- 1Department of Laboratory Medicine, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
- 2State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China
- 3Basic Medical College, Beihua University, Jilin City, China
- 4The Clinical Immunology Research Center, Beihua University, Jilin City, China
The increasing prevalence and transmission of the carbapenem resistance gene blaNDM–5 has led to a severe threat to public health. So far, blaNDM–5 has been widely detected in various species of Enterobacterales and different hosts across various cities. However, there is no report on the blaNDM–5– harboring Morganella morganii. In January 2016, the first NDM-5-producing Morganella morganii L241 was found in a stool sample of a patient diagnosed as recurrence of liver cancer in China. Identification of the species was performed using 16S rRNA gene sequencing. Carbapenemase genes were identified through both PCR and sequencing. To investigate the characteristics and complete genome sequence of the blaNDM–5-harboring clinical isolate, antimicrobial susceptibility testing, S1 nuclease pulsed field gel electrophoresis, Southern blotting, transconjugation experiment, complete genome sequencing, and comparative genomic analysis were performed. M. morganii L241 was found to be resistant to broad-spectrum cephalosporins and carbapenems. The complete genome of L241 is made up from both a 3,850,444 bp circular chromosome and a 46,161 bp self-transmissible IncX3 plasmid encoding blaNDM–5, which shared a conserved genetic context of blaNDM–5 (ΔIS3000-ΔISAba125-IS5-blaNDM–5-ble-trpF-dsbC-IS26). BLASTn analysis showed that IncX3 plasmids harboring blaNDM genes have been found in 15 species among Enterobacterales from 13 different countries around the world thus far. In addition, comparative genomic analysis showed that M. morganii L241 exhibits a close relationship to M. morganii subsp. morganii KT with 107 SNPs. Our research demonstrated that IncX3 is a key element in the worldwide dissemination of blaNDM-5 among various species. Further research will be necessary to control and prevent the spread of such plasmids.
Introduction
Morganella morganii is a facultative anaerobic Gram-negative bacterium, the representative strain of the genus Morganella (Liu et al., 2016). This bacterium tends to colonize in the intestinal tracts of humans, mammals and reptiles as part of the normal flora, and is often found in the environment (Lee et al., 2009). It is noteworthy that M. morganii is an opportunistic pathogen, but the disease spectrum associated its infections is broad, mainly including sepsis (Seija et al., 2015), abscess (Zaid et al., 2013), urinary tract infection (Jamal et al., 2015), and bacteremia (Ghosh et al., 2009). Furthermore, M. morganii can harbor ESBLs and carbapenemase, which adds resistance to multiple antibiotics and has resulted in a high mortality rate in some infections (Liu et al., 2016). There are existing reports of the detection of New Delhi metallo-β-lactamase-1 (NDM-1) (Olaitan et al., 2014), Klebsiella pneumonia carbapenemases-2 (KPC-2) (Shi et al., 2011), and Metallo-β-lactamase VIM-1 (Tsakris et al., 2007) in M. morganii. However, to date, NDM-5-producing M. morganii has not been described.
NDM-5 was first identified in Escherichia coli from a patient who had been hospitalized in India in 2011 (Hornsey et al., 2011). NDM-5 and NDM-1 are similar; the only difference they demonstrated is that two amino acids have been replaced (Val88Leu and Met154Leu), resulting in NDM-5 exhibiting a high level of resistance to carbapenems and expanded-spectrum cephalosporins, and thus posing a severe threat to public health (Hornsey et al., 2011). Since then, NDM-5 has spread globally, such as China (Zhang L.P. et al., 2016), the United States of America (Rojas et al., 2017), Australia (Wailan et al., 2015), Egypt (Soliman et al., 2016), and Italy (Giufre et al., 2018).
Worryingly, in China, NDM-5 has been detected in various species of Enterobacterales across various cities (Zhang F. et al., 2016; Li et al., 2017; Mao et al., 2018; Sun et al., 2018). In the current study, we identified a clinical M. morganii isolate producing NDM-5 and performed phylogenetic analysis. Further, we investigated the drug resistance profile and plasmid characteristic analysis to depict the potential transmission mechanisms of blaNDM–5.
Materials and Methods
Strain Screening
Since January 2016, we collected various clinical samples from patients based at the First Affiliated Hospital of Zhejiang University in Hangzhou (FAHZU). The samples were spread on the surface of MacConkey agar (OXOID, Hampshire, United Kingdom) plates that contained 2 mg/L meropenem (Meilunbio, Dalian, China) for 18–24 h at 37°C for the preliminary screening of carbapenem-resistant Enterobacterales (CRE) isolates (CLSI, 2018). The CarbaNP test and modified carbapenem inactivation method (mCIM) with EDTA-modified carbapenem inactivation method (eCIM) were used to detect carbapenemase activity (CLSI, 2018). Identification of species was performed using both matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS) (Bruker Daltonik GmbH, Bremen, Germany) and 16S rRNA gene sequencing. Carbapenemase genes (blaKPC, blaNDM, blaOXA–48, blaV IM, and blaIMP) were identified using PCR and DNA sequencing as described previously (Zheng et al., 2015). Finally, M. morganii L241 isolate was detected and its details were described in the results.
Antimicrobial Susceptibility Testing
Morganella morganii L241 isolate was tested for resistance, using the agar dilution method, against 17 antibiotics, which were piperacillin/tazobactam, cefotaxime, ceftazidime, cefepime, cefpirome, aztreonam, ertapenem, imipenem, meropenem, amikacin, tetracycline, fosfomycin, gentamicin, chloramphenicol, ciprofloxacin, levofloxacin, and trimethoprim/sulfamethoxazole. Results were interpreted following the guidelines of the CLSI document M100-S28 (2018) (CLSI, 2018). E.coli ATCC 25922 was used as a control.
Location of blaNDM–5 Gene and Transferability of Plasmid Carrying blaNDM–5
The number and size of plasmid of M. morganii L241 were determined with the S1 nuclease pulsed field gel electrophoresis (S1-PFGE) method, as described previously (Zheng et al., 2015). Southern blotting and hybridization using DIG-labeled blaNDM-specific probe were performed to estimate the location of blaNDM gene, while the transferability of NDM-carrying plasmid from the isolate was determined through the use of conjugation experiments, with rifampicin-resistant E. coli C600 as the recipient strain. Further to this, transconjugants were selected on Mueller-Hinton agar (OXOID, Hampshire, United Kingdom) plates that contained both 200 mg/L rifampicin (Meilunbio, Dalian, China) and 2 mg/L meropenem. Finally, a combination of MALDI-TOF/MS identification, blaNDM gene detection and antimicrobial susceptibility testing of the transconjugants were performed in order to confirm whether the plasmid was successfully transferred to the recipient.
Whole Genome Sequencing and in silico Analyses
Genomic DNA was extracted using the OMEGA Bacterial DNA kit (Omega Bio-tek, Norcross, United) and was then sequenced on both the llumina HiSeq 4000-PE150 (Illumina, San Diego, CA, United States) and the PacBio RS II platforms (Pacific Biosciences, California, United States). We created a complete genome sequence for M. morganii L241 using Unicycler (Wick et al., 2017) by combining our llumina sequencing reads with PacBio sequencing reads. By using Unicycler (Wick et al., 2017), raw llumina reads were assembled using SPAdes, semi-global alignment was then performed by aligning PacBio reads to the assembly data, the llumina sequenceing reads were finally used to polish the genome assembly with Bowtie2 and Pilon. Additionally, online tools1 were used to identify acquired antimicrobial resistance genes and replicon type of plasmid. This genome was annotated by the RAST server (Aziz et al., 2008), while the IS Finder database2 was used to identify transposon and IS elements. The circular image of multiple plasmids comparisons was generated by the BLAST Ring Image Generator (BRIG) (Alikhan et al., 2011). Finally, the comparison figures of the genetic context of blaNDM–5 on multiple plasmids were performed with a Python application Easyfig (Zheng et al., 2017).
Comparative Genomic Analysis
Genome sequences for 41 strains of M. morganii were downloaded from Pathogen Detection3. These genomes, plus the M. morganii L241 genomic sequence, were then analyzed using Snippy4, a process in which raw reads were mapped against the reference M. morganii genome (no.ALJX00000000) (Chen et al., 2012). A phylogenetic tree based on concatenated, qualified single nucleotide polymorphisms (SNPs) was then performed using Harvest (Treangen et al., 2014). Characteristics of all the M. morganii strains included in this study are summarized in Supplementary Table S1.
Accession Numbers
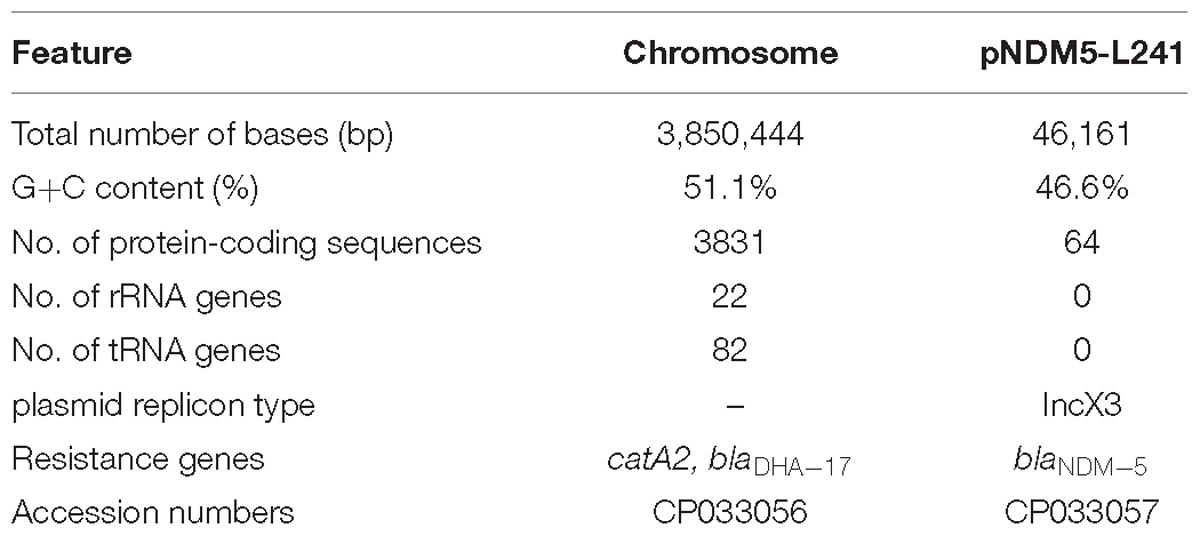
The genome sequences of both M. morganii L241 chromosome and plasmid pNDM5-L241 were deposited in the GenBank with the accession numbers CP033056 and CP033057.
Results
Isolation and Identification of NDM-5-Producing M. morganii L241 Strain
A male patient of 53 years old was admitted to surgical ward of FAHZU in January 2016 and initially diagnosed as recurrence of liver cancer. The patient received hepatectomy 2 days after hospitalization and developed acute diarrhea on the third day after surgery. Since then, diarrhea has always existed. On the sixth day after surgery, a rod shaped Gram-negative bacterium, designated as L241, was recovered from the selective medium, which was found to be positive for the CarbaNP test, and mCIM with eCIM assay. Then it was confirmed as M. morganii and found to harbor blaNDM–5 after PCR and sequencing.
Antimicrobial Susceptibility Testing
The minimum inhibitory concentration (MIC) values of antimicrobials for M. morganii L241 are shown in Table 1. M. morganii L241 exhibited resistance to almost all of the β-lactam antibiotics tested, including piperacillin/tazobactam (MIC > 128 mg/L), cefotaxime (MIC > 128 mg/L), ceftazidime (MIC > 128 mg/L), cefepime (MIC = 128 mg/L), cefpirome (MIC > 128 mg/L), ertapenem (MIC = 32 mg/L), imipenem (MIC > 32 mg/L), and meropenem (MIC > 32 mg/L), with the exception of aztreonam (MIC = 1 mg/L). In addition, the isolate also demonstrated resistance to fosfomycin (MIC = 256 mg/L) and chloramphenicol (MIC = 32 mg/L) but was shown to be susceptible to amikacin, tetracycline, gentamicin, ciprofloxacin, levofloxacin, and trimethoprim/sulfamethoxazole.
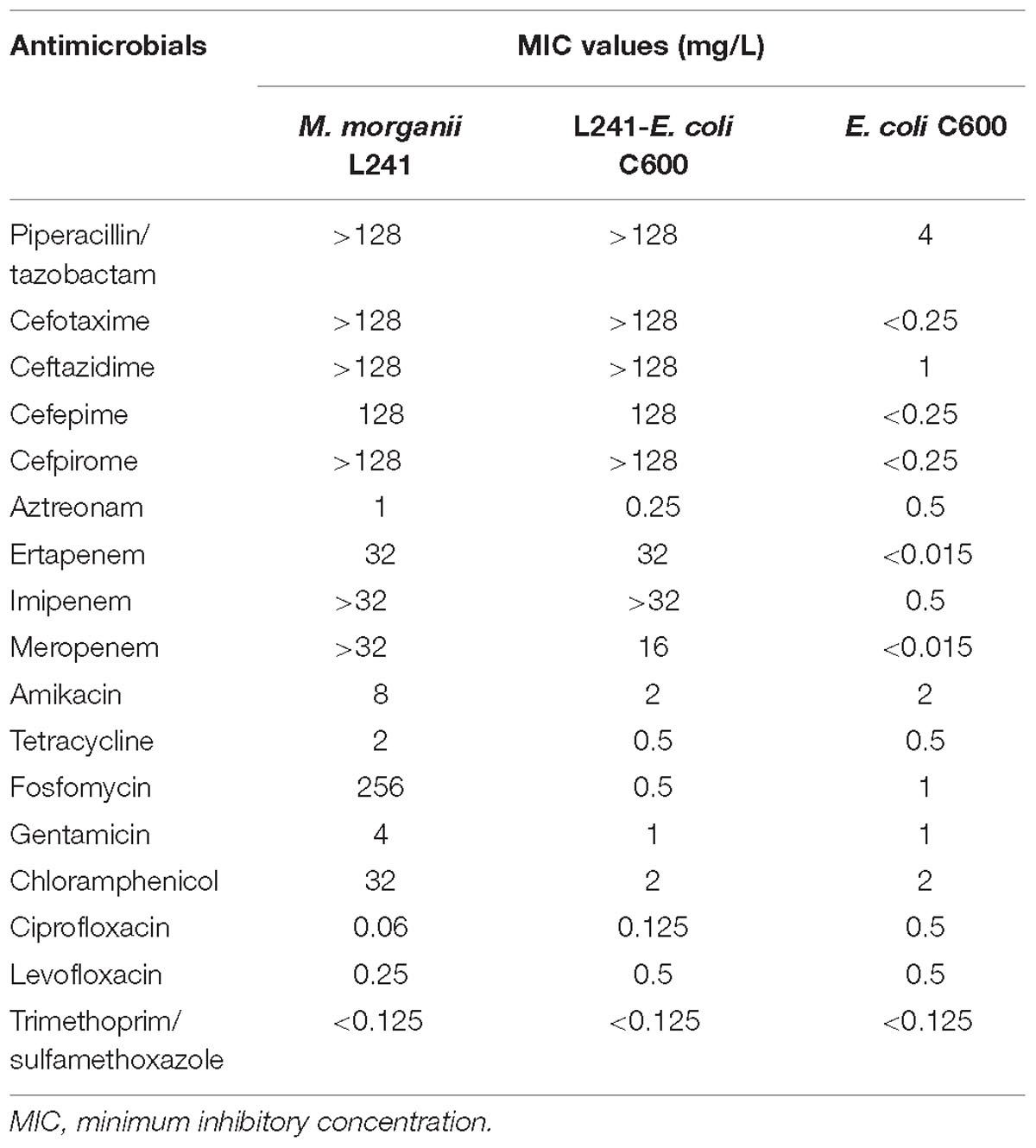
Table 1. MIC values of antimicrobials for M. morganii L241, recipient strain E. coli C600 and transconjugant L241-E. coli C600.
Genomics Features of M. morganii L241
The genomic features of the M. morganii L241 are shown in Table 2. It was found that the M. morganii L241 genome consists of a 3,850,444 bp circular chromosome with an average G+C content of 51.1% and one plasmid. Further, the chromosome contained 3,831 protein coding genes, 82 tRNAs, and 22 rRNAs. A screening for acquired resistance determinants found that chromosome possess the resistance gene catA2 encoding phenicol resistance and blaDHA–17 encoding β-lactam resistance; while the plasmid encoding acquired resistance gene confers resistance to β-lactams (blaNDM–5). This finding is consistent with the drug resistant phenotypes.
Comparative Genomic Analysis
While previous studies have reported the genomes of M. Morganii (Khatri et al., 2013; Nash and Young, 2015), there are few studies which have linked the genetic information of multiple strains of M. morganii to explore their evolutionary relationships and the internal structure of the genome. Therefore, we performed a comparative genomic analysis.
As shown in Figure 1 and Supplementary Table S1, all of M. morganii isolates were found in various specimen types, including stool, rectal swab, wound, urine, blood, sputum, abscess, pericardial fluid, lettuce leaves, cheese, phytotelma, freshwater lake and roots from different hosts (homo sapiens, animal, plant, food, and environment). At the same time, they were detected across various countries, including India, Japan, Austria, Brazil, Russia, United States, South Africa, Viet Nam, United Kingdom, Malaysia, South Korea, Portugal, Switzerland, Canada, and China from 1800 to 2017, suggesting that M. morganii isolates are widely distributed. Resistance genes present on all M. morganii isolates showed that there was a common β-lactamases resistance gene blaDHA. In addition, no plasmid replication type was detected in most M. morganii isolates.

Figure 1. A comparative genome analysis of NDM-5-producing Morganella morganii L241 and other M. morganii isolates based on SNPs. The plasmid replication types, β-lactamases resistance genes, catA genes, collection dates, locations, isolation sources, and hosts of isolates are shown. The annotation denotes the presence of plasmid replication types, β-lactamases resistance genes and catA genes as determined by online tools (http://www.genomicepidemiology.org/). The M. morganii L241 is indicated by red. Regions with blaDHA gene among isolates are shown by gray shading. The detail information of isolates included in this study is summarized in Supplementary Table S1.
In addition to M. morganii NCTC12358, M. morganii INSRALV892, M. morganii L3, M. morganii TW17014, M. morganii FAM24091, M. morganii H1r, M. morganii NLAE-zl-C84, M. morganii AR_0133, and M. morganii MRSN22709, M. morganii L241 has close genetic relationships with other M. morganii isolates, among which M. morganii L241 is clustered with M. morganii MM4, M. morganii MM190, M. morganii FDAARGOS_438, M. morganii FDAARGOS_172, M. morganii NCTC232, M. morganii NCTC12028, M. morganii E042, and M. morganii subsp. morganii KT (Chen et al., 2012). These nine clustered strains were isolated from different specimen types and were detected at different times and in different countries. Further analysis of genomic information showed that all of these strains contained blaDHA–17 and catA2 genes, but there was no common plasmid replicon type. It is noteworthy that M. morganii L241 and M. morganii subsp. morganii KT, which is the first genome sequence of M. morganii, are the most closely related isolates, differing by just 107 SNPs.
Characterization of Plasmid Bearing blaNDM–5
The S1-PFGE result showed that only a ∼46-Kb plasmid was found in M. morganii L241 (Figure 2A). Subsequently, Southern blotting revealed that the blaNDM–5 gene was located on this plasmid (Figure 2B). PCR and sequencing analysis confirmed a transconjugant as blaNDM–5–encoding E. coli C600. This transconjugant exhibited resistance to almost all β-lactams aside from aztreonam, including piperacillin/tazobactam (MIC > 128 mg/L), cefotaxime (MIC > 128 mg/L), ceftazidime (MIC > 128 mg/L), cefepime (MIC = 128 mg/L), cefpirome (MIC > 128 mg/L), ertapenem (MIC = 32 mg/L), imipenem (MIC > 32 mg/L), and meropenem (MIC = 16 mg/L) (Table 1), with considerable increases in the MICs of carbapenems when compared with the recipient strain E. coli C600. These results indicate that the blaNDM–5-encoding plasmid of M. morganii L241, designated as pNDM5-L241, was successfully transferred into recipient E. coli C600 strain. In addition, results suggest that this was a self-transmissible plasmid. Owing to M. morganii L241 only possessing plasmid pNDM5-L241, the antimicrobial resistance phenotypes of the transconjugant were acquired from pNDM5-L241.
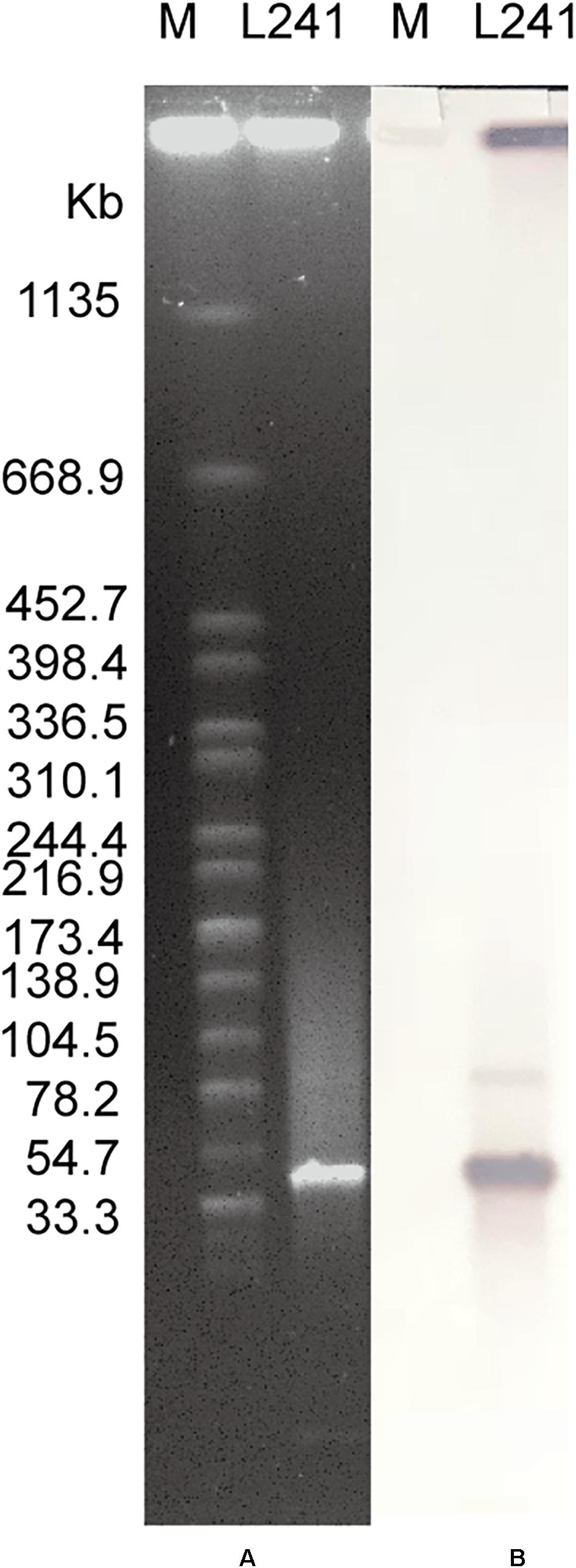
Figure 2. Plasmid profiles of M. morganii L241. (A) Plasmid size determination by S1-PFGE, with Salmonella enterica serotype Braenderup H9812 as the size marker. (B) Southern blotting hybridization with a blaNDM-specific probe.
In silico analysis identified that plasmid pNDM5-L241 is an IncX3 type plasmid, with 46,161 bp in length, 64 predicted coding sequences and a GC content of 46.6%. This plasmid was home to several types of genes, such as antimicrobial resistance genes, mobile elements genes, putative genes, genes encoding replication proteins and genes encoding proteins for plasmid stability and plasmid transfer, respectively. A search of the nr/nt database found plasmid pNDM5-L241 exhibiting 99% nucleotide identity with the IncX3 blaNDM–5 encoding plasmid pTB203 (no. CP029245) and pNDM_MGR194 (no. KF220657). In all of these cases, blaNDM–5 was the only antimicrobial resistance gene. Importantly, a conserved structure sequence (ΔIS3000-ΔISAba125-IS5-blaNDM–5-ble-trpF-dsbC-IS26) was found in the upstream and downstream of the blaNDM–5 (Figure 3B).
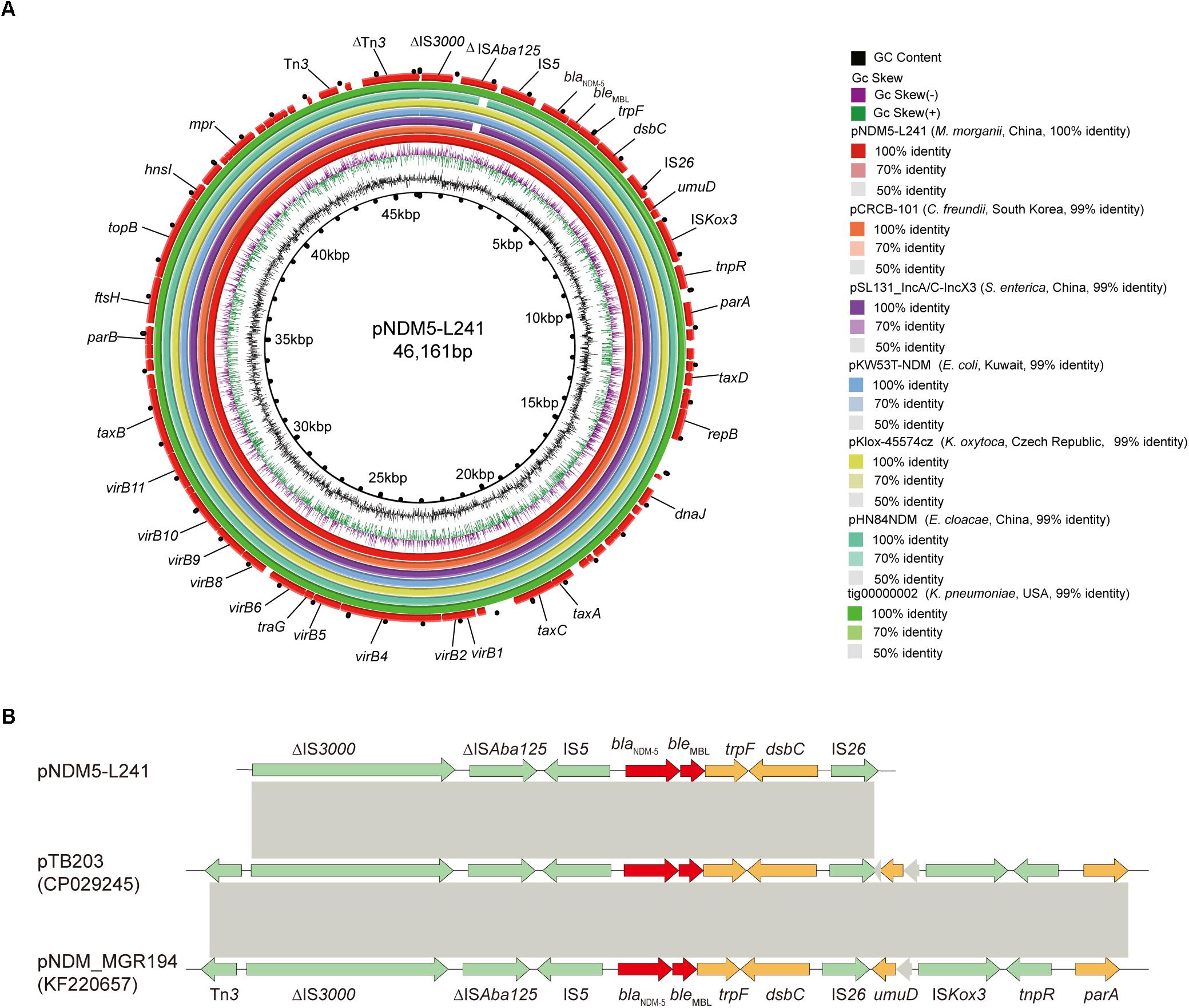
Figure 3. The genomic analysis of pNDM5-L241 plasmid. (A) Comparison of the pNDM5-L241 plasmid with the closely related plasmid based on BLASTn analyses (GenBank accession numbers from inside to outside are CP024820, MH105050, KX214669, MG833406, KY296103, and CP021759). (B) Genetic context of blaNDM–5 on pNDM5-L241, pTB203 (CP029245) and pNDM_MGR194 (KF220657). Open reading frames (ORFs) are represented by arrows and colored in accordance with their putative functions: red arrows indicate antimicrobial resistance genes, green arrows represent mobile genetic elements, while genes encoding hypothetical proteins and proteins for plasmid stability are colored as gray and brown, respectively. Regions with a high degree of homology between plasmids are shown by gray shading.
Following this, we compared pNDM5-L241 with the blaNDM–5-encoding plasmid pKlox-45574cz (Klebsiella oxytoca, Czech Republic, no. MG833406), blaNDM–7-encoding plasmid pKW53T-NDM (E. coli, Kuwait, no. KX214669) (Pal et al., 2017), blaNDM–5-encoding plasmid pCRCB-101_1 (Citrobacter freundii, South Korea, no.CP024820), blaNDM–1-encoding plasmid pSL131_IncA/C-IncX3 (Salmonella enterica subsp. enterica serovar Lomita, China, no.MH105050), blaNDM–1-encoding plasmid pHN84NDM (Enterobacter cloacae, China, no.KY296103), and blaNDM–7-encoding plasmid tig00000002 (Klebsiella pneumoniae, United States, no.CP021759), which demonstrated a sequence similarity of 99% with coverages of 100, 100, 99, 99, and 100%, respectively (Figure 3A). As shown in Figure 3A, we found that the backbone sequences of the seven plasmids were almost identical.
Discussion
Morganella morganii has been recognized as an increasingly important pathogen because of the increased frequency and a high mortality rate of its infections. In addition, according to a recent report, acquired resistance is increasingly observed in M. morganii (Liu et al., 2016). For example, M. morganii has shown resistance to β-lactams, aminoglycosides, phenicols, macrolides, tetracycline, trimethoprim, and fluoroquinolones (Liu et al., 2016). As a result of the intrinsic and acquired resistance of M. morganii, it poses a serious clinical threat which has limited treatment options. Nevertheless, there has not been too much attention on M. morganii so far.
The rapid development of gene sequencing technology has enabled us to have a deeper understanding of bacteria. As we know, the comparative genomic analysis based on SNPs combined with the plasmid replication type, antibiotic resistance gene content, time of isolation, geographical region, isolation source, and host is a valuable tool to conduct genomic epidemiological analyses. Therefore, in this study, the complete genome sequence and comparative genomic analysis were performed. Our analysis showed that M. morganii L241 is clustered with M. morganii MM4, M. morganii MM190, M. morganii FDAARGOS_438, M. morganii FDAARGOS_172, M. morganii NCTC232, M. morganii NCTC12028, M. morganii E042, and M. morganii subsp. morganii KT (Chen et al., 2012). The clustering phenomena and relatively small number of SNPs seen in these M. morganii isolates from different geographic locations over such a long time frame suggest that these isolates might be highly clonal. Further analysis showed that the clustering of M. morganii L241 with other M. morganii isolates was not determined by IncX3 type plasmid. Moreover, results showed that M. morganii L241 and M. morganii subsp. morganii KT are the most closely related isolates, prompting that M. morganii L241 may have evolved from M. morganii subsp. morganii KT. The pathogenicity-related factors of M. morganii subsp. morganii KT were identified, such as fimbrial adhesins, T3SS, TCS, iron acquisition system, IgA protease, and insecticidal and apoptotic toxins (Chen et al., 2012), implying M. morganii L241 has the similar toxicity characteristics.
To date, NDM-5 has been found in Proteus mirabilis (Zhang F. et al., 2016), K. pneumonia (Cho et al., 2015), E. coli (Soliman et al., 2016), Enterobacter aerogenes (Ahmad et al., 2018), and Salmonella enterica serovar Typhimuriumstrain (Li et al., 2017). As far as we are aware, the current study is the first report that has identified NDM-5 in M. morganii. This is a worrying development as it demonstrates the further spread of blaNDM–5 among different species of Enterobacterales.
In this work, we observed that a conserved structure sequence (ΔIS3000-ΔISAba125-IS5-blaNDM–5-ble-trpF-dsbC-IS26) was found in the upstream and downstream of the blaNDM–5 in IncX3 type plasmid. Interestingly, the conserved structure sequence is consistent with the upstream and downstream of the blaNDM–5 in IncFII type plasmid (Li et al., 2017). Recently, research has proposed that IS26 element may contribute to the vertically transfer of blaNDM–5 gene among plasmids and chromosomes (Li et al., 2017).
Horizontal gene transfer also contributed to the widespread dissemination of blaNDM–5 in Enterobacterales. Previously, the blaNDM–5 gene had been identified on various plasmid types, such as IncFII and IncX3 (Feng et al., 2018; Giufre et al., 2018). However, it has thus far been predominantly associated with IncX3 plasmids (Li et al., 2018). IncX3 plasmids carrying blaNDM–5 have spread widely among Enterobacterales worldwide (Li et al., 2018). These findings suggest that the production of blaNDM–5– harboring M. morganii may be the result of the transmission of blaNDM–5- harboring IncX3. Additionally, it has recently been shown that blaNDM–5– harboring IncX3-type plasmid isolated from raw milk and fecal samples from cows has spread among cow farms, suggesting that blaNDM–5– harboring IncX3-type plasmid also can be transmitted from animals to humans through the food chain (He et al., 2017). This is an important finding. However, even more importantly, our BLASTn analysis showed that IncX3 plasmids harboring various blaNDM genes, including blaNDM–1 (Lü et al., 2018; Zhu et al., 2018), blaNDM–4 (Espedido et al., 2015; Sugawara et al., 2017), blaNDM–5 (Krishnaraju et al., 2015; Zhu et al., 2016; Li et al., 2018; Xie et al., 2018), blaNDM–6, blaNDM–7 (Pal et al., 2017; Sugawara et al., 2017), blaNDM–13 (Lv et al., 2016), blaNDM–17 (Liu et al., 2017), blaNDM–19 (Liu et al., 2019), blaNDM–20 (Liu Z. et al., 2018), and blaNDM–21 (Liu L. et al., 2018), which have been found in 15 species among Enterobacterales [K. pneumoniae (Espedido et al., 2015; Krishnaraju et al., 2015), K. oxytoca (Paskova et al., 2018; Yoon et al., 2018), Klebsiella michiganensis, Klebsiella aerogenes, E. coli (Zhu et al., 2016; Pal et al., 2017; Sugawara et al., 2017; Li et al., 2018; Xie et al., 2018), C. freundii (Zhu et al., 2018), S. enteric, Enterobacter hormaechei, Enterobacter cloacae (Lü et al., 2018), Enterobacter asburiae (Paskova et al., 2018), Enterobacter xiangfangensis (Paskova et al., 2018), Cronobacter sakazakii, Raoultella planticola, Raoultella ornithinolytica (Paskova et al., 2018), and Kluyvera intermedia (Paskova et al., 2018)] from 13 different countries around the world thus far. These countries are China (Zhu et al., 2016; Li et al., 2018; Liu Z. et al., 2018; Xie et al., 2018), the Czech Republic (Paskova et al., 2018), Kuwait (Pal et al., 2017), Korea (Yoon et al., 2018), Oman (Pal et al., 2017), United States, Sweden, Myanmar (Sugawara et al., 2017), Vietnam, India (Krishnaraju et al., 2015), Arabian Peninsula (Pal et al., 2017), Canada, and Australia (Espedido et al., 2015). These troubling results suggest that IncX3 type plasmids have attributed to the dissemination of the NDM variant among different species around the world. Of note, the IncX3 plasmid usually also bears other β-lactamase genes (blaSHV, blaampC, blaTEM, blaOXA, and blaKPC) and encodes resistance genes which are responsible for other antibiotics, such as quinolones (qnr), sulphonamides (sul1), and tetracyclines (tet) (Dobiasova and Dolejska, 2016; Cerdeira et al., 2017; Bitar et al., 2018). Taken together, the transmission of this plasmid may lead to a severe threat to public health. It is crucial that we take urgent and effective measures to control the dissemination of the IncX3 type plasmids.
Conclusion
In summary, we first identified a blaNDM–5-positive M. morganii and reported its complete genome sequence. The blaNDM–5 gene was located on a self-transmissible IncX3 plasmid which spread among species of Enterobacterales worldwide. This study highlights the wide spread of blaNDM-encoding IncX3 plasmids, including their transmissionin to uncommon Enterobacterales strains including M. morganii. Therefore, the IncX3 plasmids must be closely monitored, and attention must be paid to uncommon Enterobacterales strains. Further research is necessary to prevent and control the spread of blaNDM-encoding IncX3 plasmids.
Data Availability
The datasets generated for this study can be found in NCBI, CP033056 and CP033057.
Ethics Statement
Written informed consent was obtained from the participants of this study.
Author Contributions
XG and BZ conceived and designed the experiments. YR, HX, TL, YC, NL, and HH performed the experiments. LG and XY analyzed the data. BZ and YR wrote the manuscript.
Funding
This study was supported by funding from the National Natural Science Foundation of China (No. 81741098); the grant from Henan UC science and Technology Department (No. 162102310509); the National Key Research and Development Program of China (No. 2016YFD0501105); and the Zhejiang Provincial Natural Science Foundation of China (No. LY17H190003).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01156/full#supplementary-material
Footnotes
- ^ http://www.genomicepidemiology.org/
- ^ https://www-is.biotoul.fr/
- ^ https://www.ncbi.nlm.nih.gov/pathogens/
- ^ https://github.com/tseemann/snippy
References
Ahmad, N., Ali, S. M., and Khan, A. U. (2018). Detection of New Delhi metallo-beta-lactamase variants NDM-4, NDM-5, and NDM-7 in Enterobacter aerogenes isolated from a neonatal intensive care unit of a north india hospital: a first report. Microb. Drug Resist. 24, 161–165. doi:10.1089/mdr.2017.0038
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Aziz, R. K., Bartels, D., Best, A. A., Dejongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bitar, I., Dagher, C., Salloum, T., Araj, G., and Tokajian, S. (2018). First report of an Escherichia coli from Lebanon carrying an OXA-181 carbapenemase resistance determinant. J. Glob. Antimicrob. Resist. 12, 113–114. doi: 10.1016/j.jgar.2018.01.002
Cerdeira, L. T., Cunha, M. P. V., Francisco, G. R., Bueno, M. F. C., Araujo, B. F., Ribas, R. M., et al. (2017). IncX3 plasmid harboring a non-Tn4401 genetic element (NTEKPC) in a hospital-associated clone of KPC-2-producing Klebsiella pneumoniae ST340/CG258. Diagn. Microbiol. Infect. Dis. 89, 164–167. doi: 10.1016/j.diagmicrobio.2017.06.022
Chen, Y. T., Peng, H. L., Shia, W. C., Hsu, F. R., Ken, C. F., Tsao, Y. M., et al. (2012). Whole-genome sequencing and identification of Morganella morganii KT pathogenicity-related genes. BMC Genomics 13(Suppl. 7):S4. doi: 10.1186/1471-2164-13-S7-S4
Cho, S. Y., Huh, H. J., Baek, J. Y., Chung, N. Y., Ryu, J. G., Ki, C. S., et al. (2015). Klebsiella pneumoniae co-producing NDM-5 and OXA-181 carbapenemases, South Korea. Emerg. Infect. Dis. 21, 1088–1089. doi: 10.3201/eid2106.150048
CLSI (2018). Performance Standards for Antimicrobial Susceptibility Testing, 28th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Dobiasova, H., and Dolejska, M. (2016). Prevalence and diversity of IncX plasmids carrying fluoroquinolone and beta-lactam resistance genes in Escherichia coli originating from diverse sources and geographical areas. J. Antimicrob. Chemother. 71, 2118–2124. doi: 10.1093/jac/dkw144
Espedido, B. A., Dimitrijovski, B., van Hal, S. J., and Jensen, S. O. (2015). The use of whole-genome sequencing for molecular epidemiology and antimicrobial surveillance: identifying the role of IncX3 plasmids and the spread of blaNDM-4-like genes in the Enterobacteriaceae. J. Clin. Pathol. 68, 835–838. doi: 10.1136/jclinpath-2015-203044
Feng, S., Shen, C., Chen, H., Zheng, X., Xia, Y., Zhong, L. L., et al. (2018). Co-production of MCR-1 and NDM-5 in Escherichia coli isolated from a colonization case of inpatient. Infect. Drug Resist. 11, 1157–1161. doi: 10.2147/IDR.S171164
Ghosh, S., Bal, A. M., Malik, I., and Collier, A. (2009). Fatal Morganella morganii bacteraemia in a diabetic patient with gas gangrene. J. Med. Microbiol. 58, 965–967. doi: 10.1099/jmm.0.008821-0
Giufre, M., Errico, G., Accogli, M., Monaco, M., Villa, L., Distasi, M. A., et al. (2018). Emergence of NDM-5-producing Escherichia coli sequence type 167 clone in Italy. Int. J. Antimicrob. Agents 52, 76–81. doi: 10.1016/j.ijantimicag.2018.02.020
He, T., Wang, Y., Sun, L., Pang, M., Zhang, L., and Wang, R. (2017). Occurrence and characterization of blaNDM-5-positive Klebsiella pneumoniae isolates from dairy cows in Jiangsu, China. J. Antimicrob. Chemother. 72, 90–94.
Hornsey, M., Phee, L., and Wareham, D. W. (2011). A novel variant, NDM-5, of the New Delhi metallo-beta-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob. Agents Chemother. 55, 5952–5954. doi: 10.1128/AAC.05108-11
Jamal, W. Y., Albert, M. J., Khodakhast, F., Poirel, L., and Rotimi, V. O. (2015). Emergence of new sequence type OXA-48 carbapenemase-producing Enterobacteriaceae in Kuwait. Microb. Drug Resist. 21, 329–334. doi: 10.1089/mdr.2014.0123
Khatri, I., Dureja, C., Raychaudhuri, S., and Subramanian, S. (2013). Draft genome sequence of the opportunistic human pathogen Morganella morganii SC01. Genome Announc. 1, e00051-12. doi: 10.1128/genomeA.00051-12
Krishnaraju, M., Kamatchi, C., Jha, A. K., Devasena, N., Vennila, R., Sumathi, G., et al. (2015). Complete sequencing of an IncX3 plasmid carrying blaNDM-5 allele reveals an early stage in the dissemination of the blaNDM gene. Indian J. Med. Microbiol. 33, 30–38. doi: 10.4103/0255-0857.148373
Lee, C. Y., Lee, H. F., Huang, F. L., and Chen, P. Y. (2009). Haemorrhagic bullae associated with a chicken scratch. Ann. Trop. Paediatr. 29, 309–311. doi: 10.1179/027249309X12547917869168
Li, X., Fu, Y., Shen, M., Huang, D., Du, X., Hu, Q., et al. (2018). Dissemination of blaNDM-5 gene via an IncX3-type plasmid among non-clonal Escherichia coli in China. Antimicrob. Resist. Infect. Control 7:59. doi: 10.1186/s13756-018-0349-6
Li, X., Jiang, Y., Wu, K., Zhou, Y., Liu, R., Cao, Y., et al. (2017). Whole-genome sequencing identification of a multidrug-resistant Salmonella enterica serovar Typhimurium strain carrying blaNDM-5 from Guangdong, China. Infect. Genet. Evol. 55, 195–198. doi: 10.1016/j.meegid.2017.09.005
Liu, H., Zhu, J., Hu, Q., and Rao, X. (2016). Morganella morganii, a non-negligent opportunistic pathogen. Int. J. Infect. Dis. 50, 10–17. doi: 10.1016/j.ijid.2016.07.006
Liu, L., Feng, Y., McNally, A., and Zong, Z. (2018). blaNDM-21, a new variant of blaNDM in an Escherichia coli clinical isolate carrying blaCTX-M-55 and rmtB. J. Antimicrob. Chemother. 73, 2336–2339. doi: 10.1093/jac/dky226
Liu, Y., Zhang, H., Zhang, X., Jiang, N., Zhang, Z., Zhang, J., et al. (2019). Characterization of an NDM-19-producing Klebsiella pneumoniae strain harboring 2 resistance plasmids from China. Diagn. Microbiol. Infect. Dis. 93, 355–361. doi: 10.1016/j.diagmicrobio.2018.11.007
Liu, Z., Li, J., Wang, X., Liu, D., Ke, Y., Wang, Y., et al. (2018). Novel variant of New Delhi metallo-beta-lactamase, NDM-20, in Escherichia coli. Front. Microbiol. 9:248. doi: 10.3389/fmicb.2018.00248
Liu, Z., Wang, Y., Walsh, T. R., Liu, D., She, Z., Zhang, R., et al. (2017). Plasmid-Mediated Novel blaNDM-17 gene encoding a carbapenemase with enhanced activity in a sequence type 48 Escherichia coli Strain. Antimicrob. Agents Chemother. 61, e2233–e2216.
Lü, Y., Liu, W., Liang, H., Zhao, S., Zhang, W., Liu, J., et al. (2018). NDM-1 encoded by a pNDM-HN380-like plasmid pNDM-BJ03 in clinical Enterobacter cloacae. Diagn. Microbiol. Infect. Dis. 90, 153–155. doi: 10.1016/j.diagmicrobio.2017.10.022
Lv, J., Qi, X., Zhang, D., Zheng, Z., Chen, Y., Guo, Y., et al. (2016). First report of complete sequence of a blaNDM-13-harboring plasmid from an Escherichia coli ST5138 clinical isolate. Front. Cell Infect. Microbiol. 6:130.
Mao, J., Liu, W., Wang, W., Sun, J., Lei, S., and Feng, Y. (2018). Antibiotic exposure elicits the emergence of colistin- and carbapenem-resistant Escherichia coli coharboring MCR-1 and NDM-5 in a patient. Virulence 9, 1001–1007. doi: 10.1080/21505594.2018.1486140
Nash, J. H., and Young, N. M. (2015). Draft whole-genome sequence of Morganella morganii serotype O:1ab. Genome Announc. 3:e00453-15. doi: 10.1128/genomeA.00453-15
Olaitan, A. O., Diene, S. M., Gupta, S. K., Adler, A., Assous, M. V., and Rolain, J. M. (2014). Genome analysis of NDM-1 producing Morganella morganii clinical isolate. Expert Rev. Anti Infect. Ther. 12, 1297–1305. doi: 10.1586/14787210.2014.944504
Pal, T., Ghazawi, A., Darwish, D., Villa, L., Carattoli, A., Hashmey, R., et al. (2017). Characterization of NDM-7 carbapenemase-producing Escherichia coli Isolates in the Arabian Peninsula. Microb. Drug Resist. 23, 871–878. doi: 10.1089/mdr.2016.0216
Paskova, V., Medvecky, M., Skalova, A., Chudejova, K., Bitar, I., Jakubu, V., et al. (2018). Characterization of NDM-Encoding plasmids from Enterobacteriaceae recovered from Czech Hospitals. Front. Microbiol. 9:1549. doi: 10.3389/fmicb.2018.01549
Rojas, L. J., Hujer, A. M., Rudin, S. D., Wright, M. S., Domitrovic, T. N., Marshall, S. H., et al. (2017). NDM-5 and OXA-181 beta-lactamases, a significant threat continues to spread in the Americas. Antimicrob. Agents Chemother. 61, e00454-17. doi: 10.1128/AAC.00454-17
Seija, V., Medina Presentado, J. C., Bado, I., Papa Ezdra, R., Batista, N., Gutierrez, C., et al. (2015). Sepsis caused by New Delhi metallo-beta-lactamase (blaNDM-1) and qnrD-producing Morganella morganii, treated successfully with fosfomycin and meropenem: case report and literature review. Int. J. Infect. Dis. 30, 20–26. doi: 10.1016/j.ijid.2014.09.010
Shi, D. S., Wang, W. P., Kuai, S. G., Shao, H. F., and Huang, M. (2011). Identification of bla KPC-2 on different plasmids of three Morganella morganii isolates. Eur. J. Clin. Microbiol. Infect. Dis. 31, 797–803. doi: 10.1007/s10096-011-1377-9
Soliman, A. M., Khalifa, H. O., Ahmed, A. M., Shimamoto, T., and Shimamoto, T. (2016). Emergence of an NDM-5-producing clinical Escherichia coli isolate in Egypt. Int. J. Infect. Dis. 48, 46–48. doi: 10.1016/j.ijid.2016.05.003
Sugawara, Y., Akeda, Y., Sakamoto, N., Takeuchi, D., Motooka, D., Nakamura, S., et al. (2017). Genetic characterization of blaNDM-harboring plasmids in carbapenem-resistant Escherichia coli from Myanmar. PLoS One 12:e0184720. doi: 10.1371/journal.pone.0184720
Sun, L., Xu, J., and He, F. (2018). Draft genome sequence of an NDM-5, CTX-M-15 and OXA-1 co-producing Escherichia coli ST167 clinical strain isolated from a urine sample. J. Glob. Antimicrob. Resist. 14, 284–286. doi: 10.1016/j.jgar.2018.08.005
Treangen, T. J., Ondov, B. D., Koren, S., and Phillippy, A. M. (2014). The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15:524. doi: 10.1186/preaccept-2573980311437212
Tsakris, A., Ikonomidis, A., Spanakis, N., Poulou, A., and Pournaras, S. (2007). Characterization of In3Mor, a new integron carrying VIM-1 metallo-beta-lactamase and sat1 gene, from Morganella morganii. J. Antimicrob. Chemother. 59, 739–741. doi: 10.1093/jac/dkm020
Wailan, A. M., Paterson, D. L., Caffery, M., Sowden, D., and Sidjabat, H. E. (2015). Draft genome sequence of NDM-5-producing Escherichia coli sequence type 648 and genetic context of blaNDM-5 in Australia. Genome Announc. 3:e00194-15. doi: 10.1128/genomeA.00194-15
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Xie, M., Li, R., Liu, Z., Chan, E. W. C., and Chen, S. (2018). Recombination of plasmids in a carbapenem-resistant NDM-5-producing clinical Escherichia coli isolate. J. Antimicrob. Chemother. 73, 1230–1234. doi: 10.1093/jac/dkx540
Yoon, E. J., Kang, D. Y., Yang, J. W., Kim, D., Lee, H., Lee, K. J., et al. (2018). New Delhi metallo-beta-lactamase-producing Enterobacteriaceae in South Korea between 2010 and 2015. Front. Microbiol. 9:571. doi: 10.3389/fmicb.2018.00571
Zaid, U. B., Bagga, H. S., Reese, A. C., and Breyer, B. N. (2013). Intratesticular abscess in a solitary testicle: the case for testicle sparing management. Case Rep. Med. 2013:184064. doi: 10.1155/2013/184064
Zhang, F., Xie, L., Wang, X., Han, L., Guo, X., Ni, Y., et al. (2016). Further Spread of bla NDM-5 in Enterobacteriaceae via IncX3 plasmids in Shanghai, China. Front. Microbiol. 7:424. doi: 10.3389/fmicb.2016.00424
Zhang, L. P., Xue, W. C., and Meng, D. Y. (2016). First report of New Delhi metallo-beta-lactamase 5 (NDM-5)-producing Escherichia coli from blood cultures of three leukemia patients. Int. J. Infect. Dis. 42, 45–46. doi: 10.1016/j.ijid.2015.10.006
Zheng, B., Yu, X., Xu, H., Guo, L., Zhang, J., Huang, C., et al. (2017). Complete genome sequencing and genomic characterization of two Escherichia coli strains co-producing MCR-1 and NDM-1 from bloodstream infection. Sci. Rep. 7:17885. doi: 10.1038/s41598-017-18273-2
Zheng, B., Zhang, J., Ji, J., Fang, Y., Shen, P., Ying, C., et al. (2015). Emergence of Raoultella ornithinolytica coproducing IMP-4 and KPC-2 carbapenemases in China. Antimicrob. Agents Chemother. 59, 7086–7089. doi: 10.1128/AAC.01363-15
Zhu, B., Ying, C., Xu, H., and Ying, J. (2018). Coexistence of NDM-1-producing Escherichia coli and Citrobacter freundii in the same patient. J. Glob. Antimicrob. Resist. 15, 79–81. doi: 10.1016/j.jgar.2018.04.013
Keywords: blaNDM–5, IncX3, Morganella morganii, complete genome sequence, comparative genomic analysis
Citation: Guo X, Rao Y, Guo L, Xu H, Lv T, Yu X, Chen Y, Liu N, Han H and Zheng B (2019) Detection and Genomic Characterization of a Morganella morganii Isolate From China That Produces NDM-5. Front. Microbiol. 10:1156. doi: 10.3389/fmicb.2019.01156
Received: 21 February 2019; Accepted: 07 May 2019;
Published: 28 May 2019.
Edited by:
Fabian Cieplik, University Medical Center Regensburg, GermanyReviewed by:
Roberto Gustavo Melano, Public Health Ontario, CanadaAgnes Sonnevend, United Arab Emirates University, United Arab Emirates
Niels Pfennigwerth, Ruhr University Bochum, Germany
Copyright © 2019 Guo, Rao, Guo, Xu, Lv, Yu, Chen, Liu, Han and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaobing Guo, gxbing928@zzu.edu.cn; Beiwen Zheng, zhengbw@zju.edu.cn
†These authors have contributed equally to this work
 Xiaobing Guo1*†
Xiaobing Guo1*† Yuting Rao
Yuting Rao Lihua Guo
Lihua Guo Hao Xu
Hao Xu Na Liu
Na Liu Beiwen Zheng
Beiwen Zheng