- Institute for Biological Sciences, University of Rostock, Rostock, Germany
Bacillus subtilis releases a broad range of volatile secondary metabolites, which are considered as long- and short distance infochemical signals mediating inter- and intra-specific processes. In addition, they often show antimicrobial or antifungal activities. This review attempts to summarize yet known volatile secondary metabolites produced and emitted by Bacillus subtilis isolates focusing on the structural diversity and distribution patterns. Using in vitro volatile-collection systems, 26 strains of B. subtilis isolated from different habitats were found to produce in total 231 volatile secondary metabolites. These volatile secondary metabolites comprised mainly hydrocarbons, ketones, alcohols, aldehydes, ester, acids, aromatics, sulfur- and nitrogen-containing compounds. Reviewed data revealed to a great extent isolate-specific emission patterns. The production and release of several volatile bioactive compounds was retained in isolates of the species B. subtilis, while volatiles without a described function seemed to be isolate-specifically produced. Detailed analysis, however, also indicated that the original data were strongly influenced by insufficient descriptions of the bacterial isolates, heterogeneous and poorly documented culture conditions as well as sampling techniques and inadequate compound identification. In order to get deeper insight into the nature, diversity, and ecological function of volatile secondary metabolites produced by B. subtilis, it will be necessary to follow well-documented workflows and fulfill state-of-the-art standards to unambiguously identify the volatile metabolites. Future research should consider the dynamic of a bacterial culture leading to differences in cell morphology and cell development. Single cell investigations could help to attribute certain volatile metabolites to defined cell forms and developmental stages.
Introduction
Bacillus subtilis is a rod-shaped, aerobic, endospore forming Gram-positive bacterium that colonizes the soil and often occurs plant-associated. Especially the rhizosphere was found to be a frequent natural habitat (Kunst et al., 1997). This habitat is also highly populated with other bacteria, fungi, and proto- and metazoa. In order to cope with this competitive environment, B. subtilis is able to release a large number of metabolites, which can modify the performance of co-habitants from the neighborhood (Romero et al., 2007; Sansinenea and Ortiz, 2011; Harwood et al., 2018). Many of these metabolites are classified as secondary metabolites, since there is no essential need for them in growth, development, or reproduction of the organisms. Nevertheless, their absence can ultimately cause damage or alter the organism or population, since secondary metabolites function as communication signals, antibiotics or siderophores (Vining, 1990; Davies, 2006; Yim et al., 2007; Ratcliff and Denison, 2011; Sansinenea and Ortiz, 2011). Due to their physico-chemical properties, secondary metabolites are grouped into non-volatile and volatile compounds. Non-volatile secondary metabolites of B. subtilis comprises lipopeptides (classes of surfactin, iturin, and fengycin), polyketides and non-ribosomal peptides. These compounds were excellently described in the past (Kakinuma et al., 1969; Peypoux et al., 1981; Hofemeister et al., 2004; Stein, 2005; Schneider et al., 2007; Sansinenea and Ortiz, 2011; Harwood et al., 2018; Caulier et al., 2019). Although no strain is able to produce a complete spectrum of non-volatile secondary metabolites, their release is found throughout the majority of investigated B. subtilis isolates (Stein, 2005).
Volatility improves the efficacy of a large number of secondary metabolites. High vapor pressure enables compounds with low molecular weight to act in the close vicinity of the producing organism but also to travel over longer distances even in soil (Rasmann et al., 2005; Effmert et al., 2012; Schulz-Bohm et al., 2018). Bacteria in general release a high diversity of volatile secondary metabolites including hydrocarbons, ketones, alcohols, sulfur- and nitrogen containing compounds, terpenes, and others (Schulz and Dickschat, 2007; Lemfack et al., 2014, 2018). These metabolites are primarily considered as long- and short distance infochemicals mediating inter- and intra-specific interactions, but they also act as antimicrobial or antifungal agents (Kai et al., 2009; Schmidt et al., 2017; Schulz-Bohm et al., 2017). Actinomycetes, for instance, produce the earthy-smelling terpene geosmin (Gerber and Lechevalier, 1965), which can be used by Drosophila flies to detect toxic bacteria (Stensmyr et al., 2012). Another well-known bacterial volatile is the multifunctional dimethyl disulfide (DMDS) produced by a wide range of bacteria including Bacillus spp., Burkholderia spp., Pseudomonas spp., Serratia spp. and Streptomyces spp. (Lemfack et al., 2018). While in insects DMDS acted insecticidal by hindering the electron transport via inhibition of cytochrome oxidase and blockage of potassium channels (Dugravot et al., 2003; Gautier et al., 2008), plants can incorporate DMDS into plant proteins leading to plant growth promotion (Meldau et al., 2013). In high doses, however, DMDS inhibited plant growth by a mechanism that is not yet known (Kai et al., 2010).
Fiddaman and Rossall (1993) reported that a B. subtilis strain releases volatiles with antifungal properties. Ten years later, Ryu et al. (2003, 2004) found out that volatile metabolites emitted by the Bacillus subtilis isolate GB03 significantly promoted plant growth and induced systemic resistance in Arabidopsis thaliana. Although the isolate GB03 was later newly classified as Bacillus amyloliquefaciens GB03 (Choi et al., 2014), these impressive results encouraged research groups to consider B. subtilis as a producer of bioactive volatiles.
The latest review on secondary metabolites of B. subtilis was published by Caulier et al. (2019). They included non-volatile and volatile compounds, but mainly focused on substances with antimicrobial properties. Yet, there is no comprehensive overview of only volatile secondary metabolites of B. subtilis regardless of their specificity or general properties and biological and ecological role. Therefore, this review attempts to summarize known volatile secondary metabolites of known B. subtilis isolates focusing on structural diversity and distribution in the various isolates.
Volatile Secondary Metabolites Emitted by Bacillus Subtilis
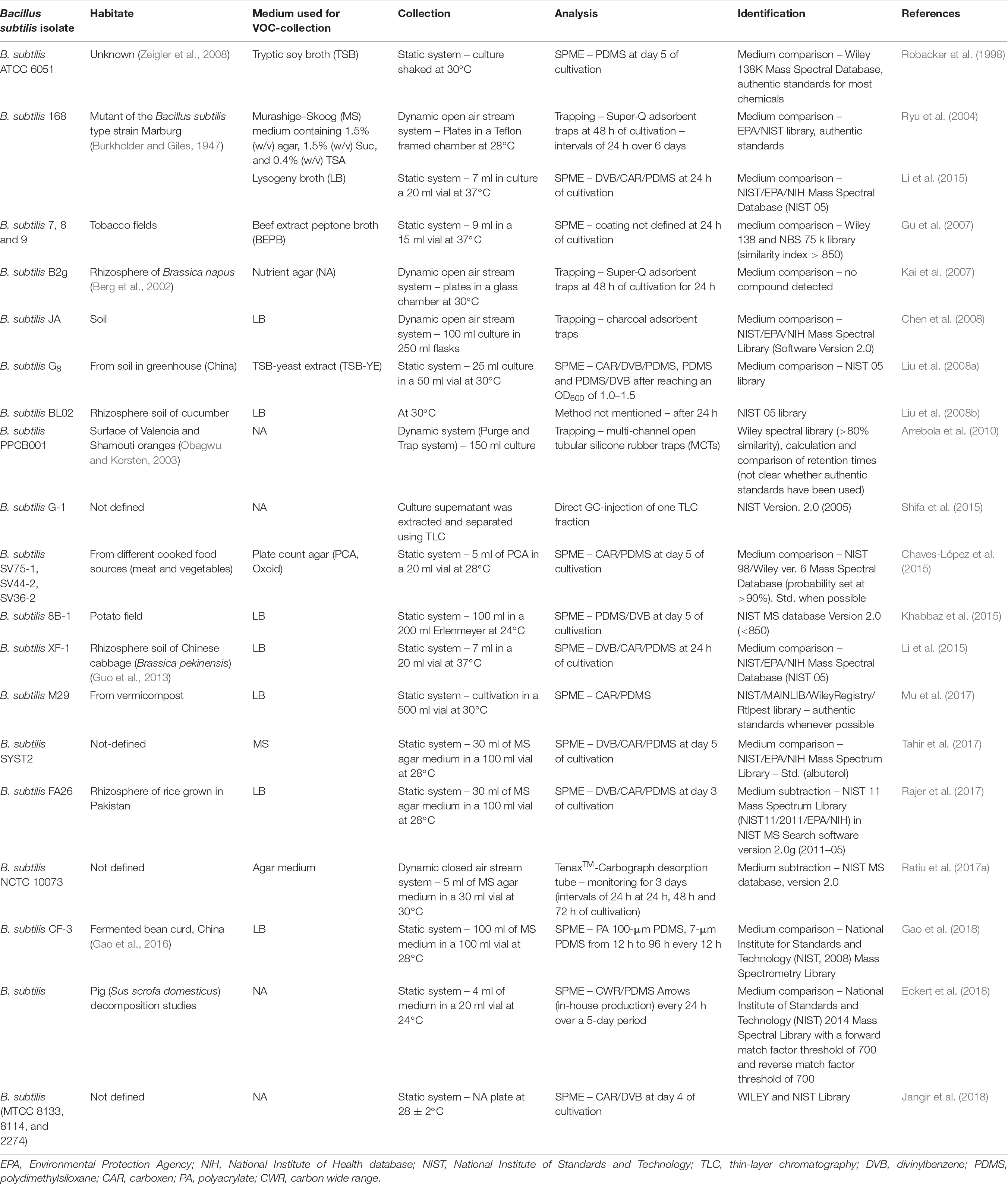
To get a comprehensive insight into the vast amount of volatile secondary metabolites emitted by B. subtilis I searched for documented B. subtilis isolates in the literature and in the database mVOC (Lemfack et al., 2018). Studies of single volatiles (e.g., acetoin and isoprene) were not considered in this review. The search revealed 26 B. subtilis strains and their corresponding volatile-profiles (Table 1) investigated in twenty studies. This included mainly isolates from different soil habitats (tobacco fields, rhizosphere of rice, cucumber or cabbage, greenhouse soil) and isolates from food sources (bean curd, oranges, and cooked food). For some isolates, the origin was not described or not known. Although several hundred wild-type strains of B. subtilis have been analyzed for non-volatile metabolites (Stein, 2005), the present review will give a first overview of volatile secondary metabolites.
Methods Used for Collection, Analysis, and Compound Identification
All volatile secondary metabolites have been collected in in vitro systems from cultivated B. subtilis strains. Noteworthy is the diversity in culture conditions of bacteria, and collection techniques for volatile metabolites. These parameters should be necessarily documented. The evaluation and interpretation of potential production and emission of bacterial metabolites in nature should always be done in close consideration of culture conditions (nutrients, temperature, oxygen availability), since they determine bacterial growth and development, which of course could influence the production of volatile secondary metabolites as it was already shown for other bacterial species (Kai et al., 2009; Blom et al., 2011; Weise et al., 2012; Ratiu et al., 2017b). Furthermore, methods to collect volatile secondary metabolites can influence the classes of volatiles that are detected. Although proper descriptions of cultivation and analysis condition are sometimes missing, the following chapters will summarize the relevant parameters.
1 – Culture Conditions During Collection of Volatile Secondary Metabolites
Bacillus subtilis isolates have mostly been cultivated on complex medium for volatile collection, including lysogeny broth (LB, often mistakenly referred to as Luria Bertani) used in seven studies and nutrient agar (NA) used in five studies (Table 1). One collection was performed using beef extract peptone broth (BEPB) and another using plate count agar (PCA, Oxoid). Murashige–Skoog medium (MS) was applied in two studies, whereby in one of these studies sucrose and tryptic soya agar (TSA) were added. Finally, in two investigations tryptic soy broth (TSB) was used. In one of these studies the authors added yeast extract (Liu et al., 2008a).
In 14 out of 19 studies, B. subtilis was cultivated in liquid medium during collection of volatile secondary metabolites, while in the remaining five investigations, B. subtilis grew on agar plates. For other bacterial species, crucial differences between cultivation on solid or in liquid medium were already demonstrated (Kai et al., 2007, 2010) most likely due to different cell numbers and varying diffusion of oxygen into the medium (Somerville and Proctor, 2013). Also, the cultivation temperature varied in the experiments in the range of 24°C and 37°C. Furthermore, collections of volatile metabolites have been performed at different time points ranging from 24 h up to 6 days of cultivation. These differences in the time of sampling play also an important role, since they reflect differences in cell numbers and developmental stages of bacterial cultures (Kai et al., 2010).
2 – Collection of Volatile Secondary Metabolites
The collection of volatiles was almost exclusively performed using static or dynamic headspace systems combined with gas chromatography/electron-ionization-mass spectrometry (GC/EI-MS) (Table 1). Only in one case the bacterial filtrate was extracted and separated applying thin-layer chromatography (TLC). TLC fractions were directly injected into the GC/EI-MS system (Shifa et al., 2015). Since this procedure does not ensure that only bacterial volatiles are captured, static and dynamic headspace systems are the preferred experimental setups (Schulz and Dickschat, 2007).
Static headspace systems
Due to their simple application, static systems are the most favored experimental systems for volatile collection. They were used in 13 out of 19 studies. Liquid medium was inoculated with the respective B. subtilis isolate and filled into a cultivation vessel. This vessel was often sealed with Parafilm® to avoid loss of volatiles during incubation. To extract volatile secondary metabolites from the headspace of the culture, solid phase micro extraction (SPME) was performed. A SPME fiber coated with different adsorbent materials was inserted into the headspace at defined time points of incubation. Coating materials included divinylbenzene (DVB), polydimethylsiloxane (PDMS), carboxen (CAR), polyacrylate (PA), and carbon wide range (CWR). These coatings have been used solely or in combination, e.g., the combination of CAR/DVB/PDMS (Table 1). PDMS extracts non-polar volatiles, while polyacrylate and PDMS/DVB are used to adsorb polar volatile compounds. Captured volatile secondary metabolites were thermally desorbed into the injector of the GC/EI-MS.
Dynamic headspace systems
The sampling of volatile secondary metabolites by streaming air over a bacterial culture changes the collection system towards a dynamic headspace system. The air was purified from pollutants and bacterial contaminations by charcoal and sterile filters, respectively. The analyzed B. subtilis culture grew on a Petri dish, which was placed in an analysis chamber (Ryu et al., 2004; Kai et al., 2007) or it grew in liquid medium in flasks (Arrebola et al., 2010; Ratiu et al., 2017a). After passing the bacterial culture, volatile secondary metabolites were trapped by an adsorbent material (SuperQ®, silicone, Tenax® and others) and thereby enriched over a certain time interval. The volatiles were solvent-based or thermally desorbed and analyzed by GC/EI-MS (Table 1).
Identification of volatile secondary metabolites
In all presented investigations, volatile secondary metabolites have been analyzed using GC/EI-MS. In order to verify the emission by B. subtilis isolates, the profiles of volatiles metabolites were compared with controls, where only medium was analyzed. Unfortunately, this important control was not always properly described in several studies. The identification of volatiles mainly based on comparison of mass spectra of the analyzed compounds with corresponding mass spectra of different mass spectral libraries, for example, various versions of the Wiley library and the National Institute of Standards and Technology (NIST) library (Table 1). Among the studies considered in this review, only Robacker et al. (1998) and Ryu et al. (2004) used authentic standards and retention indices for compound identification. Other research groups only occasionally identified single volatile compounds using authentic standards, while the remaining compounds were identified only by comparison with mass spectral databases (Tahir et al., 2017). In some cases it is not recognizable if volatiles were identified using authentic standards and/or libraries (Chaves-López et al., 2015). A clear assignment of an authentic standard confirmation was unfortunately missing. Most of the authors identified merely on the basis of database suggestions using different probabilities (from 80 to 90%, Table 1). Nevertheless, for the sake of completeness, all volatile secondary metabolites that were described to be produced by B. subtilis were included in this review.
Chemical Classification of Volatile Secondary Metabolites Emitted by B. subtilis Isolates
The individual B. subtilis isolates emitted from 5 up to 30 compounds with an average of 14 compounds per strain (Figure 1). Surprisingly, no volatile was detected in the headspace of isolate B2g (Kai et al., 2007). One study delivered only summarized information on the volatile emission of three strains, any information on individual emission was missing (Jangir et al., 2018).
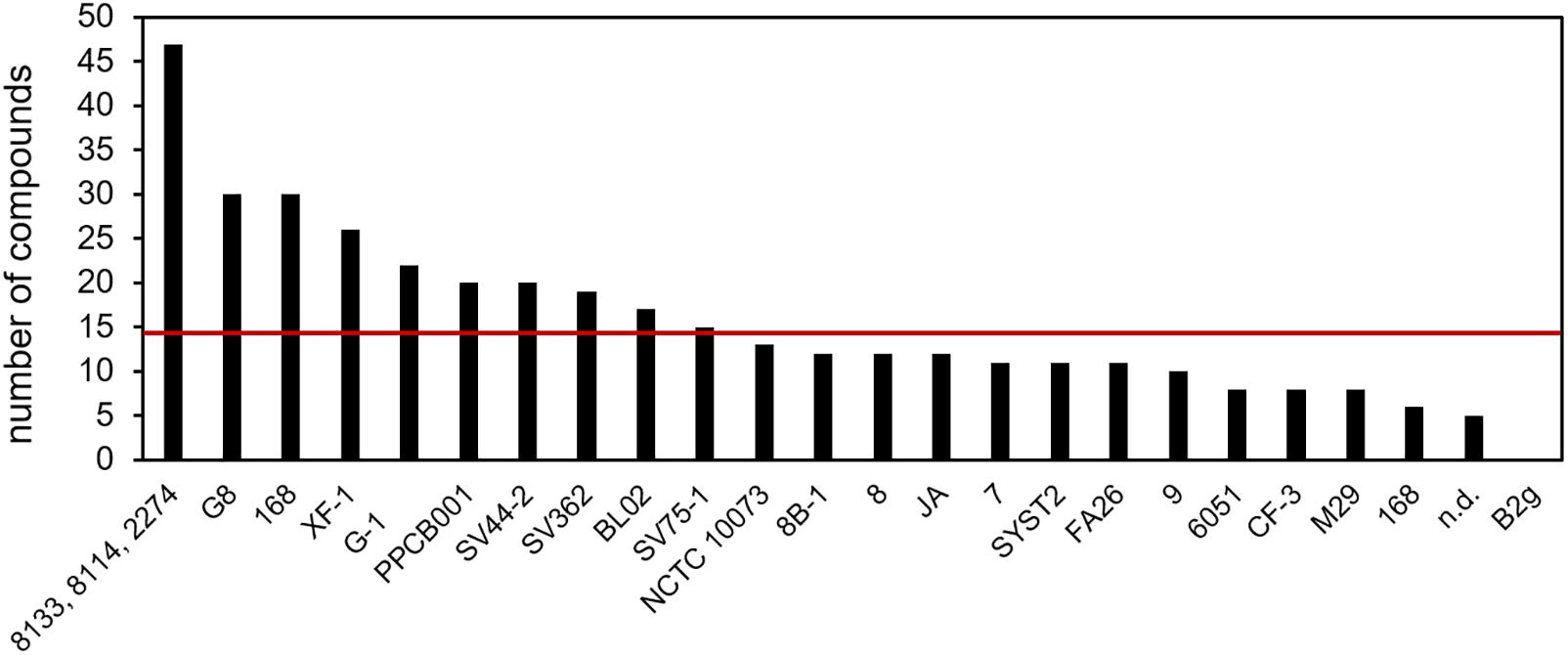
Figure 1. Detected compounds per B. subtilis isolate. Volatile secondary metabolites emitted by 26 B. subtilis isolates were summarized from 20 original scientific publication (see Table 1). n.d., isolate was not defined; red line is the arithmetic average of 14; B. subtilis 168 was analyzed twice; the results of 8133, 8114, and 2274 are aggregated in the original study.
In total, 231 different volatile secondary metabolites were found in 26 B. subtilis isolates analyzed so far (Table 2). Volatile secondary metabolites of strains, which were found in the literature and yet have not been implemented in mVOC-database (Lemfack et al., 2018), have been submitted by me to the respective database in the meantime. Recently, bacterial volatile secondary metabolites were classified according to their biosynthetic origin (Schulz and Dickschat, 2007; Caulier et al., 2019). This classification was not used in this review, since the biosynthetic pathway has not been elucidated for many individual compounds. A chemical classification seemed to be more reasonable under these circumstances. The majority of detected volatile secondary metabolites were classified as ketones (34 compounds, 15%), nitrogen-containing compounds (32 compounds, 14%), hydrocarbons (33 compounds, 14%), aromatic compounds (32 compounds, 14%), and alcohols (26 compounds, 11%) (Figure 2). Furthermore, volatile secondary metabolites included aldehydes (15 compounds, 6.5%), acids (16 compounds, 7%), and esters (15 compounds, 6.5%). To a lower extent sulfur containing compounds (8 compounds, 3.5%), silicone containing compounds (7 compounds, 3%), ethers (4 compounds, 2%), halogenated compounds (2%), naphthalenes (4 compounds, 2%), pyranones (<1%), and others (5 compounds, 2%) could be detected (Figure 2). Attention has to be paid to volatiles, which are well known artifacts, e.g., silicone containing compounds (octamethyl-cyclotetrasiloxane, dodecamethyl-2-cyclohexasiloxane), 2-ethylhexanol and phthalates; however, for the sake of completeness they have also been listed (see future aspects).
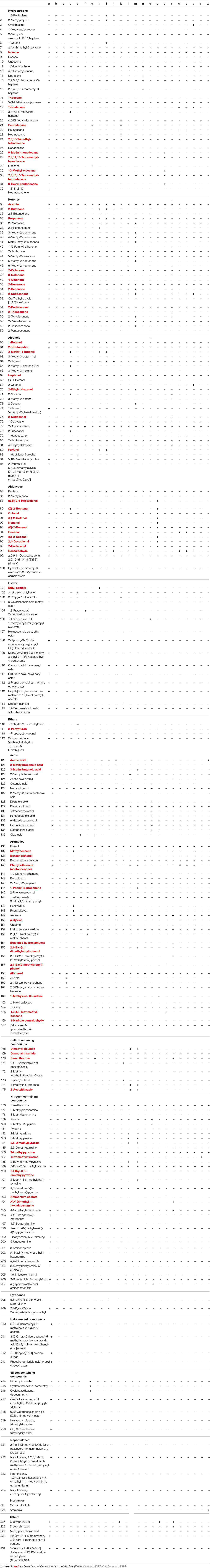
Table 2. Volatile secondary metabolites emitted by B. subtilis isolates a: 8133, 8114, 2274; b: n.d.; c: CF3; d: NCTC 10073; e: M29; f: SYST2; g: FA26; h: SV75-1; i: SV44-2; j: SV36-2; k: G-1; l: XF-1; m: 168; n: 8B-1; o: PPCB001; p: BL02; q: JA, r: G8; s: 7; t: 8; u: 9; v: 168; w: 6051.
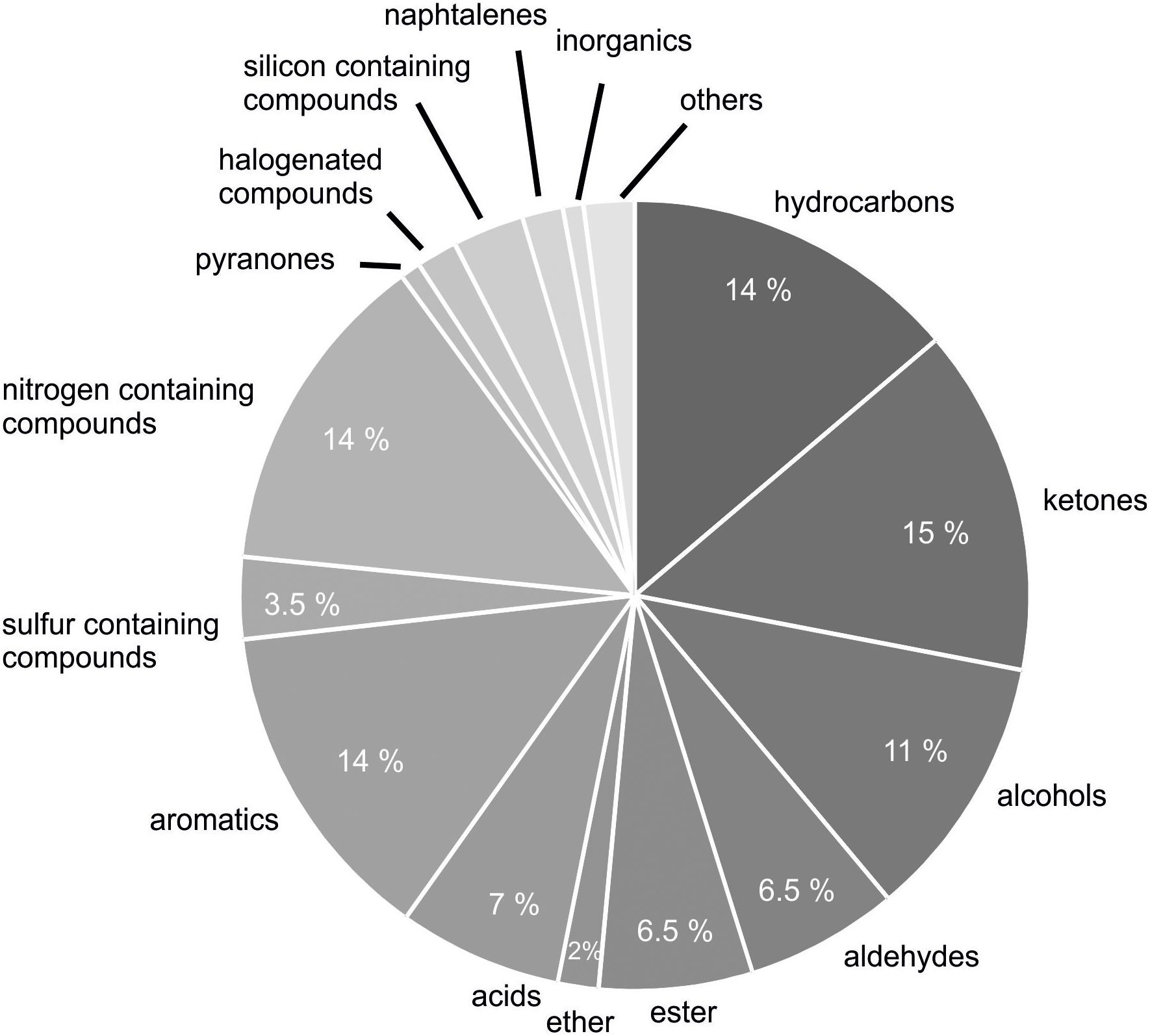
Figure 2. Classification of detected VOCs according to chemical classes. Two hundred and thirty-one volatile secondary metabolites emitted from 26 B. subtilis isolates were summarized from 20 original scientific publication (see Table 1) and grouped in chemical classes.
Distribution of Volatile Secondary Metabolites Throughout the Species B. subtilis
In order to clarify the question of how volatile secondary metabolites are distributed within the investigated strains of B. subtilis, all compounds were sorted according to the number of the strains in which they were detected. Surprisingly, 156 compounds (67% of the total number of volatiles) were detected only in one single isolate, while the production of 44 compounds (19%) was observed in two isolates. Sixteen compounds (7%) were found in three isolates, six (3%) were present in four isolates, and five (2%) were detected in five isolates. Two volatiles were present in six isolates and interestingly one single compound (acetophenone) was present in seven strains and another single compound (benzaldehyde) was even present in 10 strains (Figure 3A and Table 3).
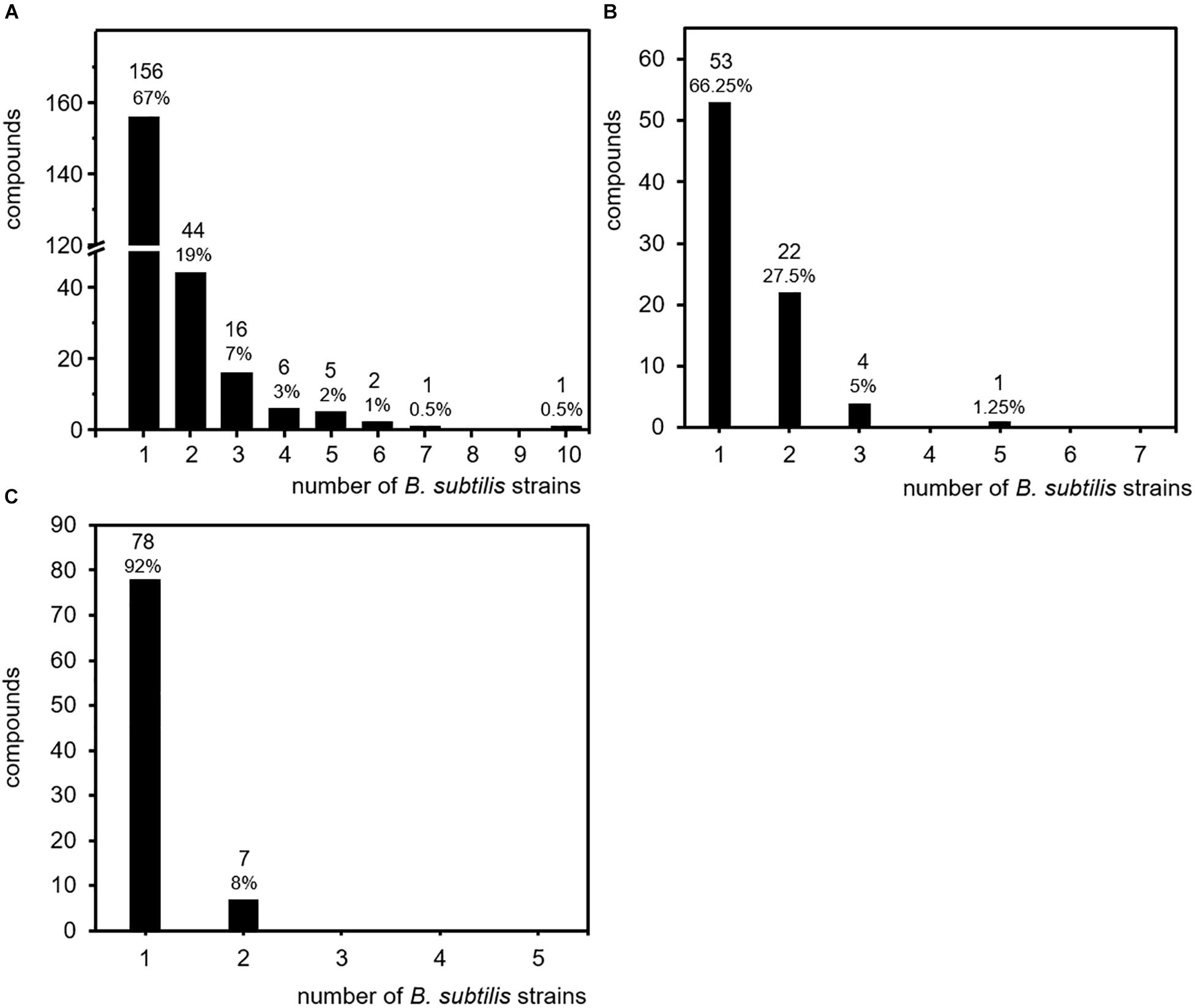
Figure 3. Specificity of volatile secondary metabolites. Volatile secondary metabolites emitted by 26 B. subtilis isolates were summarized from 20 original scientific publication (see Table 1). Compounds were related to the number of isolates responsible for emission. (A) Production frequency of all 231 compounds (entire data set of the survey). (B) Production frequency of compound emitted upon cultivation on lysogeny broth (LB). (C) Production frequency of compounds emitted upon cultivation on nutrient broth (NB).
These results revealed that 86% of the total amount of volatile secondary metabolites were emitted only by one or two out of 26 individual isolates of the species B. subtilis. These strain specific emission patterns were not expected, but may partially be explained by the diversity of culture conditions and analyses techniques. As described for other bacteria, the production and emission of secondary metabolites is influenced by nutrient supply and physico-chemical properties of the cultivation such as temperature, pH, and oxygen availability (Kai et al., 2009; Blom et al., 2011; Weise et al., 2012). Currently, there is not much data available on the nutrient dependent volatile emission of B. subtilis. Most investigations that are summarized here have been performed on nutrient rich media. When cultivated on LB, seven B. subtilis isolates released in total 80 compounds. Thereby, 53 compounds (66%) and 22 compounds (27.5%) were detected in the headspace of one or two individual B. subtilis isolates, respectively (Figure 3B). When cultivated on NA, five B. subtilis isolates released in total 85 compounds, from which 78 compounds (92%) and 7 compounds (8%) were detected in the headspace of one or two individual B. subtilis isolates, respectively (Figure 3C). These results resemble the overall impression of the studies. The influence of nutrient supply did not seem to be a decisive factor of the strain-specificity observed in B. subtilis. Although other parameters like the temperature are not yet included, these results already indicate an isolate-specific volatile emission in B. subtilis.
Distribution of Biological-Active Volatile Secondary Metabolites
In contrast to the isolate-specific emissions described above, the volatile metabolites benzaldehyde and acetophenone were found in several strains of B. subtilis. The preserved production of certain single metabolites by many strains may indicate that these compounds could provide a general advantage for B. subtilis in nature and it can be hypothesized that several isolates of the species maintained producing these compounds because they are bioactive. Some biological effects of volatile secondary metabolites emitted by B. subtilis have been already described, e.g., they can influence the growth of plants, show antimicrobial activity and affect the behavior of insects (Robacker et al., 1998; Kai et al., 2009; Caulier et al., 2019). Out of the 231 B. subtilis compounds listed here, 69 were recently described as bioactive (Table 2)
(Robacker et al., 1998; Piechulla et al., 2017; Caulier et al., 2019). In this list of bioactive compounds appears indeed benzaldehyde. This compound, which is known to inhibit the germination of spores and the mycelial growth of some fungi (Zou et al., 2007), was emitted by ten B. subtilis isolates. Also acetophenone, the single compound found in seven isolates, was described as antifungal substance and was able to directly promote plant growth (Groenhagen et al., 2013). Trimethylpyrazine and 2-undecanone, the two compounds emitted by six B. subtilis isolates, were described as bioactive metabolites and also all five volatiles present in the headspace of five different isolates revealed bioactivity. Of the six compounds emitted from four isolates and out of 16 compounds emitted from three isolates, four compounds (67%) and 11 compounds (69%) have bioactive properties, respectively. In contrast, only 33 compounds (21%) out of 156 compounds, which were released only by one of the isolates, have known biological activity (Figure 4). The result that various proven volatile bioactive compounds were produced and released in several isolates of the species B. subtilis supports the interesting conclusion that B. subtilis might have retained bioactive volatiles, which play a general role in communication or defense. In contrast, volatiles without a demonstrated bioactivity seemed to be strain-specifically produced. However, it has to be considered that a missing report of bioactivity does not mean that there is no bioactivity, since not all targets/activities may be known. To take this into account it can be additionally hypothesized that strain-specific production of a bioactive volatile might be an effect of special acclimation of B. subtilis isolates to their original habitat and thereby the volatiles might have a more specific function. Further investigations are requested in order to confirm these hypotheses. They may support the assumption that due to their biological activity, the release of specific bioactive volatile secondary metabolites is advantageous for B. subtilis in nature.
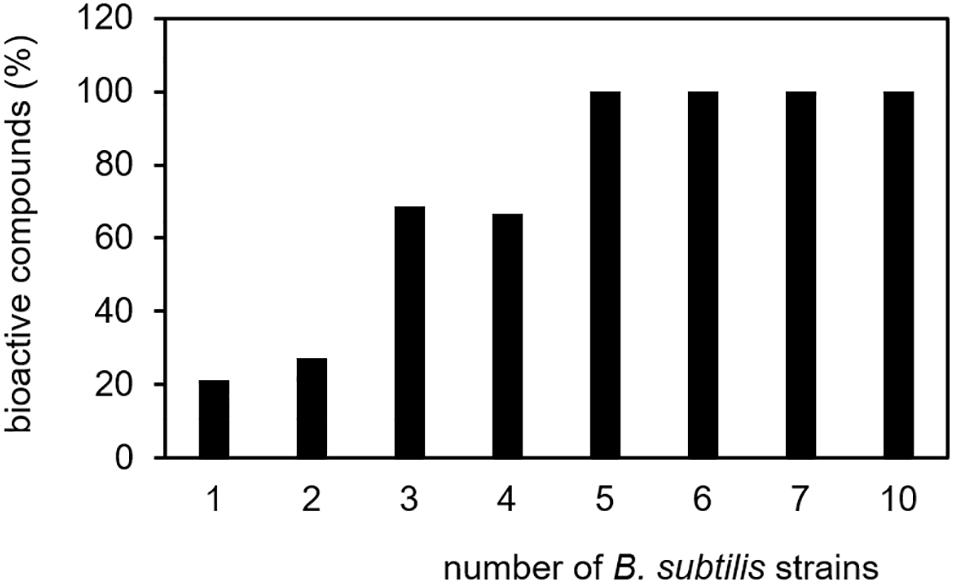
Figure 4. Specificity of biological-active volatile secondary metabolites. Volatile secondary metabolites emitted by 26 B. subtilis isolates were summarized from 20 original scientific publication (see Table 1). Biological-active compounds were related to the total number of volatiles and the number of emitting B. subtilis isolates.
Future Aspects in Volatile Secondary Metabolite Research
Specific Emission of Volatile Secondary Metabolites: A Consequence of Acclimation or Multicellularity?
In contrast to non-volatile secondary metabolites, which are produced throughout the species B. subtilis (Stein, 2005), this review suggests that the emission of the majority of volatiles secondary metabolites occurs isolate-specific. This specificity might be an effect of special acclimation of the producing B. subtilis isolates to their (original) habitat. As the investigated strains were isolated from different sources/habitats (soil, rhizospheres, and fermented food) these isolates faced other organism from which they might have horizontally acquired genes for synthesis of volatile secondary metabolites, since the plant–soil interface is considered as hot spot of horizontal gene transfer (Heuer and Smalla, 2012; Berg et al., 2014). The transferred genes in turn might fulfill a specific biological/ecological function and thereby provide an advantage for the producing B. subtilis isolate to survive in the respective habitat.
Isolate-exclusive emission of volatile secondary metabolites could also be explained by multicellularity of B. subtilis. B. subtilis exhibits a variety of multicellular behaviors including different kinds of motility (swarming, swimming, etc.), sporulation, the formation of biofilms (Kalamara et al., 2018) or oligotrophic growth stage (Gray et al., 2019). This lifestyle is of advantage for B. subtilis due to protection from predation, increased nutrient acquisition, and enhanced resistance (Lyons and Kolter, 2015; Kalamara et al., 2018). Bacterial cells in biofilms and swarms show distinct morphologies and specific patterns and levels of gene expression and metabolic contents (Lyons and Kolter, 2015). Single cell approaches, for instance, demonstrated that genetically identical B. subtilis cells divide into subpopulations releasing distinct metabolic products (Rosenthal et al., 2018). The studies evaluated for this review did not contain information about heterogeneity or multicellular “stages” of the investigated strains, however, it cannot be excluded that also the heterogeneity of B. subtilis cultures influences the strain-specific volatile production in B. subtilis. In order to address this question in future research, the monitoring and analysis of volatile secondary metabolites should be related to specific stages of growth and development of B. subtilis cells. Single cell investigations should necessarily be performed and it is particularly interesting that especially the nutrients supply is one of the most important factors that might influence cell morphology and development (Hageman et al., 1984; Fall et al., 2006; Gallegos-Monterrosa et al., 2016).
Which Volatile Secondary Metabolites Are Really Biosynthesized by B. subtilis?
The above-summarized volatiles (Table 2) have been collected from B. subtilis cultures. This includes bacterial cells but also the surrounding medium (solid or liquid, ingredients including peptides, sugars, organic acids, and amino acids). Compounds that originate from the medium should be excluded by proper controls. The remaining volatile compounds seem to be genuinely synthetized by the Bacillus cell. But it should also be considered that modification of medium-ingredients by extracellular enzymes produced by the Bacillus cell may account for newly appearing volatiles or that two individual volatiles may be reactants and synthesize ex vivo a new volatile (Kai et al., 2018). It would be very interesting to evaluate whether B. subtilis is able to volatilize nutrient substances in their surroundings by excretion of enzymes. In order to prove a genuine biosynthesis of volatile secondary metabolites by Bacillus cells, feeding experiments with selected isotopic labeled (13C, 15N) nutrient compounds (glucose, fatty acids, amino acids, or organic acids) should be performed. Isotopic labeling of certain appropriate nutrient compounds may also help to pin down volatile secondary metabolites to specific biosynthetic pathways. The biosynthesis of several microbial secondary metabolites and possible precursor substrates were already excellently described for other bacterial species (Schulz and Dickschat, 2007; Korpi et al., 2009; Scott and Piel, 2019). Nevertheless, the gap in knowledge about the biosynthesis of many volatiles in B. subtilis could be filled by using isotopic labeling of substrates. Knockout and overexpression mutants regarding specific steps in proposed pathways furthermore verify these results (Rijnen et al., 2003).
Labeling of nutrients will also help to exclude possible contaminants, which are also trapped and enriched during the sampling process. This applies to, e.g., silicone containing compounds and plasticizers. These compounds were found in some of the recent studies (Tahir et al., 2017; Jangir et al., 2018). They are highly artificial and their bacterial origin is very unlikely. In case of plasticizers, Dickschat already noted that it is essential to critically evaluate the source of each volatile (Dickschat, 2017). They already excluded phthalates in one of their earlier compilations (Schulz and Dickschat, 2007). Similarly, siloxanated compounds ought to be removed from the results. The presence of these compounds results mainly from TenaxTM-Carbotrap desorption (Ratiu et al., 2017a). Another prominent example is 2-ethylhexanol, which was present in the headspace of three B. subtilis isolates (168, JA and G8) (Chen et al., 2008; Liu et al., 2008a; Li et al., 2015). This compound is also considered as an artificial contaminant, although it was shown that microbial degradation of plasticizers represents also a likely source (Nalli et al., 2006).
Emission of Volatile Secondary Metabolites From B. subtilis in Nature
The results summarized above were all obtained using in vitro test systems. In order to understand the volatile emission and their functions in nature, experimental setups are needed that simulate the habitat as adequate as possible (Kai et al., 2016). B. subtilis colonizes food surfaces, the soil and often occurs plant associated. Nutrient supply, physico-chemical properties of the soil and environmental factors shape both growth and development of the bacteria, but also their volatile emission (Effmert et al., 2012; Burns et al., 2015). Soil aeration and oxygen availability, for instance, are crucial for the microbial activity in soil. The oxygen supply, which B. subtilis experienced in the applied in vitro set-ups, were yet not considered, although it is conceivable that the oxygen availability for a bacterial culture on solid agar plates differs compared to culture in liquid medium (Somerville and Proctor, 2013). Another fundamental factor influencing the bacterial growth and metabolism is the temperature. Colonizing the soil, B. subtilis experiences changes in temperature caused by climate and season, however, in the in vitro tests considered in these studies, temperatures were set in the range of 24 and 37°C. Future studies should consider the factor temperature and its direct influence on the emission of B. subtilis volatiles in order to evaluate regional, seasonal or climate dependencies.
Evidence for nutrient dependent production of many non-volatile secondary metabolites (Tyc et al., 2017) and volatile metabolites could be yet supplied in vitro for several species of soil bacteria when cultivated on varying nutrient compositions (Fiddaman and Rossall, 1994; Kai et al., 2009; Blom et al., 2011; Weise et al., 2012; Ratiu et al., 2017b). One of the main nutrient resources for bacteria in soil represent plants root exudates (Barber and Martin, 1975; Lynch and Whipps, 1991; Hütsch et al., 2002). So far, the emission of volatiles by B. subtilis isolates was almost exclusively studied during cultivation on complex medium containing high levels of amino acids and peptides. Plants indeed release besides excess primary metabolites and among other substances amino acids and peptides via roots into the soil (Jaeger et al., 1999; Farrar et al., 2003; Uren, 2007; Dennis et al., 2010; Sasse et al., 2018). Therefore, the artificial conditions in in vitro studies might be close to particular natural conditions in the rhizosphere of plants, however, many other soil habitats must be considered as nutrient poor (Young et al., 2008; Schulz-Bohm et al., 2015). Therefore, isolates of B. subtilis should be also investigated under conditions simulating a poor soil nutrient status as well as root exudation.
It is well known that soil-born bacterial species including B. subtilis live in complex, diverse and dynamic communities (Fierer and Jackson, 2006; Little et al., 2008; Phelan et al., 2012). Intra- and interspecific interactions within such a microbial community can strongly affect the profiles of volatile secondary metabolites released by the community (Hol et al., 2015; Schulz-Bohm et al., 2015; Tyc et al., 2015; Schmidt et al., 2017; Kai and Piechulla, 2018; Kai et al., 2018). For instance, quorum sensing systems are assumed to affect the quality and quantity of bacterial volatiles (Kesarwani et al., 2011). Since B. subtilis certainly also colonizes densely populated habitats, future studies should also consider microbial interactions when investigating volatile secondary metabolite emission.
Standards of Identification of Volatile Secondary Metabolites
The reliable and unambiguous identification of volatile secondary metabolites is the centerpiece of this research field. Schulz and Dickschat (2007) already stated that a rigorous use of the state-of-the-art methodology for compound identification could not be corroborated in all published studies. Volatile secondary metabolites are usually analyzed using GC/EI-MS. The resulting EI mass spectra are highly reproducible and show fragmentation patterns, which are characteristic for each compound. Fragmentation patterns can be submitted to databases (NIST, Whiley, etc.) resulting in a ranking of putative compounds starting with the highest score. Unequivocal identification further includes the verification of these database propositions by comparison of mass spectra and retention indices of the searched compound with the data of reference compounds (authentic standards, either commercially available or synthesized). As mentioned above, only Robacker et al. (1998) and Ryu et al. (2004) accomplished this identification workflow. Therefore, some of the compounds summarized in this review might be wrongly identified in the original paper. In future, it is recommended to follow the described workflow of identification using authentic standards. For volatile secondary metabolites, where an unambiguous identification is not possible it would be very helpful to declare them as ‘putatively identified.’ Sumner et al. (2007) suggested several confidence levels of identity for metabolomics studies (Dunn et al., 2013) (Table 4). This rating of identification should also be applicable in volatile secondary metabolite research.
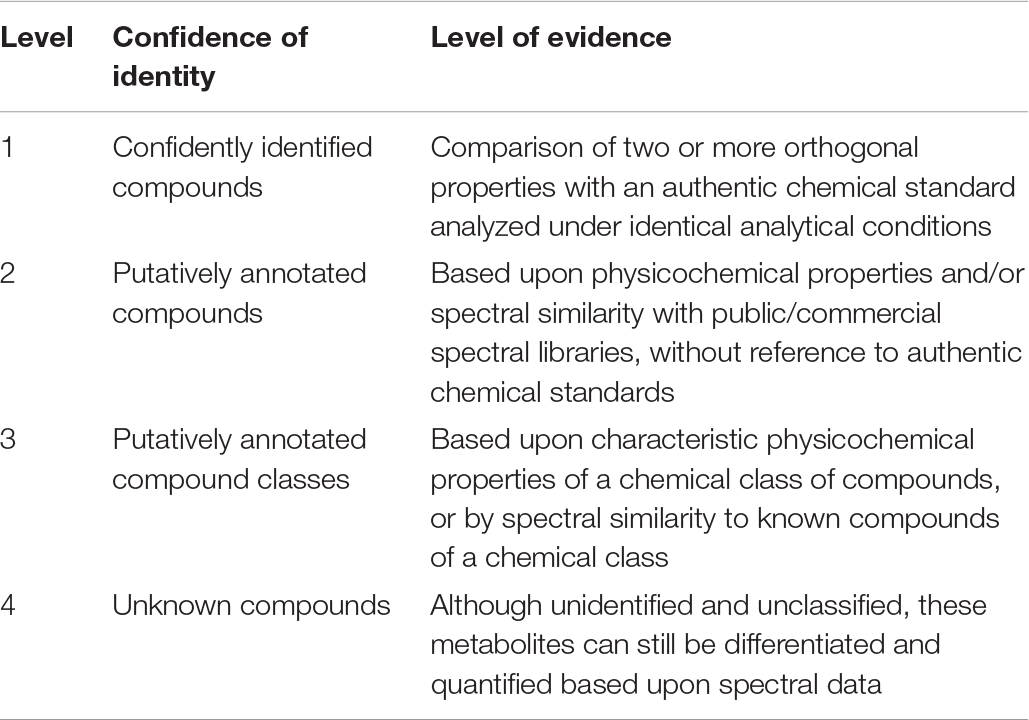
Table 4. Level of identification based on Metabolomics Standard Initiative (Sumner et al., 2007, copy from Dunn et al. (2013).
Standards of Data Acquisition
In order to compare and summarize collected data of different studies for interpretation, and also to develop follow-up experiments, or to refine hypotheses, a clear and complete documentation is essential (Fiehn et al., 2007). Reviewing the literature on volatile secondary metabolites emitted from B. subtilis isolates, it became apparent that the data acquisition was often not sufficiently documented. Farag et al. (2017) described state-of-the-art procedures for volatile secondary metabolite profiling. In 2005, the metabolomics initiative was founded in order to define minimal reporting standards of metabolomics data (metabolomics standards initiative, MSI) and best practice set of reporting standards (Fiehn et al., 2007; Sumner et al., 2007). In accordance to the protocols of volatile secondary metabolite profiling and the MSI, these reporting standards should also be applied to research on bacterial volatile secondary metabolites. These suggestions encompass both the biological context and the chemical analysis (Figure 5).
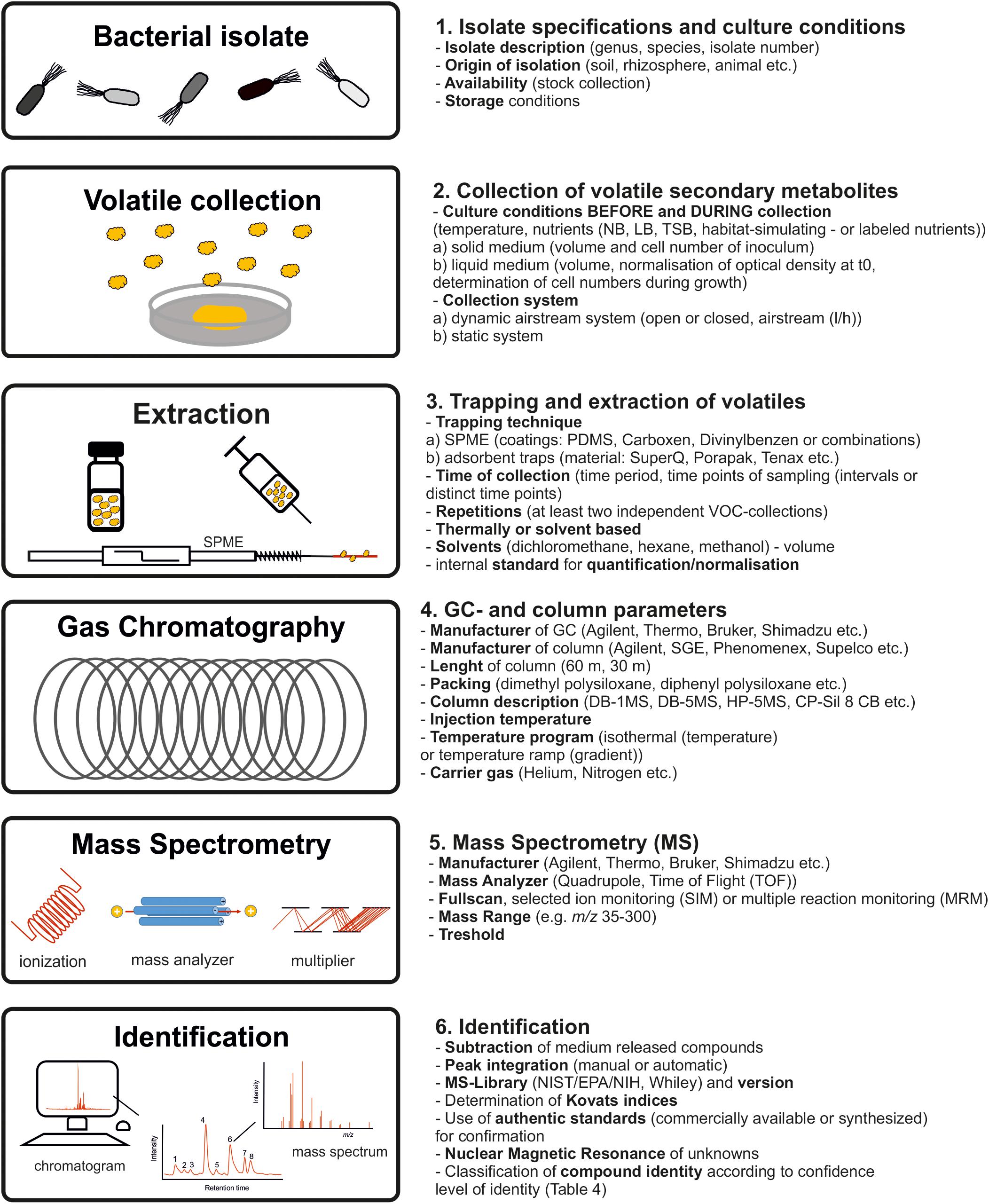
Figure 5. Best practice procedures and documentation. Workflow to obtain consistent, reliable and re-usable bacterial profiles in future studies.
The documentation should start with a comprehensive description of the B. subtilis isolate used as well as the origin of isolation and the storage conditions. The experimental design including cultivation parameter prior to volatile collection, during volatile collection and the used volatile collection system should be specified. The cell number of the isolates during collection should be always recorded. Methods of single time-point measurements or interval measurements during bacterial growth and the parameters of metabolite enrichment including adsorption and desorption techniques should be stated. Equally important is the description of GC-separation as well as the mass spectrometry parameters. This should also include parameter as mass scan range and thresholds. Compounds appearing in a proper control should be subtracted. The calculation of retention indices (Kováts, 1958) should facilitate comparisons with databases and other investigated isolates. For identification, in the case that suggestions of the MS-databases are used, authors should label these suggestions with a statement of similarity for each compound. It should be recognizable if volatiles were identified using standard reference compounds (synthesized or commercial) and/or libraries. The compound identity should be classified according to confidence level of identity (see above) (Sumner et al., 2007).
Conclusion
Bacillus subtilis isolates emit a broad range of chemically diverse volatile secondary metabolites. About 231 volatiles have been described so far. Surprisingly, on one side some volatile secondary metabolites appeared isolate-exclusive, but on the other side other volatile secondary metabolites, which are described as bioactive molecules, were emitted more generally. The observations, however, were based on original data, which were strongly influenced by insufficient descriptions of the bacterial isolates, heterogeneous and poorly documented culture conditions as well as sampling techniques and inadequate compound identification. For a better outcome and interpretability of data as well as for the development of experimental setups or refining hypotheses, future approaches should apply well-documented workflows and fulfill state-of-the-art standards to unambiguously identify the volatile metabolites. These new approaches should question whether (I) the isolate-exclusive emission represents a consequence of acclimation or multicellularity, (II) which volatile secondary metabolites are really biosynthesized by B. subtilis and (III) which effect B. subtilis volatiles exert in nature.
Author Contributions
MK conceived and designed the review, produced the figures and tables, and wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I thank Dr. Uta Effmert for critical comments that improved the manuscript, Prof. Birgit Piechulla for critical and constructive input and the University of Rostock for financial support. I am further thankful for ideas of the reviewers and appreciate their effort to improve this manuscript.
References
Arrebola, E., Sivakumar, D., and Korsten, L. (2010). Effect of volatile compounds produced by Bacillus strains on postharvest decay in citrus. Biol. Control 53, 122–128. doi: 10.1016/j.biocontrol.2009.11.010
Barber, D. A., and Martin, J. K. (1975). The release of organic substances by cereal roots into soil. New Phytol. 76, 69–80. doi: 10.1111/j.1469-8137.1976.tb01439.x
Berg, G., Grube, M., Schloter, M., and Smalla, K. (2014). Unraveling the plant microbiome: looking back and future perspectives. Front. Microbiol. 5:148. doi: 10.3389/fmicb.2014.00148
Berg, G., Roskot, N., Steidle, A., Eberl, L., Zock, A., and Smalla, K. (2002). Plant-dependent genotypic and phenotypic diversity of antagonistic rhizobacteria isolated from different Verticillium host plants. Appl. Environ. Microbiol. 68, 3328–3338. doi: 10.1128/AEM.68.7.3328-3338.2002
Blom, D., Fabbri, C., Connor, E. C., Schiestl, F. P., Klauser, D. R., Boller, T., et al. (2011). Production of plant growth modulating volatiles is widespread among rhizosphere bacteria and strongly depends on culture conditions. Environ. Microbiol. 13, 3047–3058. doi: 10.1111/j.1462-2920.2011.02582.x
Burkholder, P. R., and Giles, N. H. Jr. (1947). Induced biochemical mutations in Bacillus subtilis. Am. J. Bot. 34, 345–348. doi: 10.1002/j.1537-2197.1947.tb12999.x
Burns, J. H., Anacker, B. L., Strauss, S. Y., and Burke, D. J. (2015). Soil microbial community variation correlates most strongly with plant species identity. Followed by soil chemistry, spatial location and plant genus. AoB Plants 7:lv030. doi: 10.1093/aobpla/plv030
Caulier, S., Nannan, C., Gillis, A., Licciardi, F., Bragard, C., and Mahillon, J. (2019). Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front. Microbiol. 10:302. doi: 10.3389/fmicb.2019.00302
Chaves-López, C., Serio, A., Gianotti, A., Sacchetti, G., Ndagijimana, M., Ciccarone, C., et al. (2015). Diversity of food-borne Bacillus volatile compounds and influence on fungal growth. J. Appl. Microbiol. 119, 487–499. doi: 10.1111/jam.12847
Chen, H., Xiao, X., Wang, J., Wu, L., Zheng, Z., and Yu, Z. (2008). Antagonistic effects of volatiles generated by Bacillus subtilis on spore germination and hyphal growth of the plant pathogen, Botrytis cinerea. Biotechnol. Lett. 30, 919–923. doi: 10.1007/s10529-007-9626-9
Choi, S.-K., Jeong, H., Kloepper, J. W., and Ryu, C.-M. (2014). Genome Sequence of Bacillus amyloliquefaciens GB03, an active ingredient of the first commercial biological control product. Genome Announc. 2:e01092-14. doi: 10.1128/genomeA.01092-14
Davies, J. (2006). Are antibiotics naturally antibiotics? J. Ind. Microbiol. Biotechnol. 33, 496–499. doi: 10.1007/s10295-006-0112-5
Dennis, P. G., Miller, A. J., and Hirsch, P. R. (2010). Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol. Ecol. 72, 313–327. doi: 10.1111/j.1574-6941.2010.00860.x
Dickschat, J. S. (2017). Fungal volatiles-a survey from edible mushrooms to moulds. Nat. Prod. Rep. 34, 310–328. doi: 10.1039/c7np00003k
Dugravot, S., Grolleau, F., Macherel, D., Rochetaing, A., Hue, B., Stankiewicz, M., et al. (2003). Dimethyl disulfide exerts insecticidal neurotoxicity through mitochondrial dysfunction and activation of insect KATP channels. J. Neurophysiol. 90, 259–270. doi: 10.1152/jn.01096.2002
Dunn, W. B., Erban, A., Weber, R. J. M., Creek, D. J., Brown, M., Breitling, R., et al. (2013). Mass appeal: metabolite identification in mass spectrometry-focused untargeted Metabolomics. Metabolomics 9, S44–S66.
Eckert, K. E., Carter, D. O., and Perrault, K. A. (2018). Sampling dynamics for volatile organic compounds using headspace solid-phase microextraction arrow for microbiological samples. Separations 5:45. doi: 10.3390/separations5030045
Effmert, U., Kalderas, J., Warnke, R., and Piechulla, B. (2012). Volatile mediated interactions between bacteria and fungi in the soil. J. Chem. Ecol. 38, 665–703. doi: 10.1007/s10886-012-0135-5
Fall, R., Kearns, D. B., and Nguyen, T. (2006). A defined medium to investigate sliding motility in a Bacillus subtilis flagella-less mutant. BMC Microbiol. 6:31. doi: 10.1186/1471-2180-6-31
Farag, M. A., Song, G. C., Park, Y. S., Audrain, B., Lee, S., Ghigo, J.-M., et al. (2017). Biological and chemical strategies for exploring inter- and intra-kingdom communication mediated via bacterial volatile signals. Nat. Protoc. 12, 1359–1377. doi: 10.1038/nprot.2017.023
Farrar, J., Hawes, M., Jones, D., and Lindow, S. (2003). How roots control the flux of carbon to the rhizosphere. Ecology 84, 827–837. doi: 10.1890/0012-9658(2003)084%5B0827:hrctfo%5D2.0.co;2
Fiddaman, P. J., and Rossall, S. (1993). The production of antifungal volatiles by Bacillus subtilis. J. Appl. Bacteriol. 74, 119–126. doi: 10.1111/j.1365-2672.1993.tb03004.x
Fiddaman, P. J., and Rossall, S. (1994). Effect of substrate on the production of antifungal volatiles from Bacillus subtilis. J. Appl. Bacteriol. 76, 395–405. doi: 10.1111/j.1365-2672.1994.tb01646.x
Fiehn, O., Robertson, D., Griffin, J., van der Werf, M., Nikolau, B., Morrison, N., et al. (2007). The Metabolomics Standards Initiative (MSI). Metabolomics 3, 175–178. doi: 10.1007/s11306-007-0070-6
Fierer, N., and Jackson, R. B. (2006). The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 103, 626–631. doi: 10.1073/pnas.0507535103
Gallegos-Monterrosa, R., Mhatre, E., and Kovács, Á. T. (2016). Specific Bacillus subtilis 168 variants form biofilms on nutrient-rich medium. Microbiology 162, 1922–1932. doi: 10.1099/mic.0.000371
Gao, H., Li, P., Xu, X., Zeng, Q., and Guan, W. (2018). Research on volatile organic compounds from Bacillus subtilis CF-3: biocontrol effects on fruit fungal pathogens and dynamic changes during fermentation. Front. Microbiol. 9:456. doi: 10.3389/fmicb.2018.00456
Gao, H., Xu, X., Dai, Y., and He, H. (2016). Isolation, Identification and Characterization of Bacillus subtilis CF-3, a bacterium from fermented bean curd for controlling postharvest diseases of peach fruit. Food Sci. Technol. Res. 22, 377–385. doi: 10.3136/fstr.22.377
Gautier, H., Auger, J., Legros, C., and Lapied, B. (2008). Calcium-activated potassium channels in insect pacemaker neurons as unexpected target site for the novel fumigant dimethyl disulfide. J. Pharmacol. Exp. Ther. 324, 149–159. doi: 10.1124/jpet.107.128694
Gerber, N. N., and Lechevalier, H. A. (1965). Geosmin, an earthy-smelling substance isolated from actinomycetes. Appl. Microbiol. 13, 935–938. doi: 10.1128/aem.13.6.935-938.1965
Gray, D. A., Dugar, G., Gamba, P., Strahl, H., Jonker, M. J., and Hamoen, L. W. (2019). Extreme slow growth as alternative strategy to survive deep starvation in bacteria. Nat. Commun. 10:890. doi: 10.1038/s41467-019-08719-8
Groenhagen, U., Baumgartner, R., Bailly, A., Gardiner, A., Eberl, L., Schulz, S., et al. (2013). Production of bioactive volatiles by different Burkholderia ambifaria Strains. J. Chem. Ecol. 39, 892–906. doi: 10.1007/s10886-013-0315-y
Gu, Y.-Q., Mo, M.-H., Zhou, J. P., Zou, C.-S., and Zhang, K.-Q. (2007). Evaluation and identification of potential organic nematicidal volatiles from soil bacteria. Soil Biol. Biochem. 39, 2567–2575. doi: 10.1016/j.soilbio.2007.05.011
Guo, S., Mao, Z., Wu, Y., Hao, K., He, P., and He, Y. (2013). Genome sequencing of Bacillus subtilis Strain XF-1 with high efficiency in the suppression of Plasmodiophora brassicae. Genome Announc. 1:e0006613. doi: 10.1128/genomeA.00066-13
Hageman, J. H., Shankweiler, G. W., Wall, P. R., Franich, K., McCowan, G. W., Cauble, S. M., et al. (1984). Chemically defined sporulation medium. J. Bacteriol. 160, 438–441. doi: 10.1128/jb.160.1.438-441.1984
Harwood, C. R., Mouillon, J.-M., and Pohl, S. (2018). Secondary metabolite production and the safety of industrially important members of the Bacillus subtilis group. FEMS Microbiol. Rev. 42, 721–738. doi: 10.1093/femsre/fuy028
Heuer, H., and Smalla, K. (2012). Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol. Rev. 36, 1083–1104. doi: 10.1111/j.1574-6976.2012.00337.x
Hofemeister, J., Conrad, B., Adler, B., Hofemeister, B., Feesche, J., Kucheryava, N., et al. (2004). Genetic analysis of the biosynthesis of non-ribosomal peptide- and polyketide-like antibiotics, iron uptake and biofilm formation by Bacillus subtilis A1/3. Mol. Genet. Genomics 272, 363–378. doi: 10.1007/s00438-004-1056-y
Hol, W. H. G., Garbeva, P., Hordijk, C., Hundscheid, M. P. J., Klein Gunnewiek, P. J. A., van Agtmaal, M., et al. (2015). Non-random species loss in bacterial communities reduces antifungal volatile production. Ecology 96, 2042–2048. doi: 10.1890/14-2359.1
Hütsch, B. W., Augustin, J., and Merbach, W. (2002). Plant rhizodeposition - an important source for carbon turnover in soils. J. Plant Nutr. Soil Sci. 165, 397–407.
Jaeger, C. H. III, Lindow, S. E., Miller, W., Clark, E., and Firestone, M. K. (1999). Mapping sugar and aminon acid exudation from roots in soil using bacterial sensors of sucrose and tryptophan. Appl. Environ. Microbiol. 65, 2685–2690. doi: 10.1128/aem.65.6.2685-2690.1999
Jangir, M., Pathak, R., Sharma, A., Sharma, S., and Sharma, S. (2018). Volatiles as strong markers for antifungal activity against Fusarium oxysporum f. sp. lycopersici. Indian Phytopathol. 72, 681–687. doi: 10.1007/s42360-018-0073-4
Kai, M., Crespo, E., Cristescu, S. M., Harren, F. J. M., Francke, W., and Piechulla, B. (2010). Serratia odorifera: analysis of volatile emission and biological impact of volatile compounds on Arabidopsis thaliana. Appl. Microbiol. Biotechnol. 88, 965–976. doi: 10.1007/s00253-010-2810-1
Kai, M., Effmert, U., Berg, G., and Piechulla, B. (2007). Volatiles of bacterial antagonists inhibit mycelial growth of the plant pathogen Rhizoctonia solani. Arch. Microbiol. 187, 351–360. doi: 10.1007/s00203-006-0199-0
Kai, M., Effmert, U., Lemfack, M. C., and Piechulla, B. (2018). Interspecific formation of the antimicrobial volatile schleiferon. Sci. Rep. 8:16852. doi: 10.1038/s41598-018-35341-3
Kai, M., Effmert, U., and Piechulla, B. (2016). Bacterial-plant-interactions: approaches to unravel the biological function of bacterial volatiles in the rhizosphere. Front. Microbiol. 7:108. doi: 10.3389/fmicb.2016.00108
Kai, M., Haustein, M., Molina, F., Petri, A., Scholz, B., and Piechulla, B. (2009). Bacterial volatiles and their action potential. Appl. Microbiol. Biotechnol. 81, 1001–1012. doi: 10.1007/s00253-008-1760-3
Kai, M., and Piechulla, B. (2018). Interspecies interaction of Serratia plymuthica 4Rx13 and Bacillus subtilis B2g alters the emission of sodorifen. FEMS Microbiol. Lett. 365, fny253. doi: 10.1093/femsle/fny253
Kakinuma, A., Sugino, H., Isono, M., Tamura, G., and Arima, K. (1969). Determination of fatty acid in surfactin and elucidation of the total structure of surfactin. Agric. Biol. Chem. 33, 973–976. doi: 10.1080/00021369.1969.10859409
Kalamara, M., Spacapan, M., Mandic-Mulec, I., and Stanley-Wall, N. R. (2018). Social behaviours by Bacillus subtilis: quorum sensing, kin discrimination and beyond. Mol. Microbiol. 110, 863–878. doi: 10.1111/mmi.14127
Kesarwani, M., Hazan, R., He, J., Que, Y., Apidianakis, Y., Lesic, B., et al. (2011). A quorum sensing regulated small volatile molecule reduces acute virulence and promotes chronic infection phenotypes. PLoS Pathog. 7:e1002192. doi: 10.1371/journal.ppat.1002192
Khabbaz, S. E., Zhang, L., Cáceres, L. A., Sumarah, M., Wang, A., and Abbasi, P. A. (2015). Characterisation of antagonistic Bacillus and Pseudomonas strains for biocontrol potential and suppression of damping-off and root rot diseases. Ann. Appl. Biol. 166, 456–471. doi: 10.1111/aab.12196
Korpi, A., Järnberg, J., and Pasanen, A. L. (2009). Microbial volatile organic compounds. Crit. Rev. Toxicol. 39, 139–193. doi: 10.1080/10408440802291497
Kováts, E. (1958). Gas-chromatographische Charakterisierung Organischer Verbindungen. Teil 1: Retentionsindices Aliphatischer Halogenide, Alkohole, Aldehyde und Ketone. Helv. Chim. Acta 206, 1915–1932. doi: 10.1002/hlca.19580410703
Kunst, F., Ogasawara, N., Moszer, I., Albertini, A. M., Alloni, G., Azevedo, V., et al. (1997). The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390, 249–256.
Lemfack, M. C., Gohlke, B. O., Toguem, S. M. T., Preissner, S., Piechulla, B., and Preissner, R. (2018). MVOC 2.0: a database of microbial volatiles. Nucleic Acids Res. 46, D1261–D1265. doi: 10.1093/nar/gkx1016
Lemfack, M. C., Nickel, J., Dunkel, M., Preissner, R., and Piechulla, B. (2014). MVOC: a database of microbial volatiles. Nucleic Acids Res. 42, D744–D748. doi: 10.1093/nar/gkt1250
Li, X.-Y., Mao, Z.-C., Wu, Y.-X., Ho, H.-H., and He, Y.-Q. (2015). Comprehensive volatile organic compounds profiling of Bacillus species with biocontrol properties by head space solid phase microextraction with gas chromatography-mass spectrometry. Biocontrol Sci. Technol. 25, 132–143. doi: 10.1080/09583157.2014.960809
Little, A. E. F., Robinson, C. J., Peterson, S. B., Raffa, K. F., and Handelsman, J. (2008). Rules of engagement: interspecies interactions that regulate microbial communities. Annu. Rev. Microbiol. 62, 375–401. doi: 10.1146/annurev.micro.030608.101423
Liu, W., Mu, W., Zhu, B., and Liu, F. (2008a). Antifungal activities and components of VOCs produced by Bacillus subtilis G8. Curr. Res. Bacteriol. 1, 28–34. doi: 10.3923/crb.2008.28.34
Liu, W., Mu, W., Zhu, B.-Y., Du, Y.-C., and Feng, L. (2008b). Antagonistic activities of volatiles from four strains of Bacillus spp. and Paenibacillus spp. against soil-borne plant pathogens. Agric. Sci. China 7, 1104–1114. doi: 10.1016/s1671-2927(08)60153-4
Lynch, J. M., and Whipps, J. M. (1991). “Substrate flow in the rhizosphere,” in The Rhizosphere and Plant Growth, eds D. L. Keistercpesnm and B. Cregancpesnm (Dordrecht: Kluwer Academic Publishers), 15–24. doi: 10.1007/978-94-011-3336-4_2
Lyons, N. A., and Kolter, R. (2015). On the evolution of bacterial multicellularity nicholas. Curr. Opin. Microbiol. 24, 21–28. doi: 10.1016/j.mib.2014.12.007
Meldau, D. G., Meldau, S., Hoang, L. H., Underberg, S., Wünsche, H., and Baldwin, I. T. (2013). Dimethyl disulfide produced by the naturally associated bacterium Bacillus Sp B55 promotes Nicotiana attenuata growth by enhancing sulfur nutrition. Plant Cell 25, 2731–2747. doi: 10.1105/tpc.113.114744
Mu, J., Li, X., Jiao, J., Ji, G., Wu, J., Hu, F., et al. (2017). Biocontrol potential of vermicompost through antifungal volatiles produced by indigenous bacteria. Biol. Control 112, 49–54. doi: 10.1016/j.biocontrol.2017.05.013
Nalli, S., Horn, O. J., Grochowalski, A. R., Cooper, D. G., and Nicell, J. A. (2006). Origin of 2-Ethylhexanol as a VOC. Environ. Pollut. 140, 181–185. doi: 10.1016/j.envpol.2005.06.018
Obagwu, J., and Korsten, L. (2003). Integrated control of citrus green and blue molds using Bacillus subtilis in combination with sodium bicarbonate or hot water. Postharvest Biol. Technol. 28, 187–194. doi: 10.1016/s0925-5214(02)00145-x
Peypoux, F., Besson, F., Michel, G., and Delcambe, L. (1981). Structure of Bacillomycin D, a new antibiotic of the iturin group. Eur. J. Biochem. 118, 323–327. doi: 10.1111/j.1432-1033.1985.tb09307.x
Phelan, V. V., Liu, W.-T., Pogliano, K., and Dorrestein, P. C. (2012). Microbial Metabolic Exchange-the Chemotype-to-Phenotype Link. Nat. Chem. Biol. 8, 26–35. doi: 10.1038/nchembio.739
Piechulla, B., Lemfack, M. C., and Kai, M. (2017). Effects of discrete bioactive microbial volatiles on plants and fungi. Plant Cell Environ. 40, 2042–2067. doi: 10.1111/pce.13011
Rajer, F. U., Wu, H., Xie, Y., Xie, S., Raza, W., Tahir, H. A. S., et al. (2017). Volatile organic compounds produced by a soil-isolate, Bacillus subtilis FA26 induce adverse ultra-structural changes to the cells of Clavibacter michiganensis ssp. sepedonicus, the causal agent of bacterial ring rot of potato. Microbiology 163, 523–530. doi: 10.1099/mic.0.000451
Rasmann, S., Köllner, T. G., Degenhardt, J., Hiltpold, I., Toepfer, S., Kuhlmann, U., et al. (2005). Recruitment of entomopathogenic nematodes by insect-damaged maize roots. Nature 434, 732–737. doi: 10.1038/nature03451
Ratcliff, W. C., and Denison, R. F. (2011). Alternative actions for antibiotics. Science 332, 547–548. doi: 10.1126/science.1205970
Ratiu, I. A., Bocos-Bintintan, V., Patrut, A., Moll, V. H., Turner, M., and Thomas, C. L. P. (2017a). Discrimination of bacteria by rapid sensing their metabolic volatiles using an aspiration-type ion mobility spectrometer (a-IMS) and gas chromatography-mass spectrometry GC-MS. Anal. Chim. Acta 982, 209–217. doi: 10.1016/j.aca.2017.06.031
Ratiu, I. A., Ligor, T., Bocos-Bintintan, V., Al-Suod, H., Kowalkowski, T., Rafiñska, K., et al. (2017b). The effect of growth medium on an Escherichia coli pathway mirrored into GC/MS profiles. J. Breath Res. 11:036012. doi: 10.1088/1752-7163/aa7ba2
Rijnen, L., Yvon, M., Kranenburg, R., Courtin, P., Verheul, A., Chambellon, E., et al. (2003). Lactococcal aminotransferases AraT and BcaT are key enzymes for the formation of aroma compounds from amino acids in cheese. Int. Dairy J. 13, 805–812. doi: 10.1016/S0958-6946(03)00102-X
Robacker, D. C., Martinez, A. J., Garcia, J. A., and Bartelt, R. J. (1998). Volatiles attractive to the mexican fruit fly (Diptera: Tephritidae) from eleven bacteria taxa. Fla. Entomol. 81, 497–508. doi: 10.2307/3495948
Romero, D., de Vicente, A., Rakotoaly, R. H., Dufour, S. E., Veening, J.-W., Arrebola, E., et al. (2007). The iturin and fengycin families of lipopeptides are key factors in antagonism of Bacillus subtilis toward Podosphaera fusca. Mol. Plant Microbe Interact. 20, 430–440. doi: 10.1094/mpmi-20-4-0430
Rosenthal, A. Z., Qi, Y., Hormoz, S., Park, J., Li, S. H.-J., and Elowitz, M. B. (2018). Metabolic interactions between dynamic bacterial subpopulations. eLife 7:e33099. doi: 10.7554/elife.33099
Ryu, C.-M., Farag, M. A., Hu, C.-H., Reddy, M. S., Kloepper, J. W., and Pare, P. W. (2004). Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 134, 1017–1026. doi: 10.1104/pp.103.026583.with
Ryu, C.-M., Farag, M. A., Hu, C.-H., Reddy, M. S., Wei, H.-X., Pare, P. W., et al. (2003). Bacterial volatiles promote growth in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 100, 4927–4932. doi: 10.1073/pnas.0730845100
Sansinenea, E., and Ortiz, A. (2011). Secondary metabolites of Soil Bacillus Spp. Biotechnol. Lett. 33, 1523–1538. doi: 10.1007/s10529-011-0617-5
Sasse, J., Martinoia, E., and Northen, T. (2018). Feed your friends: Do plant exudates shape the root microbiome? Trends Plant Sci. 23, 25–41. doi: 10.1016/j.tplants.2017.09.003
Schmidt, R., de Jager, V., Zühlke, D., Wolff, C., Bernhardt, J., Cankar, K., et al. (2017). Fungal volatile compounds induce production of the secondary metabolite sodorifen in Serratia plymuthica PRI-2C. Sci. Rep. 7:862. doi: 10.1038/s41598-017-00893-3
Schneider, K., Chen, X.-H., Vater, J., Franke, P., Nicholson, G., Borriss, R., et al. (2007). Macrolactin is the polyketide biosynthesis product of the Pks2 cluster of Bacillus amyloliquefaciens FZB42. J. Nat. Prod. 70, 1417–1423. doi: 10.1021/np070070k
Schulz, S., and Dickschat, J. S. (2007). Bacterial volatiles: the smell of small organisms. Nat. Prod. Rep. 24, 814–842. doi: 10.1039/b507392h
Schulz-Bohm, K., Gerards, S., Hundscheid, M., Melenhorst, J., de Boer, W., and Garbeva, P. (2018). Calling from distance: attraction of soil bacteria by plant root volatiles. ISME J. 12, 1252–1262. doi: 10.1038/s41396-017-0035-3
Schulz-Bohm, K., Martín-Sánchez, L., and Garbeva, P. (2017). Microbial volatiles: small molecules with an important role in intra- and inter-kingdom interactions. Front. Microbiol. 8:2484. doi: 10.3389/fmicb.2017.02484
Schulz-Bohm, K., Zweers, H., de Boer, W., and Garbeva, P. (2015). A fragrant neighborhood: volatile mediated bacterial interactions in soil. Front. Microbiol. 6:1212. doi: 10.3389/fmicb.2015.01212
Scott, T. A., and Piel, J. (2019). The hidden enzymology of bacterial natural product biosynthesis. Nat. Rev. Chem. 3, 404–425. doi: 10.1038/s41570-019-0107-1
Shifa, H., Gopalakrishnan, C., and Velazhahan, R. (2015). Characterization of antifungal antibiotics produced By Bacillus subtilis G-1 antagonistic to Sclerotium rolfsii. Biochem. Cell Arch. 15, 99–104.
Somerville, G. A., and Proctor, R. A. (2013). Cultivation conditions and the diffusion of oxygen into culture media: the rationale for the flask-to-medium ratio in microbiology. BMC Microbiol. 13:9. doi: 10.1186/1471-2180-13-9
Stein, T. (2005). Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol. Microbiol. 56, 845–857. doi: 10.1111/j.1365-2958.2005.04587.x
Stensmyr, M. C., Dweck, H. K. M., Farhan, A., Ibba, I., Strutz, A., Mukunda, L., et al. (2012). A conserved dedicated olfactory circuit for detecting harmful microbes in Drosophila. Cell 151, 1345–1357. doi: 10.1016/j.cell.2012.09.046
Sumner, L. W., Amberg, A., Barrett, D., Beale, M. H., Beger, R., Daykin, C. A., et al. (2007). Proposed minimum reporting standards for chemical analysis. Metabolomics 3, 211–221. doi: 10.1007/s11306-007-0082-2
Tahir, H. A. S., Gu, Q., Wu, H., Raza, W., Hanif, A., Wu, L., et al. (2017). Plant growth promotion by volatile organic compounds produced by Bacillus subtilis SYST2. Front. Microbiol. 8:171. doi: 10.3389/fmicb.2017.00171
Tyc, O., Song, C., Dickschat, J. S., Vos, M., and Garbeva, P. (2017). the ecological role of volatile and soluble secondary metabolites produced by soil bacteria. Trends Microbiol. 25, 280–292. doi: 10.1016/j.tim.2016.12.002
Tyc, O., Zweers, H., de Boer, W., and Garbeva, P. (2015). Volatiles in inter-specific bacterial interactions. Front. Microbiol. 6:1412. doi: 10.3389/fmicb.2015.01412
Uren, N. C. (2007). “Types, amounts, and possible functions of compounds released into the rhizosphere by soil-grown plants,” in The Rhizosphere. Biochemistry and Organic Substances at the Soil-Plant Interface, 2nd Edn, eds R. Pinton, Z. Varanini, and P. Nannipiericpesnm (Boca Raton, FL: CRC Press), 1–21. doi: 10.1201/9781420005585.ch1
Weise, T., Kai, M., Gummesson, A., Troeger, A., von Reuss, S. H., Piepenborn, S., et al. (2012). Volatile organic compounds produced by the phytopathogenic bacterium Xanthomonas campestris pv. vesicatoria 85-10. Beilstein J. Org. Chem. 8, 579–596. doi: 10.3762/bjoc.8.65
Yim, G., Wang, H. H., and Davies, J. (2007). Antibiotics as signalling molecules. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 362, 1195–1200. doi: 10.1098/rstb.2007.2044
Young, I. M., Crawford, J. W., Nunan, N., Otten, W., and Spiers, A. (2008). “Microbial distribution in soils. Physics and Scaling,” in Advances in Agronomy, Vol. 100, ed. D. L. Sparkscpesnm (San Diego, CA: Elsevier Acadamic Press Inc), 81–121. doi: 10.1016/S0065-2113(08)00604-4
Zeigler, D. R., Prágai, Z., Rodriguez, S., Chevreux, B., Muffler, A., Albert, T., et al. (2008). The origin of 168, W23, and other Bacillus subtilis legacy strains. J. Bacteriol. 190, 6983–6995. doi: 10.1128/JB.00722-08
Keywords: volatiles, VOCs, secondary metabolites, Bacillus subtilis, GC/MS, GC/EI-MS, identification
Citation: Kai M (2020) Diversity and Distribution of Volatile Secondary Metabolites Throughout Bacillus subtilis Isolates. Front. Microbiol. 11:559. doi: 10.3389/fmicb.2020.00559
Received: 14 June 2019; Accepted: 16 March 2020;
Published: 08 April 2020.
Edited by:
Saskia Bindschedler, Université de Neuchâtel, SwitzerlandReviewed by:
Patrick Fink, Helmholtz Centre for Environmental Research (UFZ), GermanyHiroshi Otani, Berkeley National Laboratory (JGI), United States
Copyright © 2020 Kai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marco Kai, marcokai@hotmail.de
 Marco Kai
Marco Kai