the salt concentration increased. In general, hydrocarbon removal by the OS100-4 culture (75.2 ± 24% at 100 g/L NaCl) was higher compared to the OS100-3 culture (41.8 ± 1.1% at 50 g/L NaCl) (p < 0.05).
Salinity tolerance profile of the consortium OS-100
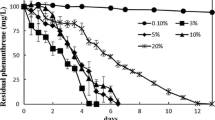
The salinity tolerance of the consortium OS-100 was tested in two different media: LB broth and CDM + oily sludge cultures inoculated with a preculture grown in the respective medium. As shown in Fig. 2, the OS-100 consortium grew over a NaCl range between 10 and 250 g/L, with an optimum growth at 50 and 100 g/L. However, it was unable to grow in medium without NaCl, indicating that OS-100 is an obligatory halophilic bacterial consortium that requires salt to grow. The salinity tolerance profile of the OS-100 consortium revealed some commonalities, as well as variations, with the cultures of the isolates. In LB cultures, the growth profiles and OD600 values of the OS-100 consortium were more similar to those of the OS100-4 isolate. Although the maximum specific growth rate in both cultures occurred in the presence of 50 g/L NaCl, the value (0.2 ± 0.01 h− 1) was similar to that of the OS100-3 culture, and much lower than that of the OS100-4 culture (0.47 ± 0.02 h− 1). In CDM + oily sludge cultures, also the overall growth profiles of the OS-100 consortium were more similar to those of the OS100-4 culture. However, the maximum biomass yield in the OS-100 cultures (0.43 ± 0.03 g/L) was higher than that attained in the cultures of the isolates. The maximum specific growth rate of the OS-100 consortium was about 0.8 ± 0.05 d− 1 in the presence of 50 g/L NaCl. This value is similar to that of the corresponding culture of OS100-3 (0.89 ± 0.05 d− 1) which occurred at 100 g/L NaCl, and higher than that of the corresponding culture of OS100-4 (0.64 ± 0.04 d− 1) which was attained at 50 g/L NaCl.
Salinity tolerance profile of the consortium OS-100 grown in CDM supplemented with oily sludge (2%, w/v) (A and B) and in LB broth (C and D). The results are represented as growth profiles (A and C) and maximum specific growth rates (A and D). In the CDM cultures, growth was monitored by measuring the biomass dry weight, while in the LB cultures the OD600 was measured
Effect of salt concentration on the degradation of oily sludge hydrocarbons by the consortium OS-100
When the oily sludge biotreatment was performed with a preculture prepared in LB broth, the hydrocarbon degradation efficiency of the OS-100 consortium increased as the salt concentration in the culture medium increased up to 100 g/L (Fig. 3A). The highest biodegradation of 78.7 ± 3.7% and 73.5 ± 1.4% was attained in the presence of 50 and 100 g/L NaCl, respectively (p > 0.05), whereas in the presence of 150 and 200 g/L NaCl, hydrocarbon biodegradation dropped significantly to 29.8 ± 5.4 and 12 ± 11.6% (p < 0.05), respectively. The results showed that hydrocarbon removal % by a preculture prepared in CDM + oily sludge was generally higher than that attained with a LB-grown preculture (Fig. S6). Nonetheless, statistical analyses revealed that there was no significant difference (p > 0.05), except in the cultures lacking NaCl (p < 0.05). Moreover, there was no statistically significant difference in hydrocarbon degradation by the LB-grown preculture of the consortium OS-100 and the OS100-4 isolate (p > 0.05). When compared to cultures of the isolate OS100-3, we noticed a significantly higher hydrocarbon degradation by the OS-100 consortium only in the presence of 50 g/L NaCl (p < 0.05). In addition, hydrocarbon degradation by a CDM-grown preculture of the OS-100 consortium was significantly higher than that of isolate OS100-3 at 50 and 100 g/L NaCl and the isolate OS100-4 at 100 g/L NaCl (p < 0.05).
Effect of temperature and medium composition on hydrocarbon removal from the oily sludge
Temperature also had a remarkable effect on oily sludge hydrocarbon removal by the OS-100 consortium. In fact, hydrocarbon degradation was the highest at 30 and 35 °C (p > 0.05) with rates of 80.7 ± 4.1 and 86.2 ± 8%, respectively. Then, the degradation activity dropped to 63.6 ± 3.7 and 23.1 ± 1.1% at 40 and 45 °C, respectively (Fig. 3B).
Evaluation of the effect of the medium composition revealed that OS-100 maintained high capacity to degrade hydrocarbons in the oily sludge even in the absence of the complementary components (FeCl2, MgCl2, CaCl2, trace elements, and vitamins). Under this condition, the hydrocarbon removal of 93.2 ± 3.2% was comparable to that observed in complete CDM (94.1 ± 3.7%) (p > 0.05) (Fig. 3C). On the contrary, the OS-100 consortium lost almost half (54.4 ± 1.3%) of its hydrocarbonoclastic efficiency in the absence of phosphate buffer, and in the absence of the nitrogen source (NH4Cl) hydrocarbon removal dropped to 5 ± 1% (Fig. 3C).
Total ion chromatograms showing the impact of salinity (A), temperature (B) and medium composition (C) on oily sludge hydrocarbons biodegradation by the OS-100 consortium. *: Hydrocarbons removal percentage. Complete medium contained all components (NH4Cl, phosphate buffer, vitamins, trace elements, FeCl2.4H2O, MgCl2.6H2O, CaCl2.2H2O). The preculture of the OS-100 consortium was prepared in LB broth
Temporal change in the degradation of oily sludge hydrocarbons by the OS-100 consortium
The ability of the bacterial consortium OS-100 to utilize oily sludge as the sole carbon, sulfur, and energy source was further evaluated by monitoring growth (biomass dry weight) during 10 days of incubation combined with GC-MS analysis (Fig. 4). During the first day of incubation, there was no noticeable change in the biomass. Starting from the 2nd day, the biomass dry weight increased until 5th day, then remained relatively constant (stationary phase) until the 10th day (stationary phase). The most pronounced increase in biomass occurred after 3 days of incubation and the maximum specific growth rate was 1.15 d− 1. Hydrocarbons removal efficiency increased with the incubation time, reaching 97.3% ± 0.2 after 10 days (Fig. 4). The highest hydrocarbon removal (approximately 70%) occurred between the 1st and 5th day. This was followed by a decline in hydrocarbon removal rate (only 27% within the remaining 5 days). Although the overall hydrocarbon degradation trend is consistent with the growth profile, hydrocarbon degradation continued to increase after 5 days despite lack of apparent increments in growth of the OS-100 consortium. Moreover, there appears to be periodical halts in hydrocarbon degradation at days 3–4, 5–6, and 7–9.
Growth profile of the OS-100 consortium on oily sludge (2%, w/v) represented as change in biomass dry weight and change in hydrocarbon degradation percentage with time. Values given represent the mean of three biological replicates ± standard deviation. vs.: versus. ap < 0.05 at day 0 vs. other incubation times; bp < 0.05 at day 1 vs. other incubation times; cp < 0.05 at day 2 vs. other incubation times; dp < 0.05 at day 3 vs. other incubation times; ep < 0.05 at day 4 vs. other incubation times; fp < 0.05 at day 5 vs. other incubation times *: Hydrocarbon removal percentage. The preculture of the OS-100 consortium was prepared in LB broth
Compositional shifts (population dynamics) in the OS-100 community structure under different culturing conditions
More than 99.8% of the sequences obtained from the bacterial culture OS-100 belonged to Gammaproteobacteria, most of which were affiliated to the genera Halomonas and Idiomarina at the average proportions of 83 ± 2 and 16 ± 2%, respectively (Fig. 5). NMDS ordination of the OTUs based on Bray-Curtis dissimilarities showed shifts in the composition of the OS-100 bacterial community when incubated at different times, salinities, temperatures and medium compositions (Fig. 6A). When the culture was incubated for 10 days, a significant decline in the relative abundance of Halomonas with time was observed (R2 = 0.73, p < 0.001, Fig. 6B). New sequences belonging to two OTUs of Alcanivorax were detectable in the cultures from 5 days of incubation onwards, of which one (i.e. Alcanivorax sp. 1) showed a significant increasing trend in proportion with time (p < 0.001) (Figs. 5A and 6B).
When incubated at different salinities, clear changes in the relative abundance of each member of the OS-100 bacterial culture were observed (Figs. 5B and 6A). While the relative abundance of Halomonas, and Chromohalobacter significantly increased with increasing salinity (p ≤ 0.001), the relative abundance of Idiomarina and Alcanivorax sp. 1 significantly decreased (p ≤ 0.001, Fig. 5B). At NaCl concentration of 50 g/L, the OS-100 culture was mostly dominated by Alcanivorax sp. 1, but this dominance changed in favor of Halomonas sp. and a newly detected species belonging to Chromohalobacter at 150 g/L NaCl (Fig. 5B).
The OS-100 consortium also exhibited significant compositional shifts at different temperatures. As the temperature increased, there was a significant increase in the proportions of Halomonas sp. and Alcanivorax sp. 1 (Fig. 6B). On the contrary, the proportion of Idiomarina sp. and Alcanivorax sp. 2 decreased followed the opposite trend (Fig. 5B). At > 40 °C, only Halomonas sp. and Alcanivorax sp. 1 dominated the culture, while the remaining species made up each < 10% of total sequences (Fig. 5C).
When the OS-100 culture was incubated in a complete CDM (containing all components), Halomonas sp., Idiomarina sp., and Alcanivorax sp. 1 were the dominant species (Fig. 5D). Their relative abundance remained more or less unchanged in the medium lacking phosphate, however, with a remarkable increase in the proportion of Alcanivorax sp. 2 to reach 25 ± 2% of total sequences. When the culture was incubated in the N-deprived medium, sequences belonging to Halomonas sp. and Idiomarina sp., over-dominated all others making up together 76 ± 1% of the total sequences (Fig. 5D).
Shifts in the relative abundance (% of the number of total sequences) of the major bacterial constituents of the OS-100 consortium (n = 3) in response to different incubation times, salinities, temperatures, and medium compositions. Complete medium contained all components (NH4Cl, phosphate buffer, vitamins, trace elements, FeCl2.4H2O, MgCl2.6H2O, CaCl2.2H2O). The preculture of the OS-100 consortium was prepared in LB broth
NMDS ordination plot of bacterial community composition of the OS-100 consortium at different incubation times, salinities, temperatures, and medium compositions (A). Linear regression of the relative abundance of different species and the biotreatment conditions (B). Complete medium contained all components (NH4Cl, phosphate buffer, vitamins, trace elements, FeCl2.4H2O, MgCl2.6H2O, and CaCl2.2H2O). The preculture of the OS-100 consortium was prepared in LB broth
Detection of functional genes in the OS-100 consortium
It was possible to amplify alkB gene fragments by PCR using two different primer sets (Fig. S7). However, no PCR products corresponding to genes of PAH-RHDα, naphthalene dioxygenase, or synthesis of osmoprotectants (betaine and ectoine) could be detected.
RT-qPCR was applied to assess changes in the expression of the alkB gene in OS-100 cultures grown on oily sludge in the presence of 100 g/L NaCl over time (7 days of incubation). Melt curve analysis confirmed the absence of unspecific PCR products (Fig. S8). The transcript level of alkB showed an upward trend from the 1st to the 3rd day of incubation (Fig. 7A). Maximum expression of alkB occurred after 3 days with 11.277-fold, then dropped to 1397-fold on the 4th day and slightly changed on the following days. The alkB expression profile was consistent with the observed increments in hydrocarbon degradation up to day 3 (Fig. 7B).
Temporal changes in the expression of the alkB gene (A) and total ion chromatograms (B) showing degradation of oily sludge hydrocarbons by the OS-100 bacterial consortium. Values given represent the mean of at least three biological replicates ± standard deviation. vs.: versus. ap < 0.05 at day 0 vs. other incubation times; bp < 0.05 at day 1 vs. other incubation times; cp < 0.05 at day 2 vs. other incubation times; dp < 0.05 at day 3 vs. other incubation times; ep < 0.05 at day 4 vs. other incubation times. The preculture of the OS-100 consortium was prepared in LB broth
Discussion
The ability of the consortium OS-100 to treat waste oily sludge under hyperosmotic conditions (up to 150 g/L) is particularly interesting since 15% NaCl is far greater than the classification criteria of “highly saline” which is 3.5% inorganic salt content (Peng et al. 2020). Isolation of two halophilic and hydrocarbon-degrading bacteria from the OS-100 consortium belonging to the genera Halomonas and Chromohalobacter is consistent with the halophilic and hydrocarbon degradation phenotypes of this consortium (Fathepure 2014; Gomes et al. 2018). It also indicates that the two isolated bacteria constitute a dominant culturable fraction of the OS-100 community and highlights a key role of these two isolates in the observed phenotypes of the OS-100 consortium. However, they might be contributing to different extents according to the prevailing culturing conditions. This postulation is based on the observed differences in salt tolerance and hydrocarbon removal capacity between the two isolates. This is further corroborated by the relatively closer similarity of salt tolerance, growth profile, and hydrocarbon degradation capacity of the OS-100 consortium to the corresponding phenotypes of the OS100-4 isolate. The compositional shifts that took place in the OS-100 consortium under different bioprocess conditions add further certainty to the validity of our reasoning.
A relatively higher biomass yield and hydrocarbon degradation by the OS-100 consortium compared to the isolates could be due to synergistic interactions between the different community members. Moreover, the OS-100 consortium appears to exhibit a degree of functional resilience. This reasoning is based on the finding that there was no significant difference in hydrocarbon degradation capacity when the oily sludge was treated with either a LB- or CDM-grown precultures. The finding that the consortium OS-100 exhibited maximum specific growth rate at 50 and 100 g/L NaCl both in LB and CDM + oily sludge cultures further suggests potential functional resilience of this consortium. Interestingly, 50 and 100 g/L NaCl were the concentrations which enabled highest hydrocarbon degradation by the OS-100 consortium regardless of the origin of the preculture, further attesting resilience. Furthermore, these were the optimum NaCl concentrations for growth and hydrocarbon degradation by the two isolates OS100-3 and OS100-4, which is in line with the proposed contribution to the functioning of the OS-100 consortium. Based on these findings, we propose that the type of the culture medium used to prepare the precultures did not remarkably alter the composition of the OS-100 consortium. However, validation of this hypothesis needs further culture-dependent and culture-independent studies, including community structure analysis of the OS-100 inoculum originating from the LB- and CDM-grown precultures, as well as CDM + oily sludge cultures inoculated with a CDM-grown preculture. This is particularly important since differences in biomass load (inoculum size), which was higher in case of the LB-grown preculture, could contribute to these findings.
The hydrocarbon degradation profile of the OS-100 consortium is similar to what we reported recently for another bacterial consortium where the highest degradation occurred within the initial days of incubation before a relative decrease which could be due to shortage of essential nutrients or depletion of readily biodegradable fraction (Hentati et al. 2022). The observed incremental upshift in hydrocarbon degradation after 5 days may be contradictory to the apparent growth arrest observed from day 5 to day 10. This apparent discrepancy may reflect attachment of biomass to oil droplets which leads to underestimation of the actual biomass. Adhesion to hydrocarbons/oils is common among hydrocarbon-degrading bacteria where it represents one strategy to facilitate access to hydrophobic substrates (Ismail et al. 2017; Obuekwe et al. 2009).
Temporal change in alkB expression indicates its key role in the biodegradation of the alkane fraction of the oily sludge by the OS-100 consortium. Alkanes represent a major family of petroleum hydrocarbons and constitute about 40–52% of oily sludge (Hu et al. 2013). Among alkane hydroxylases, AlkB is the most common and is used as a universal marker to assess bacterial alkane degradation (Koutinas et al. 2021; Akbari et al. 2021). However, there could be other alkane degradation genes (like cytochrome P450 enzymes, alkane monooxygenases AlmA or LadA) (Liu et al. 2011; Shuai et al. 2019) within the OS-100 genomic repertoire. This is because alkB expression increased up to day 3, which coincides with the increase in growth and hydrocarbon degradation during that period, but it significantly declined afterwards, despite continuous increase in hydrocarbon loss. Since we could not detect genes of PAH-RHDα, naphthalene dioxygenase, or synthesis of the osmoprotectants (betaine and ectoine), we postulate that these genes could be lacking in the genomes of the OS-100 members. Alternatively, homologues of those genes could be present, albeit the used primers or PCR conditions were not specific enough to detect them.
Detection of Halomonas, Idiomarina, Alcanivorax, and Chromohalobacter spp. as the dominant members of the OS-100 consortium is consistent with their frequently reported detection in saline petroleum-contaminated environments and their ability to metabolize organic pollutants such as petroleum hydrocarbons even in the presence of high salt levels (Fathepure 2014; Flores-Fernández et al. 2019; Gomes et al. 2018; Govarthanan et al. 2020; Paniagua-Michel and Fathepure 2018; Peng et al. 2020; Rizi et al. 2017; Rizzo et al. 2022; Srivastava et al. 2022; Zare et al. 2019) (Table S6). These findings are also consistent with the isolation of Halomonas and Chromohalobacter spp. from the OS-100 consortium.
The most remarkable shifts in the community profile of the OS-100 consortium occurred during the initial 5 days where hydrocarbon removal was the highest. This is consistent with the growth profile and suggests that the OS-100 community was most actively engaged in the degradation process during that time. It appears that different subpopulations contributed to hydrocarbon removal from the oily sludge at the different incubation times. Up to day 2, Halomonas and Idiomaria spp. could be the main hydrocarbon degraders, while Alcanivorax spp. probably contributed more to alkane degradation during the subsequent days. This can be inferred from the temporal decrease in the relative abundance of Halomonas and Idiomarina spp. which was accompanied by increase in the relative abundance of Alcanivorax spp. The increased expression of alkB, which is common in Alcanivorax spp. (Jagtap et al. 2021), is consistent with the changes in the community structure and further highlights the role of Alcanivorax spp. in the degradation of the oily sludge hydrocarbons.
Occurrence of the highest hydrocarbon degradation between 50 and 100 g/L NaCl reflects the salinity tolerance range of the OS-100 consortium. In addition, the downshift of hydrocarbon degradation at higher NaCl concentration is consistent with the known impact of high salinity on microbial growth and metabolism (Akbari et al. 2021; Hentati et al. 2021a, b; Truskewycz et al. 2019). The effect of salinity was well reflected in the compositional shifts that took place in the OS-100 consortium. It appears that at the lower end (50 g/L NaCl), Idiomarina and Alcanivorax sp. 1 were the main hydrocarbon degraders since they dominated the community of OS-100. However, toward the higher end (150 g/L NaCl), Halomonas and Chromohalobacter spp. were the main contributors to the salinity tolerance of the OS-100 consortium. This postulation is further corroborated by the ability of Chromohalobacter and Halomonas spp. (OS100-3 and OS100-4) isolated from the OS-100 consortium to thrive at NaCl concentrations up to 150 g/L. Furthermore, salinity-dependent phenotypic (growth profile and hydrocarbon degradation) differences between these two isolates suggest variable contribution to performance of the OS-100 community. Members of the genera Halomonas and Chromohalobacter have been repeatedly detected in some of the most saline habitats and implicated in petroleum hydrocarbon biodegradation (Paniagua-Michel and Fathepure 2018). The significant downshift in the abundance of Alcanivorax spp. at high salinity may explain the decrease in hydrocarbon degradation and further back our postulation that they were the key hydrocarbon degraders, as their relative abundance was maximum at 100 g/L NaCl, where the hydrocarbon degradation was also at maximum.
Temperature-dependent changes in the structure of the OS-100 consortium indicates differences in heat tolerance among the OS-100 bacterial members, which affected the hydrocarbon removal efficiency. Our results are in line with those of Ke et al. (2021) who reported that biodegradation of oily sludge hydrocarbons using a bacterial consortium increased with increasing the temperature (from 10 up 40 °C) and started to decrease at 50 °C, with an optimum between 30 and 40 °C, albeit changes in the community structure were not reported. We have recently found no remarkable change in the structure of a bacterial consortium growing on oily sludge when the temperature was shifted from 30 to 40 °C, despite significant increase in hydrocarbon degradation at 40 °C. At 50 °C, significant change in the community structure was accompanied by lower hydrocarbon removal (Hentati et al. 2022).
It was unexpected to see that the OS-100 consortium retained significant hydrocarbon removal efficiency in CDM lacking phosphate. Since significant changes in the community structure were only obvious in the relative increase in Alcanivorax sp. 2, it suggests a major contribution to hydrocarbon removal in the absence of phosphate. The decrease in hydrocarbon degradation in CDM lacking phosphate could be due to not only phosphate starvation (Imron et al. 2020), but also the lack of the buffering capacity. In either case, the OS-100 consortium probably retrieved phosphate from the oily sludge or internal polyphosphate reserves (Cappelletti et al. 2020; Presentato et al. 2018).
When the OS-100 consortium was challenged with nitrogen starvation, it lost 80% of its hydrocarbon degradation capacity, despite the higher abundance of Halomonas and Idiomarina spp. This underlies the importance of nitrogen for hydrocarbon biodegradation (Imron et al. 2020). It is, therefore, tempting to propose that loss of hydrocarbon degradation was mainly due to nitrogen deprivation not to a change in the bacterial community structure since the OS-100 community structure in CDM lacking nitrogen did not vary significantly from its composition in complete medium. In contrast to these results, we recently reported that another bacterial consortium retained almost half of its hydrocarbon removal efficiency during biotreatment of oily sludge in CDM lacking nitrogen source (Hentati et al. 2022). Moreover, lack of nitrogen was associated with remarkable shifts in the bacterial community structure.
A closer look at the population dynamics of the OS-100 community reveals that remarkable changes are not always associated with shifts in the hydrocarbonoclastic activity and vice versa. For instance, although the community structure changed significantly by increasing the salt concentration from 50 to 100 g/L, the hydrocarbon loss remained almost at the same level. Moreover, significant drop in hydrocarbon degradation occurred by upshifting the temperature from 40 to 45 °C despite the high similarity of the community structure at both temperatures. Similarly, removing the nitrogen source from the culture medium had marginal effect on the community composition, while the hydrocarbon degradation drastically declined.
It can be concluded that the bacterial consortium OS-100 has a remarkable ability to break down oily sludge hydrocarbons with a notable resistance to high-salinity stress. In addition, the OS-100 consortium was able to sustain its hydrocarbon degradation capacity regardless of the composition of culture medium used to prepare the inoculum. Hence, aged oily sludge is a good source of resilient autochthonous bacterial consortia that are well adapted to the recalcitrance of the oily sludge. The OS-100 consortium responded differentially to the tested culturing conditions which affected the hydrocarbonoclastic activity and composition of the OS-100 consortium. However, the most remarkable responses were observed for nitrogen starvation and high salt concentration. Moreover, different subpopulations of OS-100 potentially contributed to hydrocarbon degradation and salinity tolerance at different time intervals. Although our experimental approach provided insights into the structure and function of the OS-100 halophilic consortium during biotreatment of oily sludge, it did not reveal the actual role of each community member or potential interactions among the different members. Moreover, the GC-MS analysis targeted only one fraction of the oily sludge. Therefore, prospective studies should consider metagenomics/metaproteomics and deeper analysis using two-dimensional GC to better characterize the OS-100 consortium in terms of structure and function under various environmental conditions.
Data availability
The Illumina-MiSeq data set was submitted to the Sequence Read Archives under the accession number PRJNA921925.
References
Akbari A, David C, Rahim AA, Ghoshal S (2021) Salt selected for hydrocarbon-degrading bacteria and enhanced hydrocarbon biodegradation in slurry bioreactors. Water Res 202:117424. https://doi.org/10.1016/j.watres.2021.117424
Arahal DR, García MT, Vargas C, Cánovas D, Nieto JJ, Ventosa A (2001) Chromohalobacter salexigens sp. nov., a moderately halophilic species that includes Halomonas elongata DSM 3043 and ATCC 33174. Int J Syst Evol Microbiol 51:1457–1462. https://doi.org/10.1099/00207713-51-4-1457
Atashgahi S, Sánchez-Andrea I, Heipieper HJ, van der Meer JR, Stams AJM, Smidt H (2018) Prospects for harnessing biocide resistance for bioremediation and detoxification. Science 360:743–746. https://doi.org/10.1126/science.aar3778
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. https://doi.org/10.1373/clinchem.2008.112797
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583
Cao Y, Zhang B, Zhu Z, Song X, Cai Q, Chen B, Dong G, Ye X (2020) Microbial eco-physiological strategies for salinity-mediated crude oil biodegradation. Sci Total Environ 727:138723. https://doi.org/10.1016/j.scitotenv.2020.138723
Cappelletti M, Presentato A, Piacenza E, Firrincieli A, Turner RJ, Zannoni D (2020) Biotechnology of Rhodococcus for the production of valuable compounds. Appl Microbiol Biotechnol 104:8567–8594. https://doi.org/10.1007/s00253-020-10861-z
Chand P, Dutta S, Mukherji S (2022) Characterization and biodegradability assessment of water-soluble fraction of oily sludge using stir bar sorptive extraction and GCxGC-TOF MS. Environ Pollut 304:119177. https://doi.org/10.1016/j.envpol.2022.119177
Dobson J, McMEEKIN TA (1993) Phylogenetic relationships between some members of the genera Deleya, Halomonas, and Halovibrio. Int J Syst Bacteriol 43:665–673. https://doi.org/10.1099/00207713-43-4-665
Fathepure BZ (2014) Recent studies in microbial degradation of petroleum hydrocarbons in hypersaline environments. Front Microbiol 5:173. https://doi.org/10.3389/fmicb.2014.00173
Flores-Fernández CN, Chávez-Hidalgo E, Santos M, Zavaleta AI, Arahal DR (2019) Molecular characterization of protease producing Idiomarina species isolated from Peruvian saline environments. Microbiol Biotechnol Lett 47:401–411. https://doi.org/10.4014/mbl.1809.09001
Gomes MB, Gonzales-Limache EE, Sousa STP, Dellagnezze BM, Sartoratto A, Silva LCF, Gieg LM, Valoni E, Souza RS, Torres APR, Sousa MP, De Paula SO, Silva CC, Oliveira VM (2018) Exploring the potential of halophilic bacteria from oil terminal environments for biosurfactant production and hydrocarbon degradation under high-salinity conditions. Int Biodeterior Biodegradation 126:231–242. https://doi.org/10.1016/j.ibiod.2016.08.014
Govarthanan M, Khalifa AYZ, Kamala-Kannan S, Srinivasan P, Selvankumar T, Selvam K, Kim W (2020) Significance of allochthonous brackish water Halomonas sp. on biodegradation of low and high molecular weight polycyclic aromatic hydrocarbons. Chemosphere 243:125389. https://doi.org/10.1016/j.chemosphere.2019.125389
Hentati D, Chebbi A, Loukil S, Kchaou S, Godon J-J, Sayadi S, Chamkha M (2016) Biodegradation of fluoranthene by a newly isolated strain of Bacillus stratosphericus from Mediterranean seawater of the Sfax fishing harbour, Tunisia. Environ Sci Pollut Res 23:15088–15100. https://doi.org/10.1007/s11356-016-6648-7
Hentati D, Chebbi A, Mahmoudi A, Hadrich F, Cheffi M, Frikha I, Sayadi S, Chamkha M (2021a) Biodegradation of hydrocarbons and biosurfactants production by a newly halotolerant Pseudomonas sp. strain isolated from contaminated seawater. Biochem Eng J 166:107861. https://doi.org/10.1016/j.bej.2020.107861
Hentati D, Cheffi M, Hadrich F, Makhloufi N, Rabanal F, Manresa. A, Sayadi S, Chamkha M (2021b) Investigation of halotolerant marine Staphylococcus sp. CO100, as a promising hydrocarbon-degrading and biosurfactant-producing bacterium, under saline conditions. J Environ Manag 277:111480. https://doi.org/10.1016/j.jenvman.2020.111480
Hentati D, Abed RMM, Abotalib N, El Nayal AM, Ashraf I, Ismail W (2022) Biotreatment of oily sludge by a bacterial consortium: effect of bioprocess conditions on biodegradation efficiency and bacterial community structure. Front Microbiol 13:998076. https://doi.org/10.3389/fmicb.2022.998076
Hu G, Li J, Zeng G (2013) Recent development in the treatment of oily sludge from petroleum industry: a review. J Hazard Mater 261:470–490. https://doi.org/10.1016/j.jhazmat.2013.07.069
Ibrahim H, Xue P, Eltahir E (2020) Multiple salinity equilibria and resilience of Persian/Arabian Gulf basin salinity to brine discharge. Front Mar Sci 7:573. https://doi.org/10.3389/fmars.2020.00573
Imron MF, Kurniawan SB, Ismail N, ‘Izzati, Abdullah SRS (2020) Future challenges in diesel biodegradation by bacteria isolates: a review. J Clean Prod 251:119716. https://doi.org/10.1016/j.jclepro.2019.119716
Ismail WA, Mohamed ME-S, Awadh MN, Obuekwe C, El Nayal AM (2017) Simultaneous valorization and biocatalytic upgrading of heavy vacuum gas oil by the biosurfactant-producing Pseudomonas aeruginosa AK6U. Microb Biotechnol 10:1628–1639. https://doi.org/10.1111/1751-7915.12741
Jagtap CB, Ram RM, Tiwari OK, Titus S, Lodha T (2021) Genome sequence of an obligate hydrocarbonoclastic bacterium Alcanivorax Marinus NMRL4 isolated from oil polluted seawater of the Arabian Sea. Mar Genomics 60:100875. https://doi.org/10.1016/j.margen.2021.100875
Jerez S, Ventura M, Molina R, Pariente MI, Martínez F, Melero JA (2021) Comprehensive characterization of an oily sludge from a petrol refinery: a step forward for its valorization within the circular economy strategy. J Environ Manage 285:112124. https://doi.org/10.1016/j.jenvman.2021.112124
Jimoh AA, Ikhimiukor OO, Adeleke R (2022) Prospects in the bioremediation of petroleum hydrocarbon contaminants from hypersaline environments: a review. Environ Sci Pollut Res 29:35615–35642. https://doi.org/10.1007/s11356-022-19299-4
Ke C-Y, Qin F-L, Yang Z-G, Sha J, Sun W-J, Hui J-F, Zhang Q-Z, Zhang X-L (2021) Bioremediation of oily sludge by solid complex bacterial agent with a combined two-step process. Ecotoxicol Environ Saf 208:111673. https://doi.org/10.1016/j.ecoenv.2020.111673
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1. https://doi.org/10.1093/nar/gks808
Kondaveeti S, Govindarajan D, Mohanakrishna G, Thatikayala D, Abu-Reesh IM, Min B, Nambi IM, Al-Raoush RI, Aminabhavi TM (2023) Sustainable bioelectrochemical systems for bioenergy generation via waste treatment from petroleum industries. Fuel 331:125632. https://doi.org/10.1016/j.fuel.2022.125632
Koutinas M, Kyriakou M, Andreou K, Hadjicharalambous M, Kaliviotis E, Pasias D, Kazamias G, Varavvas C, Vyrides I (2021) Enhanced biodegradation and valorization of drilling wastewater via simultaneous production of biosurfactants and polyhydroxyalkanoates by Pseudomonas citronellolis SJTE-3. Bioresour Technol 340:125679. https://doi.org/10.1016/j.biortech.2021.125679
Ławniczak Ł, Woźniak-Karczewska M, Loibner AP, Heipieper HJ, Chrzanowski Ł (2020) Microbial Degradation of hydrocarbons-Basic principles for Bioremediation: a review. Molecules 25:856. https://doi.org/10.3390/molecules25040856
Li Y, Li W, Ji L, Song F, Li T, Fu X, Li Q, Xing Y, Zhang Q, Wang J (2022) Effects of salinity on the biodegradation of polycyclic aromatic hydrocarbons in oilfield soils emphasizing degradation genes and soil enzymes. Front Microbiol 12:824319. https://doi.org/10.3389/fmicb.2021.824319
Liu C, Wang W, Wu Y, Zhou Z, Lai Q, Shao Z (2011) Multiple alkane hydroxylase systems in a marine alkane degrader Alcanivorax dieselolei B-5. Environ Microbiol 13:1168–1178. https://doi.org/10.1111/j.1462-2920.2010.02416.x
Moreno R, Rojo F (2017) Enzymes for aerobic degradation of alkanes in bacteria. In: Rojo F (ed) Aerobic utilization of hydrocarbons, oils and lipids. Handbook of hydrocarbon and lipid Microbiology. Springer, Cham. https://doi.org/10.1007/978-3-319-39782-5_6-1
Muneeswari R, Swathi KV, Sekaran G, Ramani K (2023) A real time integrated approach for the treatment of petroleum industry oily waste through ancillary carbon metabolism and biocatalytic cascade induction. Process Saf Environ Prot 170:464–478. https://doi.org/10.1016/j.psep.2022.12.022
Murungi PI, Sulaimon AA (2022) Petroleum sludge treatment and disposal techniques: a review. Environ Sci Pollut Res 29:40358–40372. https://doi.org/10.1007/s11356-022-19614-z
Neifar M, Chouchane H, Najjari A, El Hidri D, Mahjoubi M, Ghedira K, Naili F, Soufi L, Raddadi N, Sghaier H, Ouzari HI, Masmoudi AS, Cherif A (2019) Genome analysis provides insights into crude oil degradation and biosurfactant production by extremely halotolerant Halomonas desertis G11 isolated from Chott El-Djerid salt-lake in Tunisian desert. Genomics 111:1802–1814. https://doi.org/10.1016/j.ygeno.2018.12.003
Obuekwe CO, Al-Jadi ZK, Al-Saleh ES (2009) Hydrocarbon degradation in relation to cell-surface hydrophobicity among bacterial hydrocarbon degraders from petroleum-contaminated Kuwait desert environment. Int Biodeterior Biodegradation 63:273–279. https://doi.org/10.1016/j.ibiod.2008.10.004
Paniagua-Michel J, Fathepure BZ (2018) Microbial consortia and biodegradation of petroleum hydrocarbons in marine environments. In: Kumar V, Kumar M, Prasad R (eds) Microbial Action on hydrocarbons. Springer, Singapore, pp 1–20. https://doi.org/10.1007/978-981-13-1840-5_1
Paparella F, D’Agostino D, Burt A J (2022) Long-term, basin-scale salinity impacts from desalination in the Arabian/Persian Gulf. Sci Rep 12:20549. https://doi.org/10.1038/s41598-022-25167-5
Peng S, Kai M, Yang X, Luo Y, Bai L (2020) Study on the osmoregulation of Halomonas socia NY-011 and the degradation of organic pollutants in the saline environment. Extremophiles 24:843–861. https://doi.org/10.1007/s00792-020-01199-5
Presentato A, Cappelletti M, Sansone A, Ferreri C, Piacenza E, Demeter MA, Crognale S, Petruccioli M, Milazzo G, Fedi S, Steinbüchel A, Turner RJ, Zannoni D (2018) Aerobic growth of Rhodococcus aetherivorans BCP1 using selected naphthenic acids as the sole carbon and energy sources. Front Microbiol 9:672. https://doi.org/10.3389/fmicb.2018.00672
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2012) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
R Core Team (2018) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Rizi MSS, Emtiazi G, Sepahi AA, Maal KB, Hosseini F (2017) The effect of nanoparticles on oil sludge biodegradation by Chromohalobacter sp. isolated from the Bahregan area in the Persian Gulf. Pet Sci Technol 35:687–695. https://doi.org/10.1080/10916466.2016.1247171
Rizzo C, Papale M, Lo Giudice A (2022) Idiomarina sp. isolates from cold and temperate environments as biosurfactant producers. J Mar Sci Eng 10:1135. https://doi.org/10.3390/jmse10081135
Saeedi R, Abtahi M, Almasi H, Jorfi S (2022) Enhanced biosurfactant-assisted composting of oily sludge using a diverse halo-tolerant consortium in the saline environment: effect of repeated inoculation and mixing ratios. Biomass Conv Bioref. https://doi.org/10.1007/s13399-022-02472-7
Sarkar J, Roy A, Sar P, Kazy SK (2020) Chap. 2 - accelerated bioremediation of petroleum refinery sludge through biostimulation and bioaugmentation of native microbiome. In: Shah MP, Rodriguez-Couto S, Şengör SS (eds) Emerging technologies in Environmental Bioremediation. Elsevier, pp 23–65. https://doi.org/10.1016/B978-0-12-819860-5.00002-X
Shuai Y, Zhou H, Mu Q, Zhang D, Zhang N, Tang J, Zhang C (2019) Characterization of a biosurfactant-producing Leclercia sp. B45 with new transcriptional patterns of alkB gene. Ann Microbiol 69:139–150. https://doi.org/10.1007/s13213-018-1409-0
Simmonds EJ, Lamboeuf M (1981) Environmental conditions in the Gulf and the Gulf of Oman and their influence on the propagation of sound. UNDP/FAO, p 62. Fish. Surv. Develop. Proj., FI: DP/RAB/71/278/12
Srivastava AK, Srivastava R, Sharma A, Bharati AP, Yadav J, Singh AK, Tiwari PK, Srivatava AK, Chakdar H, Kashyap PL, Saxena AK (2022) Transcriptome analysis to understand salt stress regulation mechanism of Chromohalobacter salexigens ANJ207. Front Microbiol 13:909276. https://doi.org/10.3389/fmicb.2022.909276
Sun W-J, Zhou J, Zhang Q-T, Xin M, Sun R, Ke C-Y, Qin F-L, Zhang Q-Z, Zhang X-L (2023) A salt-tolerant petroleum-degrading strain Pseudomonas aeruginosa SD and its remediation potential for oily sludge (preprint). In Review. https://doi.org/10.21203/rs.3.rs-2434390/v1
Taher M, Mohamed A-R, Al-Ali AKH (2012) Some ecological characteristics and ichthyofauna of surrounding Sammaliah Island, Abu Dhabi, UAE. Basrah J Sci 30:31–49
Truskewycz A, Gundry TD, Khudur LS, Kolobaric A, Taha M, Aburto-Medina A, Ball AS, Shahsavari E (2019) Petroleum hydrocarbon contamination in terrestrial ecosystems—fate and microbial responses. Molecules 24:3400. https://doi.org/10.3390/molecules24183400
US EPA (2016) A user-friendly reference document for hazardous waste listings [WWW Document]. US EPA, Washington, DC
Wang H, Kuang S, Lang Q, Wang L (2021) Bacterial community structure of aged oil sludge contaminated soil revealed by Illumina high-throughput sequencing in East China. World J Microbiol Biotechnol 37:183. https://doi.org/10.1007/s11274-021-03059-6
Yin Q, Nie H, Nie M, Guo Y, Zhang B, Wang L, Wang Y, Bai X (2023) Rapid effective treatment of waxy oily sludge using a method of dispersion combined with biodegradation in a semi-fluid state. Environ Pollut 319:120971. https://doi.org/10.1016/j.envpol.2022.120971
Zahari NZ, Tuah PM, Ali SAM, Sabullah MK (2022) Microbial growth rate and distribution of doubling time at different concentration of oil sludge medium. IOP Conf Ser: Earth Environ Sci 1022:012075. https://doi.org/10.1088/1755-1315/1022/1/012075
Zare N, Bonakdarpour B, Amoozegar MA, Shavandi M, Fallah N, Darabi S, Taromsary NB (2019) Using enriched water and soil-based indigenous halophilic consortia of an oilfield for the biological removal of organic pollutants in hypersaline produced water generated in the same oilfield. Process Saf Environ Prot 127:151–161. https://doi.org/10.1016/j.psep.2019.04.024
Acknowledgements
We would like to thank the Arabian Gulf University for funding this study.
Author information
Authors and Affiliations
Contributions
DH: Conceptualization, Data curation, Formal analysis, Methodology, Investigation, Writing - original draft; Writing - review & editing; ARR: Methodology, RMMA: Data curation, Formal analysis, Writing - review & editing. NA: Methodology. AEN: Methodology; WI: Supervision, Conceptualization, Funding acquisition, Project administration, Writing - original draft; Writing - review & editing. All authors revised the manuscript and approved the content.
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hentati, D., Ramadan, A.R., Abed, R.M.M. et al. Functional and structural responses of a halophilic consortium to oily sludge during biodegradation. Appl Microbiol Biotechnol 108, 116 (2024). https://doi.org/10.1007/s00253-023-12896-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00253-023-12896-4