Abstract
Protein glycosylation and other post-translational modifications are involved in potentially all aspects of human growth and development. Defective glycosylation has adverse effects on human physiological conditions and accompanies many chronic and infectious diseases. Altered glycosylation can occur at the onset and/or during tumor progression. Identifying these changes at early disease stages may aid in making decisions regarding treatments, as early intervention can greatly enhance survival. This review highlights some of the efforts being made to identify N- and O-glycosylation profile shifts in cancer using mass spectrometry. The analysis of single or panels of potential glycoprotein cancer markers are covered. Other emerging technologies such as global glycan release and site-specific glycosylation analysis and quantitation are also discussed.

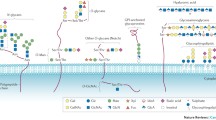
Steps involved in the biomarker discovery






Similar content being viewed by others
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90.
Ferlay J, Autier P, Boniol M, Heanue M, Colombet M, Boyle P. Estimates of the cancer incidence and mortality in Europe in 2006. Ann Oncol. 2007;18(3):581–92.
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–917.
Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58(2):71–96.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30.
Sawyers CL. The cancer biomarker problem. Nature. 2008;452(7187):548–52.
Kohn EC, Azad N, Annunziata C, Dhamoon AS, Whiteley G. Proteomics as a tool for biomarker discovery. Dis Markers. 2007;23(5/6):411–7.
Diamandis EP, van der Merwe D-E. Plasma protein profiling by mass spectrometry for cancer diagnosis: opportunities and limitations. Clin Cancer Res. 2005;11(3):963–5.
Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol. 2008;5(10):588–99.
van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AAM, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–6.
Espina V, Woodhouse EC, Wulfkuhle J, Asmussen HD, Petricoin Iii EF, Liotta LA. Protein microarray detection strategies: focus on direct detection technologies. J Immunol Methods. 2004;290(1/2):121–33.
Wulfkuhle JD, Liotta LA, Petricoin EF. Proteomic applications for the early detection of cancer. Nat Rev Cancer. 2003;3(4):267–75.
Nagler RM. Saliva as a tool for oral cancer diagnosis and prognosis. Oral Oncol. 2009;45(12):1006–10.
Laxman B, Morris DS, Yu J, Siddiqui J, Cao J, Mehra R, et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res. 2008;68(3):645–9.
Alexander H, Stegner AL, Wagner-Mann C, Du Bois GC, Alexander S, Sauter ER. Proteomic analysis to identify breast cancer biomarkers in nipple aspirate fluid. Clin Cancer Res. 2004;10(22):7500–10.
Teng P-N, Bateman NW, Hood BL, Conrads TP. Advances in proximal fluid proteomics for disease biomarker discovery. J Proteome Res. 2010;9(12):6091–100.
Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452(7187):571–9.
Hassanein M, Callison JC, Callaway-Lane C, Aldrich MC, Grogan EL, Massion PP. The state of molecular biomarkers for the early detection of lung cancer. Cancer Prev Res. 2012;5(8):992–1006.
Polanski M, Anderson NL. A list of candidate cancer biomarkers for targeted proteomics. Biomarker Insights. 2006;1:1.
Chen X, Shan Q, Jiang L, Zhu B, Xi X. Quantitative proteomic analysis by iTRAQ for identification of candidate biomarkers in plasma from acute respiratory distress syndrome patients. Biochem Biophys Res Commun. 2013;441(1):1–6.
Lam YW, Mobley JA, Evans JE, Carmody JF, Ho SM. Mass profiling-directed isolation and identification of a stage-specific serologic protein biomarker of advanced prostate cancer. Proteomics. 2005;5(11):2927–38.
Palmblad M, Tiss A, Cramer R. Mass spectrometry in clinical proteomics—from the present to the future. Proteom Clin Appl. 2009;3(1):6–17.
Miller RA, Spellman DS. Mass spectrometry-based biomarkers in drug development. In: Woods AG, Darie CC (Ed.) Advancements of mass spectrometry in biomedical research, vol 806, pp 341–359. Advances in Experimental Medicine and Biology. Switzerland: Springer International Publishing; 2014.
Pusch W, Flocco MT, Leung SM, Thiele H, Kostrzewa M. Mass spectrometry-based clinical proteomics. Pharmacogenomics. 2003;4(4):463–76.
Zhang Y, Jiao J, Yang P, Lu H. Mass spectrometry-based N-glycoproteomics for cancer biomarker discovery. Clin Proteom. 2014;11(1):18.
Jimenez CR, Verheul HMW. Mass spectrometry-based proteomics: from cancer biology to protein biomarkers, drug targets, and clinical applications. Am Soc Clinl Oncol educational book/ASCO Am Soc Clin Oncol Meeting. e504–510. 2014.
Yin H, Lin Z, Nie S, Wu J, Tan Z, Zhu J, et al. Mass-selected site-specific core-fucosylation of ceruloplasmin in alcohol-related hepatocellular carcinoma. J Proteome Res. 2014;13(6):2887–96.
Griffiths WJ, Wang Y. Mass spectrometry: from proteomics to metabolomics and lipidomics. Chem Soc Rev. 2009;38(7):1882–96.
Ly M, Laremore TN, Linhardt RJ. Proteoglycomics: recent progress and future challenges. OMICS J Integrative Biol. 2010;14(4):389–99.
Wuhrer M, Catalina MI, Deelder AM, Hokke CH. Glycoproteomics based on tandem mass spectrometry of glycopeptides. J Chromatogr B. 2007;849(1):115–28.
Zaia J. Mass spectrometry and the emerging field of glycomics. Chem Biol. 2008;15(9):881–92.
Hua S, Lebrilla C, An HJ. Application of nano-LC-based glycomics towards biomarker discovery. Bioanalysis. 2011;3(22):2573–85.
Alley WR, Madera M, Mechref Y, Novotny MV. Chip-based reversed-phase liquid chromatography-mass spectrometry of permethylated N-linked glycans: a potential methodology for cancer-biomarker discovery. Anal Chem. 2010;82(12):5095–106.
Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5(11):976–89.
Kronewitter SR, De Leoz MLA, Strum JS, An HJ, Dimapasoc LM, Guerrero A, et al. The glycolyzer: automated glycan annotation software for high performance mass spectrometry and its application to ovarian cancer glycan biomarker discovery. Proteomics. 2012;12(15/16):2523–38.
Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis, and treatment selection. Nat Rev Cancer. 2005;5(11):845–56.
Yin BWT, Lloyd KO. Molecular Cloning of the CA125 Ovarian Cancer Antigen: identification as a new mucin, MUC16. J Biol Chem. 2001;276(29):27371–5.
Jacobs I, Bast RC. The CA 125 tumor-associated antigen: a review of the literature. Hum Reprod. 1989;4(1):1–12.
Miralles C, Orea M, España P, Provencio M, Sánchez A, Cantos B, et al. Cancer antigen 125 associated with multiple benign and malignant pathologies. Ann Surg Oncol. 2003;10(2):150–4.
Adamczyk B, Tharmalingam T, Rudd PM. Glycans as cancer biomarkers. Biochim Biophys Acta - General Subjects. 2012;1820(9):1347–53.
Duffy M, Shering S, Sherry F, McDermott E, O'higgins N. CA 15-3: a prognostic marker in breast cancer. Int J Biol Markers. 1999;15(4):330–3.
Drake RR, Jones EE, Powers TW, Nyalwidhe JO. Chapter 10-altered glycosylation in prostate cancer. Adv Cancer Res. 2015;126:345–82.
Catalona W, Richie J, Ahmann F, Hudson M, Scardino P, Flanigan R, et al. Comparison of digital rectal examination and serum prostate specific antigen in the early detection of prostate cancer: results of a multicenter clinical trial of 6,630 men. J Urol. 1994;151(5):1283–90.
Hammarström S. The carcinoembryonic antigen (CEA) family: structures, suggested functions, and expression in normal and malignant tissues. Semin Cancer Biol. 1999;9(2):67–81.
Benchimol S, Fuks A, Jothy S, Beauchemin N, Shirota K, Stanners CP. Carcinoembryonic antigen, a human tumor marker, functions as an intercellular adhesion molecule. Cell. 1989;57(2):327–34.
Moertel CG, Fleming TR, Macdonald JS, Haller DG, Laurie JA, Tangen C. An evaluation of the carcinoembryonic antigen (CEA) test for monitoring patients with resected colon cancer. JAMA. 1993;270(8):943–7.
Thompson S, Turner G. Elevated levels of abnormally-fucosylated haptoglobins in cancer sera. Br J Cancer. 1987;56(5):605.
Okuyama N, Ide Y, Nakano M, Nakagawa T, Yamanaka K, Moriwaki K, et al. Fucosylated haptoglobin is a novel marker for pancreatic cancer: a detailed analysis of the oligosaccharide structure and a possible mechanism for fucosylation. Int J Cancer. 2006;118(11):2803–8.
Miyoshi E, Nakano M. Fucosylated haptoglobin is a novel marker for pancreatic cancer: detailed analyses of oligosaccharide structures. Proteomics. 2008;8(16):3257–62.
Zhao C, Annamalai L, Guo C, Kothandaraman N, Koh SCL, Zhang H, et al. Circulating haptoglobin is an independent prognostic factor in the sera of patients with epithelial ovarian cancer. Neoplasia (New York, NY). 2007;9(1):1–7.
Ahmed N, Barker G, Oliva KT, Hoffmann P, Riley C, Reeve S, et al. Proteomic-based identification of haptoglobin-1 precursor as a novel circulating biomarker of ovarian cancer. Br J Cancer. 2004;91(1):129–40.
Huang HL, Stasyk T, Morandell S, Dieplinger H, Falkensammer G, Griesmacher A, et al. Biomarker discovery in breast cancer serum using 2-D differential gel electrophoresis/MALDI-TOF/TOF and data validation by routine clinical assays. Electrophoresis. 2006;27(8):1641–50.
Ralhan R, DeSouza LV, Matta A, Tripathi SC, Ghanny S, Gupta SD, et al. Discovery and verification of head-and-neck cancer biomarkers by differential protein expression analysis using iTRAQ labeling, multidimensional liquid chromatography, and tandem mass spectrometry. Mol Cell Proteom. 2008;7(6):1162–73.
Patz EF, Campa MJ, Gottlin EB, Kusmartseva I, Guan XR, Herndon JE. Panel of serum biomarkers for the diagnosis of lung cancer. J Clin Oncol. 2007;25(35):5578–83.
Drake RR, Schwegler EE, Malik G, Diaz J, Block T, Mehta A, et al. Lectin capture strategies combined with mass spectrometry for the discovery of serum glycoprotein biomarkers. Mol Cell Proteom. 2006;5(10):1957–67.
Zhao J, Simeone DM, Heidt D, Anderson MA, Lubman DM. Comparative serum glycoproteomics using lectin selected sialic acid glycoproteins with mass spectrometric analysis: application to pancreatic cancer serum. J Proteome Res. 2006;5(7):1792–802.
Kaji H, Saito H, Yamauchi Y, Shinkawa T, Taoka M, Hirabayashi J, et al. Lectin affinity capture, isotope-coded tagging, and mass spectrometry to identify N-linked glycoproteins. Nat Biotech. 2003;21(6):667–72.
Wu J, Xie X, Liu Y, He J, Benitez R, Buckanovich RJ, et al. Identification and confirmation of differentially expressed fucosylated glycoproteins in the serum of ovarian cancer patients using a lectin array and LC-MS/MS. J Proteome Res. 2012;11(9):4541–52.
Alvarez-Manilla G, Warren NL, Atwood III J, Orlando R, Dalton S, Pierce M. Glycoproteomic analysis of embryonic stem cells: identification of potential glycobiomarkers using lectin affinity chromatography of glycopeptides. J Proteome Res. 2010;9(5):2062–75.
Kuno A, Kato Y, Matsuda A, Kaneko MK, Ito H, Amano K, et al. Focused differential glycan analysis with the platform antibody-assisted lectin profiling for glycan-related biomarker verification. Mol Cell Proteom. 2009;8(1):99–108.
Li Y, Wen T, Zhu M, Li L, Wei J, Wu X, et al. Glycoproteomic analysis of tissues from patients with colon cancer using lectin microarrays and nanoLC-MS/MS. Mol Biosyst. 2013;9(7):1877–87.
Dennis JW, Laferte S, Waghorne C, Breitman ML, Kerbel RS. Beta 1-6 branching of Asn-linked oligosaccharides is directly associated with metastasis. Science. 1987;236(4801):582–5.
Ihara S, Miyoshi E, Ko JH, Murata K, Nakahara S, Honke K, et al. Prometastatic effect of N-acetylglucosaminyltransferase V is due to modification and stabilization of active matriptase by adding β1–6 GlcNAc branching. J Biol Chem. 2002;277(19):16960–7.
Buckhaults P, Chen L, Fregien N, Pierce M. Transcriptional regulation of N-acetylglucosaminyltransferase V by the src oncogene. J Biol Chem. 1997;272(31):19575–81.
Abbott KL, Aoki K, Lim J-M, Porterfield M, Johnson R, O'Regan RM, et al. Targeted glycoproteomic identification of biomarkers for human breast carcinoma. J Proteome Res. 2008;7(4):1470–80.
Thompson A, Schäfer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75(8):1895–904.
Nie S, Lo A, Wu J, Zhu J, Tan Z, Simeone DM, et al. Glycoprotein biomarker panel for pancreatic cancer discovered by quantitative proteomics analysis. J Proteome Res. 2014;13(4):1873–84.
Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat Biotech. 2003;21(3):255–61.
Pagel O, Loroch S, Sickmann A, Zahedi RP. Current strategies and findings in clinically relevant post-translational modification-specific proteomics. Expert Rev Proteom. 2015;12(3):235–53.
Christiansen MN, Chik J, Lee L, Anugraham M, Abrahams JL, Packer NH. Cell surface protein glycosylation in cancer. Proteomics. 2014;14(4/5):525–46.
Holst S, Wuhrer M, Rombouts Y (2015) Chapter 6 – Glycosylation characteristics of colorectal cancer. In: Richard RD, Lauren EB (Eds.) Advances in Cancer Research, vol 126, pp 203–256. Academic Press.
Walsh G, Jefferis R. Post-translational modifications in the context of therapeutic proteins. Nat Biotech. 2006;24(10):1241–52.
An HJ, Froehlich JW, Lebrilla CB. Determination of glycosylation sites and site-specific heterogeneity in glycoproteins. Curr Opin Chem Biol. 2009;13(4):421–6.
Shriver Z, Raguram S, Sasisekharan R. Glycomics: a pathway to a class of new and improved therapeutics. Nat Rev Drug Discov. 2004;3(10):863–73.
Dube DH, Bertozzi CR. Glycans in cancer and inflammation—potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;4(6):477–88.
Reis CA, Osorio H, Silva L, Gomes C, David L. Alterations in glycosylation as biomarkers for cancer detection. J Clin Pathol. 2010;63(4):322–9.
Varki A, Kannagi R, Toole BP. Glycosylation changes in cancer. 2009.
An HJ, Kronewitter SR, de Leoz MLA, Lebrilla CB. Glycomics and disease markers. Curr Opin Chem Biol. 2009;13(5/6):601–7.
Guo H, Abbott KL (2015) Chapter 8 – functional impact of tumor-specific N-linked glycan changes in breast and ovarian cancers. In: Richard RD, Lauren EB (Eds.) Advances in Cancer Research, vol 126, pp 281–303. Academic Press.
Drake PM, Cho W, Li B, Prakobphol A, Johansen E, Anderson NL, et al. Sweetening the pot: adding glycosylation to the biomarker discovery equation. Clin Chem. 2010;56(2):223–36.
Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15:540–55.
de Leoz MLA, Young LJT, An HJ, Kronewitter SR, Kim J, Miyamoto S, et al. High-mannose glycans are elevated during breast cancer progression. Mol Cell Proteom. 2011;10(1).
Kailemia MJ, Ruhaak LR, Lebrilla CB, Amster IJ. Oligosaccharide analysis by mass spectrometry: a review of recent developments. Anal Chem. 2013;86(1):196–212.
Ruhaak LR, Miyamoto S, Lebrilla CB. Developments in the identification of glycan biomarkers for the detection of cancer. Mol Cell Proteom. 2013;12(4):846–55.
Budnik BA, Lee RS, Steen JAJ. Global methods for protein glycosylation analysis by mass spectrometry. Biochim Biophys Acta. 2006;1764(12):1870–80.
An HJ, Lebrilla CB (2010) A glycomics approach to the discovery of potential cancer biomarkers. In: Functional glycomics, 199–213. Springer.
JooáAn H. The prospects of glycan biomarkers for the diagnosis of diseases. Mol Biosyst. 2009;5(1):17–20.
An HJ, Miyamoto S, Lancaster KS, Kirmiz C, Li B, Lam KS, et al. Profiling of glycans in serum for the discovery of potential biomarkers for ovarian cancer. J Proteome Res. 2006;5(7):1626–35.
Bharti A, Ma PC, Maulik G, Singh R, Khan E, Skarin AT, et al. Haptoglobin α-subunit and hepatocyte growth factor can potentially serve as serum tumor biomarkers in small-cell lung cancer. Anticancer Res. 2004;24(2C):1031–8.
Bones J, Mittermayr S, O'Donoghue N, Guttman A, Rudd PM. Ultra performance liquid chromatographic profiling of serum N-glycans for fast and efficient identification of cancer associated alterations in glycosylation. Anal Chem. 2010;82(24):10208–15.
Wada Y, Azadi P, Costello CE, Dell A, Dwek RA, Geyer H, et al. Comparison of the methods for profiling glycoprotein glycans—HUPO Human Disease Glycomics/Proteome Initiative multi-institutional study. Glycobiology. 2007;17(4):411–22.
Ressom HW, Varghese RS, Goldman L, An Y, Loffredo CA, Abdel-Hamid M, et al. Analysis of MALDI-TOF mass spectrometry data for discovery of peptide and glycan biomarkers of hepatocellular carcinoma. J Proteome Res. 2008;7(2):603–10.
Rodrigo MAM, Zitka O, Krizkova S, Moulick A, Adam V, Kizek R. MALDI-TOF MS as evolving cancer diagnostic tool: a review. J Pharm Biomed Anal. 2014;95:245–55.
Powers TW, Neely BA, Shao Y, Tang H, Troyer DA, Mehta AS, et al. MALDI imaging mass spectrometry profiling of N-glycans in formalin-fixed paraffin embedded clinical tissue blocks and tissue microarrays. PLoS One. 2014;9(9).
Kyselova Z, Mechref Y, Al Bataineh MM, Dobrolecki LE, Hickey RJ, Vinson J, et al. Alterations in the serum glycome due to metastatic prostate cancer. J Proteome Res. 2007;6(5):1822–32.
Kyselova Z, Mechref Y, Kang P, Goetz JA, Dobrolecki LE, Sledge GW, et al. Breast cancer diagnosis and prognosis through quantitative measurements of serum glycan profiles. Clin Chem. 2008;54(7):1166–75.
Balog CIA, Stavenhagen K, Fung WLJ, Koeleman CA, McDonnell LA, Verhoeven A, et al. N-glycosylation of colorectal cancer tissues. Mol Cell Proteom. 2012;11(9):571–85.
de Leoz MLA, An HJ, Kronewitter S, Kim J, Beecroft S, Vinall R, et al. Glycomic approach for potential biomarkers on prostate cancer: Profiling of N-linked glycans in human sera and pRNS cell lines. Dis Markers. 2008;25(4/5):243–58.
Hecht ES, Scholl EH, Walker SH, Taylor AD, Cliby WA, Motsinger-Reif AA, et al. Relative quantification and higher-order modeling of the plasma glycan cancer burden ratio in ovarian cancer case-control samples. J Proteome Res. 2015;14(10):4394–401.
Kirmiz C, Li B, An HJ, Clowers BH, Chew HK, Lam KS, et al. A serum glycomics approach to breast cancer biomarkers. Mol Cell Proteom. 2007;6(1):43–55.
Alley WR, Vasseur JA, Goetz JA, Svoboda M, Mann BF, Matei DE, et al. N-linked glycan structures and their expressions change in the blood sera of ovarian cancer patients. J Proteome Res. 2012;11(4):2282–300.
Saldova R, Royle L, Radcliffe CM, Abd Hamid UM, Evans R, Arnold JN, et al. Ovarian cancer is associated with changes in glycosylation in both acute-phase proteins and IgG. Glycobiology. 2007;17(12):1344–56.
Kanoh Y, Mashiko T, Danbara M, Takayama Y, Ohtani S, Imasaki T, et al. Analysis of the Oligosaccharide chain of human serum immunoglobulin G in patients with localized or metastatic cancer. Oncology. 2004;66(5):365–70.
Ruhaak LR, Stroble C, Dai J, Barnett MJ, Taguchi A, Goodman GE, et al. Serum glycans as risk markers for non-small cell lung cancer. Cancer Prev Res. 2016. doi:10.1158/1940-6207.capr-15-0033.
Hua S, An HJ, Ozcan S, Ro GS, Soares S, DeVere-White R, et al. Comprehensive native glycan profiling with isomer separation and quantitation for the discovery of cancer biomarkers. Analyst. 2011;136(18):3663–71.
Ruhaak LR, Deelder A, Wuhrer M. Oligosaccharide analysis by graphitized carbon liquid chromatography-mass spectrometry. Anal Bioanal Chem. 2009;394(1):163–74.
Stavenhagen K, Kolarich D, Wuhrer M. Clinical glycomics employing graphitized carbon liquid chromatography-mass spectrometry. Chromatographia. 2014;78(5):307–20.
Chu CS, Niñonuevo MR, Clowers BH, Perkins PD, An HJ, Yin H, et al. Profile of native N-linked glycan structures from human serum using high performance liquid chromatography on a microfluidic chip and time-of-flight mass spectrometry. Proteomics. 2009;9(7):1939–51.
Song T, Aldredge D, Lebrilla CB. A method for in-depth structural annotation of human serum glycans that yields biological variations. Anal Chem. 2015;87(15):7754–62.
Ruhaak LR, Barkauskas DA, Torres J, Cooke CL, Wu LD, Stroble C, et al. The serum immunoglobulin G glycosylation signature of gastric cancer. EuPA Open Proteom. 2015;6:1–9.
Strum JS, Nwosu CC, Hua S, Kronewitter SR, Seipert RR, Bachelor RJ, et al. Automated Assignments of N- and O-site specific glycosylation with extensive glycan heterogeneity of glycoprotein mixtures. Anal Chem. 2013;85(12):5666–75.
Nwosu CC, Seipert RR, Strum JS, Hua SS, An HJ, Zivkovic AM, et al. Simultaneous and extensive site-specific N- and O-glycosylation analysis in protein mixtures. J Proteome Res. 2011;10(5):2612–24.
Ueda K, Takami S, Saichi N, Daigo Y, Ishikawa N, Kohno N, et al. Development of serum glycoproteomic profiling technique; simultaneous identification of glycosylation sites and site-specific quantification of glycan structure changes. Mol Cell Proteom. 2010;9(9):1819–28.
Ueda K. Glycoproteomic strategies: From discovery to clinical application of cancer carbohydrate biomarkers. Proteom Clin App. 2013;7(9/10):607–17.
Bern M, Kil YJ, Becker C. Byonic: advanced peptide and protein identification software. Curr Protoc Bioinform. 2012;40:13.20.11–13.20.14.
Ozohanics O, Krenyacz J, Ludányi K, Pollreisz F, Vékey K, Drahos L. GlycoMiner: a new software tool to elucidate glycopeptide composition. Rapid Commun Mass Spectrom. 2008;22(20):3245–54.
He L, Xin L, Shan B, Lajoie GA, Ma B. GlycoMaster DB: Software to assist the automated identification of N-linked glycopeptides by tandem mass spectrometry. J Proteome Res. 2014;13(9):3881–95.
Mayampurath A, Yu C-Y, Song E, Balan J, Mechref Y, Tang H. Computational framework for identification of intact glycopeptides in complex samples. Anal Chem. 2014;86(1):453–63.
Mayampurath AM, Wu Y, Segu ZM, Mechref Y, Tang H. Improving confidence in detection and characterization of protein N‐glycosylation sites and microheterogeneity. Rapid Commun Mass Spectrom. 2011;25(14):2007–19.
Khatri K, Staples GO, Leymarie N, Leon DR, Turiák L, Huang Y, et al. Confident assignment of site-specific glycosylation in complex glycoproteins in a single step. J Proteome Res. 2014;13(10):4347–55.
Go EP, Rebecchi KR, Dalpathado DS, Bandu ML, Zhang Y, Desaire H. GlycoPep DB: a tool for glycopeptide analysis using a “Smart Search”. Anal Chem. 2007;79(4):1708–13.
Woodin CL, Maxon M, Desaire H. Software for automated interpretation of mass spectrometry data from glycans and glycopeptides. Analyst. 2013;138(10):2793–803.
Reid GE, Stephenson JL, McLuckey SA. Tandem mass spectrometry of ribonuclease A and B: N-linked glycosylation site analysis of whole protein ions. Anal Chem. 2002;74(3):577–83.
Siuti N, Kelleher NL. Decoding protein modifications using top-down mass spectrometry. Nat Methods. 2007;4(10):817–21.
Ito S, Hayama K, Hirabayashi J (2009) Enrichment strategies for glycopeptides. In: Packer N, Karlsson N (Eds.) Glycomics, vol 534, pp 194–203. Methods in Molecular Biology. Humana Press.
Wuhrer M, Deelder AM, Hokke CH. Protein glycosylation analysis by liquid chromatography–mass spectrometry. J Chromatogr B. 2005;825(2):124–33.
Zhu Z, Go EP, Desaire H. Absolute quantitation of glycosylation site occupancy using isotopically labeled standards and LC-MS. J Am Soc Mass Spectrom. 2014;25(6):1012–7.
Dalpathado DS, Desaire H. Glycopeptide analysis by mass spectrometry. Analyst. 2008;133(6):731–8.
Gbormittah FO, Bones J, Hincapie M, Tousi F, Hancock WS, Iliopoulos O. Clusterin glycopeptide variant characterization reveals significant site-specific glycan changes in the plasma of clear cell renal cell carcinoma. J Proteome Res. 2015;14(6):2425–36.
Tan Z, Yin H, Nie S, Lin Z, Zhu J, Ruffin MT, et al. Large-scale identification of core-fucosylated glycopeptide sites in pancreatic cancer serum using mass spectrometry. J Proteome Res. 2015;14(4):1968–78.
Lin Z, Yin H, Lo A, Ruffin MT, Anderson MA, Simeone DM, et al. Label-free relative quantification of α-2-macroglobulin site-specific core-fucosylation in pancreatic cancer by LC-MS/MS. Electrophoresis. 2014;35(15):2108–15.
Saldova R, Fan Y, Fitzpatrick JM, Watson RWG, Rudd PM. Core fucosylation and α2-3 sialylation in serum N-glycome is significantly increased in prostate cancer comparing to benign prostate hyperplasia. Glycobiology. 2011;21(2):195–205.
Miyoshi E, Moriwaki K, Nakagawa T. Biological function of fucosylation in cancer biology. J Biochem. 2008;143(6):725–9.
Ruhaak LR, Lebrilla C. Applications of multiple reaction monitoring to clinical glycomics. Chromatographia. 2015;78(5/6):335–42.
Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Meth. 2012;9(6):555–66.
Song E, Pyreddy S, Mechref Y. Quantification of glycopeptides by multiple reaction monitoring liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2012;26(17):1941–54.
Hong Q, Lebrilla CB, Miyamoto S, Ruhaak LR. Absolute quantitation of immunoglobulin G and its glycoforms using multiple reaction monitoring. Anal Chem. 2013;85(18):8585–93.
Sanda M, Pompach P, Brnakova Z, Wu J, Makambi K, Goldman R. Quantitative liquid chromatography-mass spectrometry-multiple reaction monitoring (LC-MS-MRM) analysis of site-specific glycoforms of haptoglobin in liver disease. Mol Cell Proteom. 2013;12(5):1294–305.
Yuan W, Sanda M, Wu J, Koomen J, Goldman R. Quantitative analysis of immunoglobulin subclasses and subclass specific glycosylation by LC-MS-MRM in liver disease. J Proteom. 2015;116:24–33.
Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. Mass spectrometric quantitation of peptides and proteins using stable isotope standards and capture by anti-peptide antibodies (SISCAPA). J Proteome Res. 2004;3(2):235–44.
Ahn YH, Lee JY, Lee JY, Kim Y-S, Ko JH, Yoo JS. Quantitative analysis of an aberrant glycoform of TIMP1 from colon cancer serum by L-PHA-enrichment and SISCAPA with MRM mass spectrometry. J Proteome Res. 2009;8(9):4216–24.
Hong Q, Ruhaak LR, Stroble C, Parker E, Huang J, Maverakis E, et al. A method for comprehensive glycosite-mapping and direct quantitation of serum glycoproteins. J Proteome Res. 2015;14(12):5179–92.
Ruhaak LR, Kim K, Stroble C, Taylor SL, Hong Q, Miyamoto S, et al. Protein-specific differential glycosylation of immunoglobulins in serum of ovarian cancer patients. J Proteome Res. 2016;15(3):1002–10.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared no conflict of interest.
Additional information
Published in the topical collection Glycomics, Glycoproteomics and Allied Topics with guest editors Yehia Mechref and David Muddiman.
Rights and permissions
About this article
Cite this article
Kailemia, M.J., Park, D. & Lebrilla, C.B. Glycans and glycoproteins as specific biomarkers for cancer. Anal Bioanal Chem 409, 395–410 (2017). https://doi.org/10.1007/s00216-016-9880-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-016-9880-6