- 1Clinical Laboratory of Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, China
- 2Clinical Laboratory of Taian City Central Hospital, Tai’an, China
NDM-7, a variant of New Delhi metallo-beta-lactamases (NDM), has the highest carbapenem-hydrolyzing activity. NDM-7-producing enterobacteria have been reported in many countries. In this study, we reported NDM-7 production in ST11 Klebsiella pneumoniae isolated from a boy hospitalized in the pediatric intensive care unit of a teaching hospital in China. The isolate exhibited resistance to β-lactam antimicrobials, quinolones, and trimethoprim/sulfamethoxazole, and it harbored blaNDM–7, blaCTX–M–15, qnrA, qnrB, and qnrS. The serotype of the isolated K. pneumoniae was assigned as K1, and it contained three virulence genes, including kfuBC, uge, and fim. The blaNDM–7 gene was located on a conjugative IncX3 plasmid designated as pB14NDM-7. This plasmid was fully sequenced and compared with the available blaNDM–7-harboring IncX3 plasmids. pB14NDM-7 contained a conserved genetic context of ISkox3-umuD-IS26-ΔTn125-IS5-ΔTn125-IS3000-ΔTn2. pB14NDM-7 showed 99% nucleotide identity and the same genetic context with three blaNDM–7-harboring IncX3 plasmids obtained from Escherichia coli in China. Our results indicate that IncX3 plasmid may contribute to the prevalence of blaNDM–7 in China. The high prevalence of NDM variants worldwide highlights the critical need for careful monitoring and control of the rapid dissemination of blaNDM.
Introduction
The prevalence of carbapenem-resistant Klebsiella pneumoniae has increased and becomes a serious public health threat since the early 2000s. Several mechanisms are responsible for carbapenem resistance of K. pneumoniae, among which carbapenemase production remains the most clinically relevant (Pitout et al., 2015). The most common metallo-β-lactamases (MBLs) identified in K. pneumoniae are New Delhi metallo-beta-lactamases (NDM), while other metallolactamases are relatively rare in this species (Gregson et al., 2016). NDM was first described in Sweden in 2009 in a patient who had received medical care in India. Thereafter, it has been reported in more than 40 countries worldwide, even in cases with no epidemiological links with India, such as in the Balkan states or the Middle East (Yong et al., 2009; Pérez-Vázquez et al., 2019).
The gene encoding NDM is often carried by plasmids and is easily transferred to other microorganisms through horizontal gene transfer. Owing to this reason, the emergence of carbapenem-resistant strains of pathogenic microorganisms has increased rapidly (Rolain et al., 2010). NDM-1 is the most described NDM and has emerged as a global health threat. Similar to other MBLs, NDM-1 can hydrolyze all beta-lactams except aztreonam (Yong et al., 2009). Many NDM variants have evolved in enterobacteriaceae by single and/or double amino acid residue substitutions at different positions, and a total of 24 known variants of NDM have been identified so far (Khalid et al., 2020).
NDM-7 was first discovered in Escherichia coli in Germany in 2013 (Cuzon et al., 2013). NDM-7 contains Asp-130-Asn and Met-154-Leu substitutions and has the greatest carbapenem-hydrolyzing activity (Yoon et al., 2018). To date, NDM-7-producing enterobacteria have been reported in more than 10 countries (Solgi et al., 2017; Ahmad et al., 2018; Yoon et al., 2018; Mouftah et al., 2019). In China, NDM-7 has been detected and described in E. coli but not in K. pneumoniae (Wang et al., 2016; Bi et al., 2018; Hao et al., 2018; Xu and He, 2019). In this study, we isolated a ST11 K. pneumoniae harboring blaNDM–7 from a boy hospitalized in the pediatric intensive care unit of a Chinese Hospital. Genotypic and phenotypic characterization of the IncX3 plasmid carrying blaNDM–7 gene was performed. In 2015, an IncX3 plasmid carrying blaNDM–7 was isolated from E. coli in the same hospital (Hao et al., 2018). To analyze the evolution of IncX3 plasmid carrying blaNDM–7, we constructed a phylogenetic tree from 16 plasmids based on homologous proteins.
Materials and Methods
Bacterial Strains
Carbapenem-resistant K. pneumoniae was isolated from a 4-year-old boy who was diagnosed with autoimmune encephalitis and hospitalized in the pediatric intensive care unit of a teaching hospital in Shandong Province of China in 2018. We designated this isolate as B14. During hospitalization, arterial catheterization was performed. B14 was obtained from blood cultures 2 weeks after hospitalization; however, no pathogen was isolated from the sputum, cerebrospinal fluid, and ascites. The patient was treated with ceftriaxone before blood culture, following which his condition improved, and he was discharged before the antibiotic sensitivity results were obtained.
The patient had no history of traveling abroad. Informed consent was signed by the family member of the patient involved in this study. The methods in this study were approved by the Ethics Committee of Shandong Provincial Hospital and were carried out in accordance with the approved guidelines. The isolate was identified as K. pneumoniae using Vitek-2 compact system and confirmed using Vitek-MS system (BioMérieux, France). Phenotypic detection of carbapenemases was performed using carbapenem inactivation method (CIM) and EDTA-modified CIM (eCIM) test.
Antibiotic Susceptibility Assay
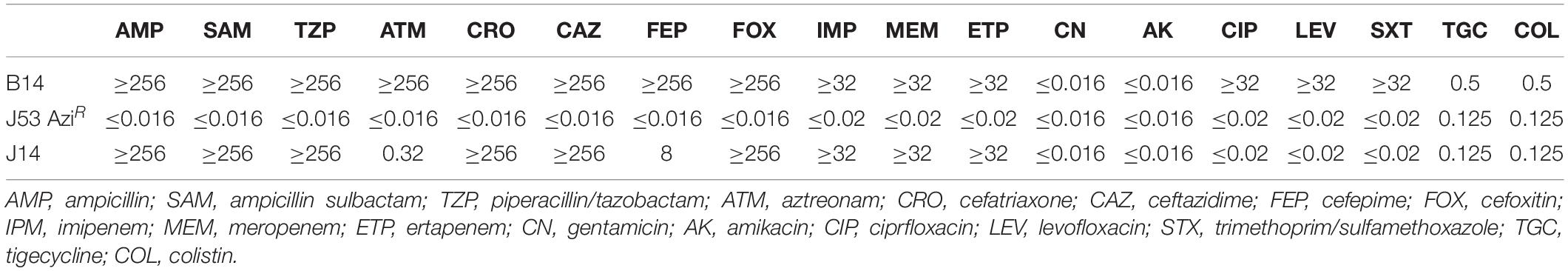
Antimicrobial susceptibility testing was performed on Mueller Hinton agar plates using E-test strips (BioMérieux, France) (Table 1). The minimum inhibitory concentrations of tigecycline and colistin were determined using broth microdilution method (Bio-kont, China). E. coli ATCC25922 and K. pneumoniae ATCC700603 served as the quality controls. All antibiotics were administered according to the approved standard of the 2019 European Committee on Antimicrobial Susceptibility Testing breakpoint.1
PCR and DNA Sequence Analysis of Drug Resistance Genes, Serotype, and Virulence Genes
As described previously, a variety of antimicrobial resistance genes were screened using PCR and DNA sequencing (Zhu et al., 2016). These genes included carbapenem resistance genes (blaKPC, blaSME, blaIMI, blaNMC, blaGES, blaIMP, blaVIM, blaGIM, blaSIM, blaSPM, blaNDM, and blaOXA–48like), extended-spectrum β-lactamase genes (ESBLs) (blaCTX–M, blaTEM, and blaSHV), AmpC β-lactamase genes (blaMOX, blaFOX, blaDHA, blaCIT, and blaEBC), and plasmid-mediated quinolone resistance genes [qnrA, qnrB, qnrC, qnrD, qnrS, qepA, and aac(6)-Ib-cr]. Subsequently, the isolate was serotyped for K1, K2, K5, K20, K54, and K57 serotypes, and 12 virulence-associated genes (rmpA, aerobactin, wcaG, ybtA, iucB, iroNB, ureA, uge, kfuBC, fim, wabG, and allS) were screened using PCR as previously described (Wang et al., 2017).
Multilocus Sequence Typing
Multilocus sequence typing (MLST) of K. pneumoniae was performed according to protocols available on the MLST Pasteur website2. Seven conserved housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) were amplified, sequenced, and compared with those in the MLST databases.
Analysis of blaNDM-Carrying Plasmids
Conjugation was performed using the mixed broth method. Briefly, E. coli J53 AziR was used as the recipient strain, and B14 served as the donor. Transconjugants were selected on Mueller Hinton agar supplemented with meropenem (0.5 μg/ml) and sodium azide (100 μg/ml). Antimicrobial susceptibility test of the transconjugant was carried out as described for the clinical strain. In order to evaluate the stability of the plasmid in the recipient E. coli J53 after conjugation, antibiotic susceptibility test was performed after the recipient cells containing the plasmid were sequentially subcultured 20 times on blood agar plate without any antibiotics.
The size and amount of plasmids carried by the clinical isolate and transconjugant were evaluated using S1-pulsed-field gel electrophoresis (S1-PFGE) as previously described (Hao et al., 2018). The genome of Salmonella H9812 digested with XbaI was used as the marker.
Plasmid Sequencing
The plasmid carrying blaNDM–7 was defined as pB14NDM-7. To better understand the characteristics of pB14NDM-7, its complete sequence was determined. The plasmid was extracted and sequenced using an Illumina Hiseq platform and assembled using SOAPdenovo at MajorBio Co., (Shanghai, China). The gaps were closed through PCR and Sanger Sequencing at Sangon Biotech (Shanghai, China). The plasmid sequences were annotated using basic local alignment search tool (BLAST) against the non-redundant protein database. Plasmid Finder was used for detection and typing of the plasmid.
Phylogenetic Tree Construction
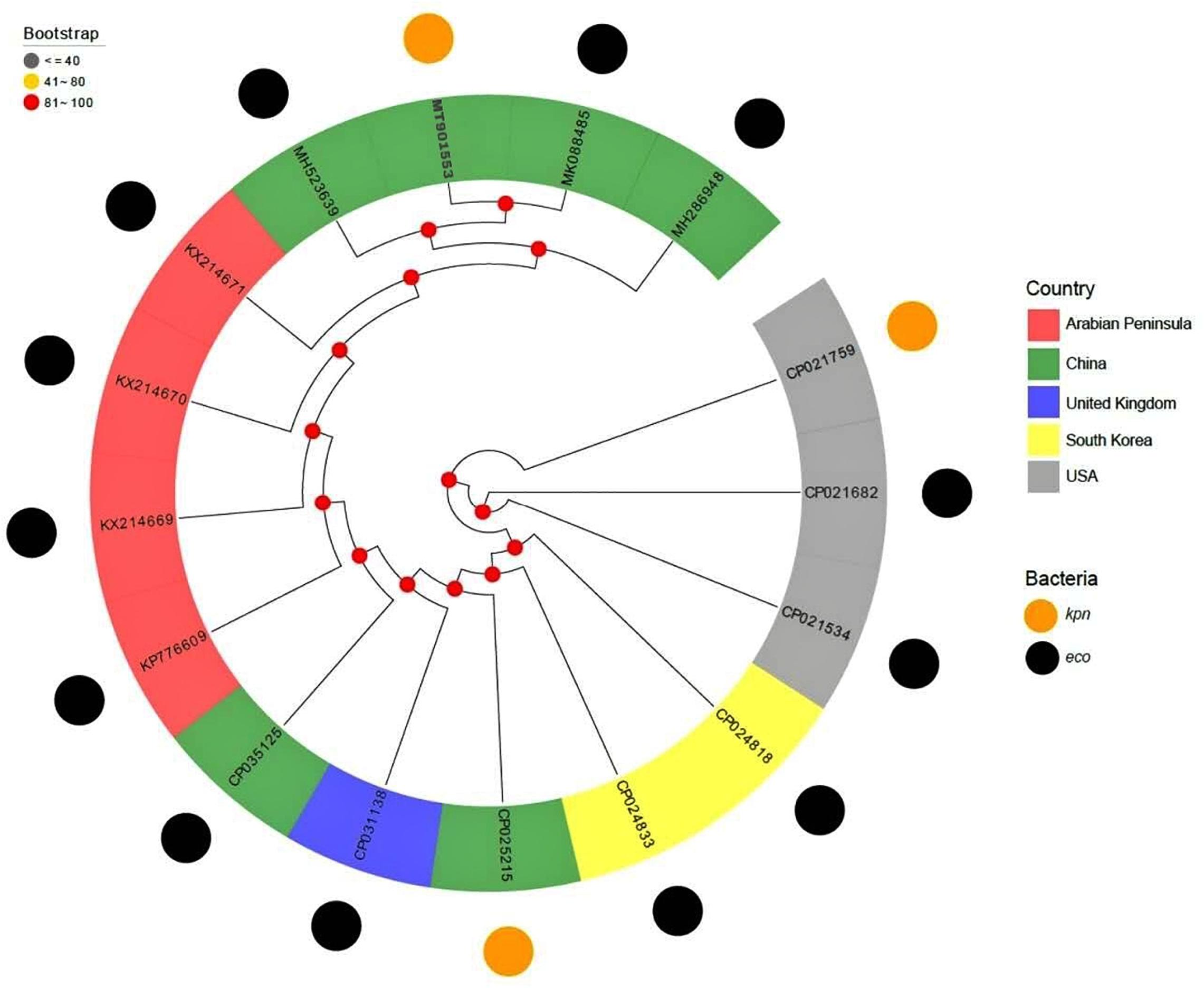
The plasmids carrying blaNDM–7 were retrieved from National Center for Biotechnology Information (NCBI) database for the query “NDM-7 and plasmid.” Out of 49 plasmids, 15 belonged to the IncX3 incompatibility group. The sequence of pB14NDM-7 was compared with the 15 blaNDM–7-harboring IncX3 plasmids. The plasmid sequences were annotated for encoded proteins using Prodigal, a prokaryote genome annotation tool3. The STAG algorithm of OrthoFinder was used to construct a phylogenetic tree from 16 plasmids based on homologous proteins.
Results
Resistance Profile of B14 Strain
Clinical K. pneumoniae B14 was identified as an MBL-producing strain using carbapenem inactivation method (CIM) and EDTA-modified CIM (eCIM) test. B14 was resistant to aztreonam, ampicillin, ampicillin/sulbactam, piperacillin/tazobactam, cephalosporins, carbapenems, quinolones, and trimethoprim/sulfamethoxazole and susceptible to aminoglycosides, colistin, and tigecycline (Table 1).
MLST, Serotype, Resistance, and Virulence Genotyping
PCR amplification and sequencing confirmed that B14 harbored blaNDM–7 and blaCTX–M–15. These two genes were responsible for the resistance to β-lactam antibiotics. In addition, B14 carried qnrA, qnrB, and qnrS, which may lead to quinolones resistance. However, high-level fluoroquinolone resistance was probably mediated by chromosomal mutations in gyrA and/or parC genes. MLST revealed that the sequence type of B14 was ST11. Based on PCR amplification results, the serotype of B14 was determined to be K1. Furthermore, B14 contained three virulence genes, which included kfuBC, uge, and fim.
Transferability and Stability of Plasmid
blaNDM–7-harboring plasmid of B14 was successfully transferred into E. coli J53 by conjugation. The transconjugant was identified as E. coli using Vitek-2 compact system and confirmed using Vitek-MS system. The presence of blaNDM–7 in the transconjugant was confirmed using PCR. The transconjugant was resistant to carbapenems and cephalosporin but susceptible to aztreonam, aminoglycosides, quinolones, colistin, and tigecycline. After 20 passages, the antibiotic susceptibility patterns of the transconjugant showed no changes. S1-pulsed-field gel electrophoresis showed that B14 harbored two plasmids, and the transconjugant J14 contained a single plasmid, which was approximately 46 kb (Supplementary Figure S1).
Characterization of blaNDM–7-Harboring Plasmid
pB14NDM-7 is a 46,133-bp plasmid belonging to the IncX3 incompatibility group. The complete sequence of pB14NDM-7 was submitted to GenBank under accession number MT901553. In pB14NDM-7, blaNDM–7 was preceded by ISkox3-umuD-IS26-ΔTn125 (ΔdsbC-trpF-bleMBL) in the upstream region and succeeded by IS5-ΔTn125-IS3000-ΔTn2 in the downstream region (Figure 1).
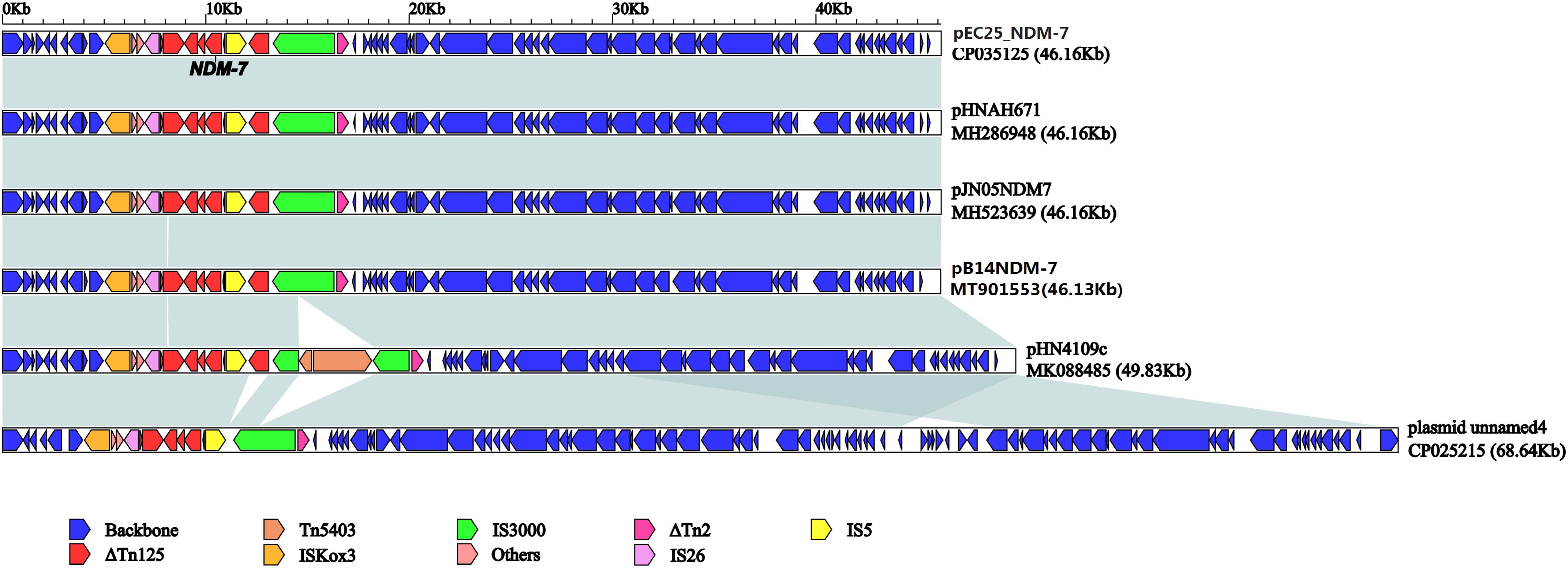
Figure 1. Linear comparison of pB14NDM-7 with five blaNDM–7-harboring IncX3 plasmids from China. A linear comparison was carried out for the complete DNA sequences of pB14NDM-7 (MT901553, this study), pJN05NDM7 (MH523639), pHNAH671 (MH286948), pEC25_NDM-7 (CP035125), pHN4109c (MK088485), and plasmid unnamed 4 (CP025215). Genes, mobile elements, and other features are colored based on function classification. Shading denotes regions of homology (>95% nucleotide identity).
The full published sequences of 15 IncX3 plasmids harboring blaNDM–7 were downloaded and compared with pB14NDM-7 (Figure 2). Only 3 of the 16 plasmids were isolated from K. pneumoniae, and the other 13 were from E. coli. Sequence alignments revealed more than 80% homology among the 16 plasmids. The sequence of pB14NDM-7 showed more than 99% homology with three plasmids, including pHN4109c (accession no. MK088485), pJN05NDM-7 (accession no. MH523639), and pHNAH671 (accession no. MH286948). All four plasmids were from China, with pB14NDM-7 being the only plasmid to be isolated from K. pneumoniae. To determine the detailed structural differences among six blaNDM–7-harboring IncX3 plasmids from China, including pB14NDM-7 (MT901553, this study), pJN05NDM7 (MH523639), pHNAH671 (MH286948), pEC25_NDM-7 (CP035125), pHN4109c (MK088485), and plasmid unnamed 4 (CP025215), additional linear comparative genomics analysis of these six plasmids was performed by BLAST. The six plasmids showed similar genomic contents. pB14NDM-7, pJN05NDM7, pEC25_NDM-7, and pHNAH671 had the same genetic structure. In plasmid pHN4109c, the insertion of Tn5403 resulted in the interruption of IS3000 (Figure 1).
Plasmids pB14NDM-7 and pJN05NDM-7 were from the same hospital and differed only in gene dsbC. pJN05NDM-7 is a 46,161-bp circular IncX3-type plasmid. In dsbC gene, there was a 60-bp repetitive sequence in the region between the positions 8084 and 8144, corresponding to the 8023–8083 regions in plasmid pJN05NDM-7. In addition, there was a 88-bp deletion in pB14NDM-7 corresponding to the 8085–8172 region in gene dsbC of plasmid pJN05NDM-7.
Discussion
NDM-7-producing K. pneumoniae are relatively rare as compared to NDM-7-producing E. coli. NDM-7 was first reported in K. pneumoniae in 2014 in Minnesota (Lee et al., 2014). Herein, we described the emergence of ST11 K. pneumoniae carrying blaNDM–7 on IncX3 plasmid in China. Although NDM-7 production has been reported in diverse clones of K. pneumoniae, including ST138, ST273, ST278, ST437, and ST654 (Shankar et al., 2019), our study is the first to report the presence of blaNDM–7 in ST11 K. pneumoniae in China. This strain was isolated from a 4-year-old boy with no history of traveling abroad.
The K serotypes and phenotypes are responsible for the invasive nature of certain K. pneumoniae. Serotypes K1, K2, K4, and K5 are highly virulent and may cause severe infections in humans (Wang et al., 2017). The serotype of B14 was found to be K1. The carbapenem-resistant K1 K. pneumoniae can cause infections at multiple sites, including liver abscesses, pneumonia, meningitis, and blood stream infections. Additionally, B14 contained three virulence genes, which included kfuBC, uge, and fim. ST11 is the major sequence type of hypervirulent carbapenem-resistant K. pneumoniae from Asia, especially China (Zhao et al., 2019). Therefore, it is speculated that B14 has certain virulence traits. However, it is gratifying that the patient’s condition improved, following which he was discharged. As this study was retrospective, we could not perform in-depth analysis of the patient’s surrounding environment and the source of the strain.
B14 was highly resistant to broad spectrum cephalosporins, monobactam, and carbapenems. However, it is known that NDM does not decompose aztreonam (Paul et al., 2017). Therefore, resistance of B14 to aztreonam in the present study might be due to the coexistence of blaCTX–M–15 gene that encodes extended spectrum β-lactamase. In addition, PCR results showed that B14 also carried quinolone-resistance genes, including qnrA, qnrB, and qnrS. Conjugation test revealed that only blaNDM–7 could be horizontally transferred, and the sequencing results showed that plasmid IncX3 harbored by B14 does not carry antimicrobial resistance genes other than blaNDM–7. This finding corroborates the results of Hao et al. (2018).
IncX3 plasmids carrying blaNDM variants have been increasingly reported all over the world in recent years. A previous study has proved that IncX3 plasmid can transfer blaNDM between different enterobacterial species over a wide range of temperatures (Liu et al., 2019). IncX3 has also been reported to be an important carrier of blaNDM–7 (Paul et al., 2017). In order to further understand the evolutionary relationship of blaNDM–7-harboring IncX3 plasmid, we downloaded the nucleotide sequences of 15 IncX3 plasmids from NCBI and analyzed their homology with pB14NDM-7. The results showed that all the 16 plasmids were closely related to each other, suggesting parallel evolution of these plasmids. It should be noted that pB14NDM-7 showed 99% nucleotide identity with three plasmids obtained from E. coli in China, which suggests that IncX3-type plasmids are popular vectors in mediating dissemination of blaNDM–7 in China (Wang et al., 2016; Bi et al., 2018).
In plasmids, genes are frequently associated with mobile genetic elements such as transposons (Tn) and insertion sequences (IS). pB14NDM-7 contained a conserved genetic context of ISkox3-umuD-IS26-ΔTn125-IS5-ΔTn125-IS3000-ΔTn2. The blaNDM genetic structure is common in enterobacteriaceae for the horizontal transfer of blaNDM and has been reported in the transmission of blaNDM–7 and blaNDM–5 (Zhang et al., 2016; Guo et al., 2019). In addition, plasmids pB14NDM-7 and pJN05NDM-7 were from the same hospital. However, pJN05NDM-7 was isolated from E. coli, and the two patients were not in the same hospital area. These two plasmids differed only in gene dsbC. Disulfide bond formation is a crucial step in the folding process of a protein and is catalyzed by bacterial proteins of the Dsb system. DsbC is responsible for rearranging incorrect disulfides introduced between cysteine residues (Banaś et al., 2020).
Conclusion
In summary, our study described the emergence of blaNDM–7 in ST11 K. pneumoniae in China for the first time. Our results indicate that IncX3 plasmid may contribute to the prevalence of blaNDM–7 in China. The high prevalence of NDM variants worldwide highlights the critical need for careful monitoring and control of the rapid dissemination of blaNDM.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Shandong Provincial Hospital. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin. Written informed consent was obtained from the minor(s)’ legal guardian/next of kin for the publication of any potentially identifiable images or data included in this article.
Author Contributions
YJ designed the experiments and revised the manuscript. CS carried out the experiments and wrote the manuscript. YH analyzed the data. YW and MJ contributed to experiment conception. All authors contributed to the article and approved the submitted version.
Funding
This study is funded by the National Natural Science Foundation of China (nos. 81401696 and 81902119).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.576823/full#supplementary-material
Abbreviations
MBL, metallo- β-lactamases; MLST, multilocus sequence typing; NDM, New Delhi metallo-beta-lactamases.
Footnotes
- ^ www.eucast.org/clinical-breakpoint
- ^ http://www.pasteur.fr/recherche/genopole/PF8/mlst/K. pneumoniae.html
- ^ http://compbio.ornl.gov/prodigal/
References
Ahmad, N., Khalid, S., Ali, S. M., and Khan, A. U. (2018). Occurrence of blaNDM variants among Enterobacteriaceae from a neonatal intensive care unit in a Northern India Hospital. Front. Microbiol. 9:407. doi: 10.3389/fmicb.2018.00407
Banaś, A. M., Bocian-Ostrzycka, K. M., Plichta, M., Dunin-Horkawicz, S., Ludwiczak, J., Płaczkiewicz, J., et al. (2020). C8J_1298, a bifunctional thioloxid oreductase of Campylobacterjejuni, affects Dsb(disulfidebond) network functioning. PLoS One 15:e0230366. doi: 10.1371/journal.pone.0230366
Bi, R., Kong, Z., Qian, H., Jiang, F., Kang, H., Gu, B., et al. (2018). High prevalence of bla NDM Variants among carbapenem-resistant Escherichia coli in Northern Jiangsu Province, China. Front. Microbiol. 9:2704. doi: 10.3389/fmicb.2018.02704
Cuzon, G., Bonnin, R. A., and Nordmann, P. (2013). First identification of novel NDM carbapenemase, NDM-7, in Escherichia coli in France. PLoS One 8:e61322. doi: 10.1371/journal.pone.0061322
Gregson, D. B., Church, D. L., Conly, J., Kreiswirth, B. N., and Pitout, J. D. (2016). Molecular evolution of Klebsiella pneumoniae ST27 8Isolate harboring blaNDM-7 and involved in nosocomia transmission. J. Infect. Dis. 214, 798–806. doi: 10.1093/infdis/jiw240
Guo, X., Rao, Y., Guo, L., Xu, H., Lv, T., Yu, X., et al. (2019). Detection and genomic characterization of a Morganella morganii isolate from China that produces NDM-5. Front. Microbiol. 10:1156. doi: 10.3389/fmicb.2019.01156
Hao, Y., Shao, C., Bai, Y., and Jin, Y. (2018). Genotypic and phenotypic characterization of IncX3 plasmid carrying blaNDM-7 in Escherichia coli sequence Type 167 isolated from a patient with urinary tract infection. Front. Microbiol. 9:2468. doi: 10.3389/fmicb.2018.02468
Khalid, S., Ahmad, N., Ali, S. M., and Khan, A. U. (2020). Outbreak of efficiently transferred carbapenem-resistant blaNDM-producing gram-negative Bacilli isolated from neonatal intensive care unit of an Indian Hospital. Microb. Drug Resist. 26, 284–289. doi: 10.1089/mdr.2019.0092
Lee, C. S., Vasoo, S., Hu, F., Patel, R., and Doi, Y. (2014). Klebsiella pneumoniae ST147 coproducing NDM-7 carbapenemase and RmtF 16S rRNA methyltransferase in Minnesota. J. Clin. Microbiol. 52, 4109–4110. doi: 10.1128/JCM.01404-14
Liu, Z., Xiao, X., Li, Y., Liu, Y., Li, R., and Wang, Z. (2019). Emergence of IncX3 plasmid-harboring blaNDM-5 dominated by Escherichia coli ST48 in a goose farm in Jiangsu, China. Front. Microbiol. 10:2002. doi: 10.3389/fmicb.2019.02002
Mouftah, S. F., Pál, T., Darwish, D., Ghazawi, A., Villa, L., Carattoli, A., et al. (2019). Epidemic IncX3 plasmids spreading carbapenemase genes in the United Arab emirates and wordwide. Infect. Drug Resist. 12, 1729–1742. doi: 10.2147/IDR.S210554
Paul, D., Bhattacharjee, A., Ingti, B., Choudhury, N. A., Maurya, A. P., Dhar, D., et al. (2017). Occurrence of blaNDM-7 within IncX3-type plasmid of Escherichia coli from India. J. Infect. Chemother. 23, 206–210. doi: 10.1016/j.jiac.2016.12.009
Pérez-Vázquez, M., Sola Campoy, P. J., Ortega, A., Bautista, V., Monzón, S., Ruiz-Carrascoso, G., et al. (2019). Emergence of NDM-producing Klebsiella pneumoniae and Escherichia coli in Spain: phylogeny, resistome, virulence and plasmids encoding blaNDM-like genes as determined by WGS. J. Antimicrob. Chemother. 74, 3489–3496. doi: 10.1093/jac/dkz366
Pitout, J. D., Nordmann, P., and Poirel, L. (2015). Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob. Agents Chemother. 59, 5873–5884. doi: 10.1128/AAC.01019-15
Rolain, J. M., Parola, P., and Cornaglia, G. (2010). New Delhimetallo-beta-lactamase (NDM) towards a new pandemia? Clin. Microbiol. Infect. 16, 1699–1701. doi: 10.1111/j.1469-0691.2010.03385.x
Shankar, C., Kumar, S., Venkatesan, M., and Veeraraghavan, B. (2019). Emergence of ST147 Klebsiella pneumoniae carryin gblaNDM-7 on IncA/C2 with ompK35 and ompK36 mutationsin India. J. Infect. Public Health 12, 741–743. doi: 10.1016/j.jiph.2019.03.023
Solgi, H., Badmasti, F., Aminzadeh, Z., Giske, C. G., Pourahmad, M., Vaziri, F., et al. (2017). Molecular characterization of intestinal carriage of carbapenem-resistant Enterobacteriaceae among inpatients at two Iranian university hospitals: first report of co-production of blaNDM-7and blaOXA-48. Eur. J. Clin. Microbiol. Infect. Dis. 36, 2127–2135. doi: 10.1007/s10096-017-3035-3
Wang, J. L., Shang, Y. Y., Guo, S. Y., Diao, F. F., Yu, J. Y., Wei, X. H., et al. (2017). Serotype and virulence gene of Klebsiella pneumoniae isolated from mink and its pathogenesis in mice and mink. Sci. Rep. 7:17291. doi: 10.1038/s41598-017-17681-8
Wang, L. H., Liu, P. P., Wei, D. D., Liu, Y., Wan, L. G., Xiang, T. X., et al. (2016). Clinical isolates of uropathogenic Escherichia coli ST131 producing NDM-7 metallo-β-lactamase in China. Int. J. Antimicrob. Agents 48, 41–45. doi: 10.1016/j.ijantimicag
Xu, J., and He, F. (2019). Characterization of a NDM-7 carbapenemase-producing Escherichia coli ST410 clinical strain isolated from a urinary tract infection in China. Infect Drug Resist. 12, 1555–1564. doi: 10.2147/IDR.S206211
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/AAC.00774-09
Yoon, E. J., Kang, D. Y., Yang, J. W., Kim, D., Lee, H., Lee, K. J., et al. (2018). New Delhi Metallo-beta-lactamase-producing Enterobacteriaceae in South Korea between 2010 and 2015. Front. Microbiol. 9:571. doi: 10.3389/fmicb.2018.00571
Zhang, F., Xie, L., Wang, X., Han, L., Guo, X., Ni, Y., et al. (2016). Further Spread of bla NDM-5 in Enterobacteriaceae via IncX3 Plasmids in Shanghai, China. Front. Microbiol. 7:424. doi: 10.3389/fmicb.2016.00424
Zhao, Y. J., Zhang, X. C., Von Vergel, L. T., Liu, H. Y., Rocker, A., Zhang, Y. Z., et al. (2019). An Outbreak of carbapenem-resistant and hypervirulent Klebsiella pneumoniae in an intensive care Unit of a major teaching Hospital in Wenzhou, China. Front. Public Health 7:229. doi: 10.3389/fpubh.2019.00229
Keywords: NDM-7, K. pneumoniae, ST11, IncX3, China
Citation: Shao C, Hao Y, Wang Y, Jiang M and Jin Y (2020) Genotypic and Phenotypic Characterization of blaNDM–7-Harboring IncX3 Plasmid in a ST11 Klebsiella pneumoniae Isolated From a Pediatric Patient in China. Front. Microbiol. 11:576823. doi: 10.3389/fmicb.2020.576823
Received: 27 June 2020; Accepted: 07 September 2020;
Published: 02 October 2020.
Edited by:
Miklos Fuzi, Semmelweis University, HungaryReviewed by:
Kwan Soo Ko, Sungkyunkwan University School of Medicine, South KoreaÁkos Tóth, National Public Health Institute (OKI), Hungary
Copyright © 2020 Shao, Hao, Wang, Jiang and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Jin, sdjinyan@163.com
 Chunhong Shao
Chunhong Shao Yingying Hao
Yingying Hao Yong Wang1
Yong Wang1