Abstract
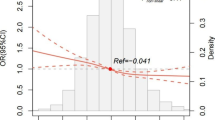
Experimental models suggest an important role for mitochondrial dysfunction in the pathogenesis of chronic kidney disease (CKD) and acute kidney injury (AKI), but little is known regarding the impact of common mitochondrial genetic variation on kidney health. We sought to evaluate associations of inherited mitochondrial DNA (mtDNA) variation with risk of CKD and AKI in a large population-based cohort. We categorized UK Biobank participants who self-identified as white into eight distinct mtDNA haplotypes, which were previously identified based on their associations with phenotypes associated with mitochondrial DNA copy number, a measure of mitochondrial function. We used linear and logistic regression models to evaluate associations of these mtDNA haplotypes with estimated glomerular filtration rate by serum creatinine and cystatin C (eGFRCr-CysC, N = 362,802), prevalent (N = 416 cases) and incident (N = 405 cases) end-stage kidney disease (ESKD), AKI defined by diagnostic codes (N = 14,170 cases), and urine albumin/creatinine ratio (ACR, N = 114,662). The mean age was 57 ± 8 years and the mean eGFR was 90 ± 14 ml/min/1.73 m2. MtDNA haplotype was significantly associated with eGFR (p = 2.8E−12), but not with prevalent ESKD (p = 5.9E−2), incident ESKD (p = 0.93), AKI (p = 0.26), or urine ACR (p = 0.54). The association of mtDNA haplotype with eGFR remained significant after adjustment for diabetes mellitus and hypertension (p = 1.2E−10). When compared to the reference haplotype, mtDNA haplotypes I (β = 0.402, standard error (SE) = 0.111; p = 2.7E−4), IV (β = 0.430, SE = 0.073; p = 4.2E−9), and V (β = 0.233, SE = 0.050; p = 2.7E−6) were each associated with higher eGFR. Among self-identified white UK Biobank participants, mtDNA haplotype was associated with eGFR, but not with ESKD, AKI or albuminuria.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chronic kidney disease (CKD) and acute kidney injury (AKI) are major public health problems with tremendous impact on morbidity and mortality worldwide (Rhee and Kovesdy 2015). Mitochondrial dysfunction has been implicated in the pathogenesis of CKD and AKI due to the roles of mitochondria in oxidative phosphorylation, reactive oxygen species generation, and programmed cell death (Che et al. 2014; Eirin et al. 2014; Small et al. 2012; Hall and Unwin 2007; Tran et al. 2011, 2016). Within the kidney, mitochondria are particularly important for the tubules, where oxygen consumption by mitochondria is driven by the need for active solute transport (Weinberg et al. 2000; Hansell et al. 2013).
Mammalian mitochondria possess a maternally inherited, intronless 16,569 base-pair genome (mtDNA) containing 37 genes that encode polypeptides, tRNAs, and rRNAs (Chinnery and Hudson 2013; Taanman 1999). The majority of these genes are essential for the machinery that converts metabolic energy into adenosine triphosphate. Recent studies within the UK Biobank and the Biobank Japan Project reported associations of mtDNA variants with kidney function, assessed by estimated glomerular filtration rate (eGFR) (Yonova-Doing et al. 2021; Yamamoto et al. 2020). Specific mtDNA variants have also been associated with tubulointerstitial kidney disease, IgA nephropathy, and focal segmental glomerulosclerosis (Douglas et al. 2014; Connor et al. 2017; Doleris et al. 2000; Hotta et al. 2001). To our knowledge, no prior study has examined associations of mitochondrial genetic variants with the risk of ESKD, AKI or albuminuria.
We previously assessed the impact of mitochondrial genetic variation on mitochondrial DNA copy number (mtDNA-CN), a measure of mitochondrial abundance, along with 41 additional traits associated with mtDNA-CN within the UK Biobank Study (Longchamps et al. 2022). This prior analysis identified six mtDNA single nucleotide polymorphisms (mtSNPs) with independent effects that correspond to eight haplotypes with minor allele frequency > 0.005. Given the haploid nature of the mitochondrial genome, haplotypes can capture potential synergistic or compensatory effects between mtSNPs. In this study, we investigated associations of these haplotypes with eGFR, end-stage kidney disease (ESKD) risk, AKI risk, and albuminuria in participants of the UK Biobank. Among the subset of participants with whole-genome sequencing data, we also evaluated associations of common mtSNPs with the kidney outcomes of interest.
Materials and methods
Study design and population
The UK Biobank is a large-scale cohort with medical and genetic data collected from 502,419 participants residing in the United Kingdom (Bycroft et al. 2018). Briefly, participants aged 37–73 years of age were recruited from 22 centers between 2006 and 2010. The assessment visit included a standardized questionnaire with detailed sociodemographic, lifestyle and medical information, a physical examination, and collection of biological specimens. The database enables linkage to a range of health-related records and outcomes, including data obtained from hospitalizations and from national death and cancer registries. The present study was conducted in accordance with the Declaration of Helsinki. Ethical approval for the UK Biobank was provided by the National Health Service National Research Ethics Service (21/NW/0157). All participants provided written informed consent for data use in research studies.
The present study included all unrelated (“used.in.pca.calculation = 1”) self-identified white participants with mtDNA genotyping data available and without prevalent end-stage renal disease. Other racial and ethnic groups were excluded from the analysis due to their small sample sizes. Among these 381,039 participants, eight distinct mtDNA haplotypes accounted for 379,432 participants. At the assessment visit, serum creatinine and cystatin C measurements were available for estimation of glomerular filtration rate in 362,802 participants, and albuminuria measurements were available in 114,662 participants.
Predictors
MtDNA haplotypes were identified within the UK Biobank based on association studies performed with approximately 150 mtSNPs (minor allele frequency, MAF > 0.5%) with mtDNA-CN and 41 additional traits as described previously (Longchamps et al. 2022). Briefly, mtDNA genotype phasing and imputation were performed using the 1000 Genomes Project phase 3 mtDNA variants as the reference panel (Genomes Project C et al. 2015). The SusieR package was used to identify potential causal variants for each independent locus associated with the outcomes of interest, and resulted in a total of six independent mtSNPs across four traits (MT73, MT7028, MT10238, MT12612, MT13617, MT15257) (Wang et al. 2020). Haplotypes were constructed by concatenating the six SNPs, resulting in eight haplotypes with minor allele frequency > 0.5%, with the most prevalent haplotype set as the reference (Supplemental Table 1). The primary analysis focused on using haplotypes to minimize the multiple-testing burden from a single SNP approach and to account for multiple functional variants within an individual. Secondary analyses were performed using whole-genome sequencing data which were available from 144,741/381,039 participants.
Outcomes
Estimated glomerular filtration rate (eGFR) was calculated by the 2012 CKD-EPI equation for serum creatinine and cystatin C (eGFRCr-CysC) (Inker et al. 2012). In sensitivity analyses, we calculated eGFR by the 2012 CKD-EPI equation for serum cystatin C (eGFRCysC) alone. We analyzed eGFR as a continuous outcome and as a dichotomous outcome, eGFR < 60 ml/min/1.73 m2. Prevalent and incident ESKD events were ascertained algorithmically as defined previously by UK Biobank (UK Biobank Outcome Adjudication Group 2022) and included individuals with Stage 5 CKD who received renal replacement therapy and those who received a kidney transplant. The AKI outcome was defined by ICD9 and ICD10 codes (Supplemental Table 2) and included past and incident events. Albuminuria was quantified as the albumin-to-creatinine ratio (ACR, mg/g) in spot urine specimens collected at the assessment visit, and was evaluated as a continuous outcome and as a dichotomous outcome, ACR > 30 mg/g. Participants with ESKD at baseline were excluded from analyses of eGFR, AKI, and ACR.
Covariates
Covariates for adjustment included age, sex, center, genotyping principal components, and genotyping array (Bycroft et al. 2018). Diabetes mellitus was defined by any one of the following: self-report, ICD9 or ICD10 diagnostic codes (Supplemental Table 2), insulin use, or hemoglobin A1c > = 48 mmol/mol. Hypertension was defined by any one of the following: antihypertensive medication use, systolic blood pressure > = 140 mm Hg, or diastolic blood pressure > = 90 mm Hg.
Statistical analysis
Baseline characteristics were compared across mtDNA haplotypes. Linear and logistic regression analyses were used to evaluate associations of mtDNA haplotypes and mtSNPs with the outcomes of interest. Individuals with missing data were excluded. To explore the potential effects of ancestry-related haplogroups on associations of haplotype with eGFR, we conducted regression analyses with and without haplogroup in the model. Due to its right-skewed distribution, urine ACR was log-transformed for analyses. Statistical significance was evaluated by ANOVA between regression models with and without the haplotypes, using the most prevalent haplotype as the reference group. Primary analyses were adjusted for age, age squared, sex, center, 40 principal components, and genotyping array. In exploratory analyses, we adjusted for diabetes mellitus and hypertension as potential mediators. Cox proportional hazards models were used to evaluate associations of mtDNA haplotypes with the outcome of incident ESKD. To account for multiple testing with five independent tests (eGFR, prevalent and incident ESKD, AKI and ACR), we used a Bonferroni corrected p value < 0.01 as the study-wide threshold for statistical significance. All analyses were performed using R version 4.0.3.
Results
MtDNA haplotypes and baseline characteristics of UK Biobank participants
Among the 381,039 participants with available mtDNA genotyping data, eight distinct mtDNA haplotypes accounted for 379,432 participants (99.6%). The most prevalent mtDNA haplotypes accounted for 40% and 28% of participants, respectively (Table 1). The remaining mtDNA haplotypes were much less common, each accounting for fewer than 10% of participants.
Clinical characteristics were similar across mtDNA haplotypes. The mean (SD) age was 57 ± 8 years and 54% of participants were female. Diabetes mellitus was present in 6% of the cohort, whereas hypertension was prevalent in 54% of participants. The mean eGFRCr-CysC was 90 ± 14 ml/min/1.73 m2 and mean eGFRCysC was 88 ± 16 ml/min/1.73 m2. Only 2% of participants had an eGFRCr-CysC < 60 ml/min/1.73 m2, whereas 5% had an eGFRCysC < 60 ml/min/1.73 m2. The mean urine ACR was 28 mg/g and 14% of participants had a urine ACR > 30 mg/g.
Associations of mtDNA haplotypes with CKD and ESKD
MtDNA haplotype was significantly associated with the continuous outcome of eGFRCr-CysC (Table 2; p value for model, 2.8E-12). When compared to the reference haplotype, mtDNA haplotypes I (β = 0.402, standard error (SE) = 0.111; p = 2.7E−4), IV (β = 0.430, SE = 0.073; p = 4.2E−9), and V (β = 0.233, SE = 0.050; p = 2.7E−6) were each associated with higher eGFRCr-CysC. When we adjusted additionally for diabetes mellitus and hypertension, the association of mtDNA haplotype with eGFR remained robust (p = 1.2E−10). In sensitivity analyses, addition of haplogroup, which largely captures mitochondrial genetic variation due to genetic ancestry, as a covariate to a regression model with haplotype did not improve the model (p = 0.34). By contrast, addition of haplotype into a model with haplogroup significantly improved the model (p = 1.26E−5), indicating the observed associations are driven by haplotype rather than ancestral haplogroup or population substructure.
In a secondary analyses to identify potential associations not captured by the haplotype analyses, we conducted single SNP analyses among the subset of participants with whole-genome sequencing data available (N = 144,741). We tested 3,189 variants with minor allele count ≥ 20, using rank normal transformed eGFR to protect against outlier measures unduly influencing rare variant results (Supplemental Table 3). The most significant association observed for a rare variant was mt16214C > T (p < 7.65 × 10–5, MAF = 0.001). The most significant variant with MAF ≥ 0.01 was mt3010G > A (p < 0.0007, MAF = 0.13), which has previously been associated with eGFR (Yonova-Doing et al. 2021). Neither of these variants passed Bonferroni correction for multiple testing.
Findings were similar when we examined associations of mtDNA haplotype with eGFRCysC (Supplemental Table 4; p value for model, p = 2.2E−7). MtDNA haplotypes I (β = 0.351, SE = 0.126; p = 5.4E−3), IV (β = 0.405, SE = 0.083; p = 1.2E−6), and V (β = 0.186, SE = 0.056; p = 1.0E−3) were each associated with higher eGFRCysC as compared to the reference haplotype. By contrast, mtDNA haplotype was not associated with the dichotomous outcome of eGFR < 60 ml/min/1.73m2 when assessed either by creatinine and cystatin C (p = 0.93) or by cystatin C alone (p = 0.69).
There were 416 cases of ESKD at the baseline assessment, and an additional 405 ESKD events occurred over approximately 12 years of follow-up (Table 3). There were no significant associations of mtDNA haplotype with prevalent or incident ESKD (p = 5.9E−2 and p = 0.93, respectively).
Associations of mtDNA haplotypes with AKI and albuminuria
Among the 379,432 UK Biobank participants in this analysis, there were a total of 14,170 cases of AKI with 13,693 occurring after the baseline assessment. There were no significant associations of mtDNA haplotype with risk of AKI (Table 4; p value for model, 0.26). Similarly, there were no significant associations of mtDNA haplotype with urine ACR (p = 0.54) or ACR > 30 mg/g (p = 0.35) in the overall models (Supplemental Table 5).
Discussion
The kidney is a metabolically active organ with the highest oxygen consumption rate after the heart (Pagliarini et al. 2008; O'Connor Oct 2006; Wang et al. 2010). Healthy mitochondrial function is, therefore, required for the bioenergetic processes that drive solute transport, elimination of toxins, acid–base homeostasis, and regulation of volume status. As evolutionary descendants of bacterial endosymbionts, mitochondria contain their own circular mtDNA that encodes the translational machinery and proteins critical for oxidative phosphorylation (Andersson et al. 1998; Giles et al. 1980; Anderson et al. 1981). We hypothesized that variability in this maternally inherited genome could contribute to the risk of kidney dysfunction and injury. In this population-based cohort study, we found that mtDNA haplotype is associated with eGFR, but not with risk of ESKD, AKI or albuminuria. The associations of mtDNA haplotype with eGFR remained robust even after adjustment for the presence of diabetes mellitus and hypertension, suggesting the effects of mtDNA haplotype are not driven by mediation through these known risk factors.
Prior studies within the UK Biobank and the Biobank Japan Project reported associations of mtDNA variants with eGFR (Yonova-Doing et al. 2021; Yamamoto et al. 2020). The present study builds upon existing literature by examining associations of mtDNA haplotypes with not only eGFR, but also ESKD, AKI, and albuminuria. We previously found that mtDNA haplotypes are causally associated with mtDNA copy number, a measure of mitochondrial abundance, and several associated traits (Longchamps et al. 2022). Here, we observed independent associations of mtDNA haplotype with eGFR, and compared to the reference haplotype, three mtDNA haplotypes with population fractions of 4%, 9%, and 28%, respectively, had significantly higher eGFR of 0.23–0.43 ml/min/1.73 m2 in magnitude. In our prior work, we did not observe patterns that would suggest mediation by other phenotypic traits for the associations of mtDNA haplotypes and kidney function (Longchamps et al. 2022). Adjustment for blood counts, liver enzymes, or mtDNA copy number also did not attenuate associations of the haplotypes with eGFR (data not shown), suggesting the mtDNA genome has direct effects on kidney health. Further studies are needed to determine the functional impact of specific mtDNA haplotypes, including identifying the individual underlying causal variants, and to uncover the mechanisms underlying their associations with kidney risk.
Although we did not detect significant associations of mtDNA haplotype with ESKD risk, possible explanations include our limited power and the potential impact of survival bias, as individuals with CKD often die prior to the development of ESKD. Additionally, our evaluation of common variants may have reduced our ability to detect variants that are under negative selection due to their impact on ESKD risk. The lack of association of mtDNA haplotype with AKI was unexpected because of experimental evidence linking mitochondrial function with both protection from and recovery following AKI (Emma et al. 2016). Despite our large sample size overall, our use of diagnostic codes to identify AKI cases may have reduced accuracy for this outcome. We were unable to use changes in serum creatinine to define AKI due to the unavailability of these data within the UK Biobank database.
Strengths of this study include the large sample size with mtDNA data, our ascertainment of eGFR by both serum creatinine and cystatin C, our inclusion of albuminuria, and our use of an algorithmically defined ESKD outcome. There are important limitations to this study. First, as a study of UK Biobank participants who self-identified as white, our findings may not be generalizable to other populations. Second, the UK Biobank is healthier than the sampling population with evidence of a “healthy volunteer” selection bias (Fry et al. 2017). Hence, the burden of CKD and CKD risk factors was lower than would be expected for the general population, and may have reduced power for the outcome of prevalent CKD. Third, this study was not designed to evaluate changes in eGFR over time. Fourth, we did not have access to the etiology of ESKD or kidney transplant status, so we could not determine whether associations might differ by underlying disease or transplant status.
In conclusion, among UK Biobank participants who self-identified as white, mtDNA haplotype was associated with eGFR, but not with ESKD risk, AKI risk or albuminuria. Compared to the most prevalent haplotype, mtDNA haplotypes I, IV, and V were associated with higher eGFR. Future studies are needed to provide insight into the biological mechanisms underlying these associations.
Data availability
The dataset analyzed in the current study is available from the UK Biobank to approved researchers.
References
Anderson S, Bankier AT, Barrell BG et al (1981) Sequence and organization of the human mitochondrial genome. Nature 290(5806):457–465. https://doi.org/10.1038/290457a0
Andersson SG, Zomorodipour A, Andersson JO et al (1998) The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396(6707):133–140. https://doi.org/10.1038/24094
Bycroft C, Freeman C, Petkova D et al (2018) The UK Biobank resource with deep phenotyping and genomic data. Nature 562(7726):203–209. https://doi.org/10.1038/s41586-018-0579-z
Che R, Yuan Y, Huang S, Zhang A (2014) Mitochondrial dysfunction in the pathophysiology of renal diseases. Am J Physiol Renal Physiol 306(4):F367–F378. https://doi.org/10.1152/ajprenal.00571.2013
Chinnery PF, Hudson G (2013) Mitochondrial genetics. Br Med Bull 106:135–159. https://doi.org/10.1093/bmb/ldt017
Connor TM, Hoer S, Mallett A et al (2017) Mutations in mitochondrial DNA causing tubulointerstitial kidney disease. PLoS Genet 13(3):e1006620. https://doi.org/10.1371/journal.pgen.1006620
Doleris LM, Hill GS, Chedin P et al (2000) Focal segmental glomerulosclerosis associated with mitochondrial cytopathy. Kidney Int 58(5):1851–1858. https://doi.org/10.1111/j.1523-1755.2000.00356.x
Douglas AP, Vance DR, Kenny EM, Morris DW, Maxwell AP, McKnight AJ (2014) Next-generation sequencing of the mitochondrial genome and association with IgA nephropathy in a renal transplant population. Sci Rep 4:7379. https://doi.org/10.1038/srep07379
Eirin A, Ebrahimi B, Zhang X et al (2014) Mitochondrial protection restores renal function in swine atherosclerotic renovascular disease. Cardiovasc Res 103(4):461–472. https://doi.org/10.1093/cvr/cvu157
Emma F, Montini G, Parikh SM, Salviati L (2016) Mitochondrial dysfunction in inherited renal disease and acute kidney injury. Nat Rev Nephrol 12(5):267–280. https://doi.org/10.1038/nrneph.2015.214
Fry A, Littlejohns TJ, Sudlow C et al (2017) Comparison of sociodemographic and health-related characteristics of UK biobank participants with those of the general population. Am J Epidemiol 186(9):1026–1034. https://doi.org/10.1093/aje/kwx246
Genomes Project C, Auton A, Brooks LD et al (2015) A global reference for human genetic variation. Nature 526(7571):68–74. https://doi.org/10.1038/nature15393
Giles RE, Blanc H, Cann HM, Wallace DC (1980) Maternal inheritance of human mitochondrial DNA. Proc Natl Acad Sci USA 77(11):6715–6719
UK Biobank Outcome Adjudication Group (2022) UK Biobank Definitions of End Stage Renal Disease. https://biobank.ctsu.ox.ac.uk/crystal/ukb/docs/alg_outcome_esrd.pdf. Accessed 7 Jan 2022
Hall AM, Unwin RJ (2007) The not so “mighty chondrion”: emergence of renal diseases due to mitochondrial dysfunction. Nephron Physiol 105(1):p1-10. https://doi.org/10.1159/000096860
Hansell P, Welch WJ, Blantz RC, Palm F (2013) Determinants of kidney oxygen consumption and their relationship to tissue oxygen tension in diabetes and hypertension. Clin Exp Pharmacol Physiol 40(2):123–137. https://doi.org/10.1111/1440-1681.12034
Hotta O, Inoue CN, Miyabayashi S, Furuta T, Takeuchi A, Taguma Y (2001) Clinical and pathologic features of focal segmental glomerulosclerosis with mitochondrial tRNALeu(UUR) gene mutation. Kidney Int 59(4):1236–1243. https://doi.org/10.1046/j.1523-1755.2001.0590041236.x
Inker LA, Schmid CH, Tighiouart H et al (2012) Estimating glomerular filtration rate from serum creatinine and cystatin C. N Engl J Med 367(1):20–29. https://doi.org/10.1056/NEJMoa1114248
Longchamps RJ, Yang SY, Castellani CA et al (2022) Genome-wide analysis of mitochondrial DNA copy number reveals loci implicated in nucleotide metabolism, platelet activation, and megakaryocyte proliferation. Hum Genet 141(1):127–146. https://doi.org/10.1007/s00439-021-02394-w
O’Connor PM (2006) Renal oxygen delivery: matching delivery to metabolic demand. Clin Exp Pharmacol Physiol 33(10):961–967. https://doi.org/10.1111/j.1440-1681.2006.04475.x
Pagliarini DJ, Calvo SE, Chang B et al (2008) A mitochondrial protein compendium elucidates complex I disease biology. Cell 134(1):112–123. https://doi.org/10.1016/j.cell.2008.06.016
Rhee CM, Kovesdy CP (2015) Epidemiology: spotlight on CKD deaths-increasing mortality worldwide. Nat Rev Nephrol 11(4):199–200. https://doi.org/10.1038/nrneph.2015.25
Small DM, Coombes JS, Bennett N, Johnson DW, Gobe GC (2012) Oxidative stress, anti-oxidant therapies and chronic kidney disease. Nephrology (carlton) 17(4):311–321. https://doi.org/10.1111/j.1440-1797.2012.01572.x
Taanman JW (1999) The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta 1410(2):103–123. https://doi.org/10.1016/s0005-2728(98)00161-3
Tran M, Tam D, Bardia A et al (2011) PGC-1alpha promotes recovery after acute kidney injury during systemic inflammation in mice. J Clin Investig 121(10):4003–4014. https://doi.org/10.1172/JCI58662
Tran MT, Zsengeller ZK, Berg AH et al (2016) PGC1alpha drives NAD biosynthesis linking oxidative metabolism to renal protection. Nature 531(7595):528–532. https://doi.org/10.1038/nature17184
Wang Z, Ying Z, Bosy-Westphal A et al (2010) Specific metabolic rates of major organs and tissues across adulthood: evaluation by mechanistic model of resting energy expenditure. Am J Clin Nutr 92(6):1369–1377. https://doi.org/10.3945/ajcn.2010.29885
Wang G, Sarkar A, Carbonetto P, Stephens M (2020) A simple new approach to variable selection in regression, with application to genetic fine mapping. J R Stat Soc Ser B (stat Methodol) 82(5):1273–1300. https://doi.org/10.1111/rssb.12388
Weinberg JM, Venkatachalam MA, Roeser NF, Nissim I (2000) Mitochondrial dysfunction during hypoxia/reoxygenation and its correction by anaerobic metabolism of citric acid cycle intermediates. Proc Natl Acad Sci USA 97(6):2826–2831
Yamamoto K, Sakaue S, Matsuda K et al (2020) Genetic and phenotypic landscape of the mitochondrial genome in the Japanese population. Commun Biol 3(1):104. https://doi.org/10.1038/s42003-020-0812-9
Yonova-Doing E, Calabrese C, Gomez-Duran A et al (2021) An atlas of mitochondrial DNA genotype-phenotype associations in the UK Biobank. Nat Genet 53(7):982–993. https://doi.org/10.1038/s41588-021-00868-1
Acknowledgements
This research has been conducted using the UK Biobank Resource under Application Number 17731. We thank the volunteer participants of the UK Biobank and the UK Biobank researchers. This study was supported by funding from the National Institute of Health (R01HL144569 for DEA, SYY; R35HL138424 for SMP; R01AG027002 for VJ, MGS, MJS, SMP; and R01HL085757, UH3DK114866, U01DK106962 and R01DK093770 for CRP).
Funding
This work was supported by NHLBI with grant number R01HL144569 and NIA with grant number R01AG027002.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The results presented in this paper have not been published previously in whole or part, except in abstract format. M.G.S. has served on advisory boards for AstraZeneca, Bayer, and Boerhinger Ingelheim, and receives research support from Bayer. J.H.I. holds an investigator-initiated research grant from Baxter International Inc., serves as a member of a data safety monitoring board for Sanifit Therapeutics, is a member of the scientific advisory board for Alpha Young, and has served on advisory boards for AstraZeneca and Ardelyx. S.M.P has served as a consultant or on advisory boards for Astellas, AstraZeneca, Boehringer-Ingelheim, Janssen, Merck, Pfizer, Casma, Mission Therapeutics, Entrada, and Cytokinetics in the last 24 months. C.R.P. is a member of the advisory board of and owns equity in RenalytixAI. He also serves as a consultant for Genfit and Novartis.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jotwani, V., Yang, S.Y., Thiessen-Philbrook, H. et al. Mitochondrial genetic variation and risk of chronic kidney disease and acute kidney injury in UK Biobank participants. Hum. Genet. 143, 151–157 (2024). https://doi.org/10.1007/s00439-023-02615-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-023-02615-4