K+ Nutrition Exchange in the Serendipita-Arabidopsis Symbiosis: Study of the Fungal K+ Transporters Involved
- 1Centro de Biotecnología y Genómica de Plantas (CBGP), Universidad Politécnica de Madrid (UPM), Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA), Madrid, Spain
- 2Departamento de Biotecnología-Biología Vegetal, Escuela Técnica Superior de Ingeniería Agronómica, Alimentaria y de Biosistemas, Universidad Politécnica de Madrid (UPM), Madrid, Spain
There is mounting evidence that the root-colonizing endosymbiotic fungus Serendipita indica improves plant growth. The beneficial effects have been observed when plants are growing in optimal conditions or under nutritionally deficient soils (e.g., phosphate poor soil) or exposed to stressful environmental conditions such as drought or salinity. However, until now its role in the nutrition of other plant essential macronutrient, such as K+, has not been fully clarified. Here, we study the role of the fungus in the K+ nutrition of Arabidopsis thaliana plants, during growth under K+ limiting conditions. As a first step, we studied the high-affinity K+ uptake of the plant and fungus when growing separately and in symbiosis. In the search for putative fungal actors involved in K+ nutrition, we also have cloned and functionally characterized the K+ transporters of S. indica SiHAK1, SiTRK1, SiTRK2, and SiTOK1, among which it has been shown that SiHAK1 is the main transporter involved in the K+ uptake in the high affinity range of concentrations. In addition, a gene expression study of these transporters and other candidates that could participate in the K+ homeostasis of the fungus has been carried out. The results indicated that, contrary to what happens with P nutrition, S. indica seems not to improve neither the growth nor the plant K+ reserves during K+ starvation. Instead, this nutritionally restrictive condition favored fungal colonization, suggesting that the fungus obtains the greatest benefit in K+ supply during symbiosis.
Introduction
The Sustainable Development Goals established by the United Nations constitute one of the challenges to be faced by agriculture by 2050.1 It consists of increasing plant productivity while reducing the use of agrochemicals. These two main goals are to improve plant nutrition and simultaneously optimize fertilization schemes and reduce the use of pesticides and herbicides as much as possible. This threefold challenge entails a better understanding of the molecular, physiological, and developmental plant mechanisms for optimizing nutrient uptake and use. In addition, this challenge is vertiginously raising interest in the use of biofertilizers and specifically learning about the role of microorganisms living associated with plant roots, with the aim of exploring their contribution to plant nutrition.
There are many beneficial microorganisms living around and inside plant roots, establishing intimate relationships in which a mutual positive effect is produced. Well-known beneficial microorganisms for legume nutrition involve N nutrition through nitrogen fixation processes carried out by bacteria. Mycorrhiza are also well known for improving Pi absorption by plants. However, not so much is known about the effect of soil and endophytic microorganisms on the nutrition of K+ in plants.
Potassium is the most abundant cation in plant cells, where it reaches cytoplasmic concentrations ranging between 80 and 100 mM to fulfill diverse essential physiological functions (Leigh and Wyn Jones, 1984; Marschner, 1995; Rodríguez-Navarro, 2000). To ensure this cellular concentration, plants take up K+ from the soil solution by root epidermal and cortical cells though K+ transport systems, among which HAK transporters and AKT1 channels are the main systems for K+ uptake in the high-affinity range of concentrations, the former being strongly induced under K+ limiting conditions (Nieves-Cordones et al., 2016). High-affinity transporters are generally referred to as those transporters that uptake K+ when it is available at low concentrations, although this “affinity concept,” which has been used for the classification of transporters based on enzyme kinetics, has recently been questioned (Dreyer and Michard, 2020).
Pioneer studies on the putative role of symbionts in plant K+ nutrition have been conducted in the ectomycorrhizal Hebeloma cylindrosporum and Pinus pinaster (Corratgé et al., 2007; Garcia et al., 2014; Guerrero-Galán et al., 2018). Other studies carried out on the symbiosis between Medicago truncatula and the arbuscular mycorrhizal (AM) association with Rhizofagus irregularis, have proposed that AM provides some tolerance to K+ deprivation in host plants (Garcia et al., 2017). So far, the most significant effect of the mycorrhizal pine plants on improving plant growth under K+ limiting conditions has been observed when the fungal K+ transporter named HcTRK1 was overexpressed (Garcia et al., 2014). In any case, these studies focused on unraveling the fungal and plant K+ transportome to identify those transporters that have a leading role in capturing the nutrient from the soil and in the exchange between the two partners (Garcia and Zimmermann, 2014; Garcia et al., 2016; Liu et al., 2019).
Regarding fungal K+ transporters, there are two highly conserved families of proteins named TRK and HAK transporters that are involved in K+ acquisition (Benito et al., 2011). TRK transporters are H+/K+ symporters, although they eventually can co-transport Na+ with K+ (Corratgé et al., 2007), whereas most of the HAK transporters identified so far have shown a high affinity only for K+ (Bañuelos et al., 1995). Fungi can be equipped with more than one mechanism of transport, and more than one representative may be present within a family of transporters. In examples of fungi in which TRK and HAK proteins are present, both systems have finely coordinated functions depending on growth conditions (Cabrera et al., 2012). Kinetic studies of these transporters indicate that HAK1 shows a high affinity for K+ and is strongly inhibited by NH4+ and Cs+ (Bañuelos et al., 1995; Nieves-Cordones et al., 2016), while the K+ affinity of TRK1 is two orders of magnitude lower and is not inhibited by these cations. Furthermore, TRK1 transporters are not transcriptionally regulated, while HAK1 transporters are strongly induced in response to very low K+ concentrations and are negatively regulated by the presence of K+ (Haro et al., 1999; Cabrera et al., 2012; Rivetta et al., 2013). With K+ transport function but opposite directional flow, there are other proteins in fungal membranes that mediate K+ efflux from cells. Among other proteins, TOK and SKC (the latter only identified so far in the basidiomycetes and basal fungi), are K+ channels that can cover this function (Casieri et al., 2013; Garcia et al., 2016). Most of the information currently available on the function of K+ transporters families in fungi has been obtained from non-symbiotic fungi, and information on the functional characterization of K+ transporters in fungi associated with plants is scarce, especially among endophytic fungi.
To gain insight into whether endosymbiotic fungi have any impact on the K+ nutrition of plants and to find the K+ transportome with which this group of microorganisms are equipped, the basidiomycete Serendipita indica was chosen for this study.
Serendipita indica (formerly named Piriformospora indica) is a promiscuous endosymbiotic fungus that colonizes a wide range of plant species, from monocots to dicots, and promotes their growth and fitness. It has demonstrated beneficial effects on plant growth under both biotic and abiotic conditions such as drought and saline conditions (Waller et al., 2005; Ghaffari et al., 2016; Jogawat et al., 2016; Abdelaziz et al., 2017; Li et al., 2017; Zhang et al., 2018; Ghorbani et al., 2019). However, although recently progress is being made in the knowledge of the nutrition of certain micronutrients such as magnesium, iron and sulfur (Prasad et al., 2019; Narayan et al., 2021; Verma et al., 2021), there is not much information available on the contribution of this endophyte to the plant nutrition of the main macronutrients, as previous studies to determine its role in plant P nutrition have obtained contradictory results. Although there are data indicating that S. indica does not play any role in the supply of Pi in the symbiosis with barley (Achatz et al., 2010), it has been found that S. indica enhances plant growth by transferring phosphate to host maize plants (Yadav et al., 2010). In Arabidopsis, it has been shown that the fungus activates a variety of plant genes under low P and promotes plant growth and P uptake, which is not mediated by plant transporters of P (Bajaj et al., 2018).
In this article, we studied the K+ nutrition of S. indica in free living conditions and examined whether it contributes to the K+ nutrition in host plants. For this purpose, we used Arabidopsis thaliana as the host plant. We also investigated the function and putative contribution of fungal K+ transport systems for fungal and plant K+ supply. The results indicated that under K+ limiting conditions, the fungus had no impact, neither on host fitness nor on plant K+ content, in contrast to the clear improvement of plant growth observed when Arabidopsis plants were infected under P limiting conditions. Instead, symbiosis may benefit the fungus in term of guaranteeing its K+ supply obtained from the plant.
Materials and Methods
Plant, Fungi, Bacteria, and Yeast Strains
Arabidopsis thaliana accession Col-0 was used in this study together with the derived hak5 and hak5/akt1 mutant plants, that have been published previously (Rubio et al., 2010a). Serendipita indica strain DSM 11827, DSMZ, Braunschweig, Germany, was also used throughout this study. The Escherichia coli strain DH5α was routinely used for the propagation of plasmids. In addition, the Saccharomyces cerevisiae strain WΔ6 (Mat a ade2 ura3 trp1 trk1Δ:LEU2 trk2Δ:HIS3), which is deficient in the endogenous K+ uptake systems TRK1 and TRK2 (Haro and Rodríguez-Navarro, 2003) was used for complementation studies and K+ uptake kinetics. Arabidopsis seeds were surface sterilized for 5 min in 5% sodium hypochlorite and subsequently washed thrice with sterile water. After stratification for 2 days at 4°C in the dark, the seeds were germinated on MS-agar plates adjusted to pH 5.7 placed vertically for 1 week. The plants were grown under control conditions in a greenhouse with an 16 h light/8 h dark regime at 23°C. S. indica was grown on arginine phosphate (AP) medium pH 6.5 (Rodriguez-Navarro and Ramos, 1984) supplemented with 2% glucose and the indicated K+ concentrations.
K+ Starvation of Serendipita indica in Free Living Conditions
For experiments carried out in free living conditions, 1.5 × 103 spores were inoculated in 100 ml of liquid AP medium with 1 mM K+ and the suspension was incubated at 28°C for 1 week. After that, the mycelium was centrifuged, and transferred to fresh AP medium without KCl in which it was incubated for another week. During this time, a clear growth of the fungus was observed forming clumps and small cottony balls. After that, the K+-starved fungal culture was used to perform high-affinity K+ uptake kinetics or for total RNA extraction.
K+ Starvation of Infected Plants
Several pools of twenty Arabidopsis seeds were first germinated in MS medium for 1 week. After that, the seedlings grown in pools were transferred to sterile square petri dishes containing modified Hoagland agar medium adjusted to pH 5.7 (Álvarez-Aragón et al., 2016) lacking KCl and glucose. One week later, pools of roots were inoculated with 250 μl of a chlamydospore suspension which contained 1.5 × 106 ml–1. Non-inoculated plants were grown under the same conditions as control. The incubation of plants was performed by placing the plates vertically in a Conviron phyto-chamber with a day/night cycle of 16/8 h (light 21 intensity: 110 μmol m–2 s–1) at a temperature of 23°C. After 1 week of co-cultivation, the plants were dissected into roots and shoots and used for cation content determination. Alternatively, roots of K+-starved plants were used to perform high-affinity K+ uptake kinetics.
For the extraction of total RNA from roots, the seeds were sown directly on modified Hoagland medium containing the K+ concentration specified in each case. After 1 week, the plants were inoculated with spores as described above and 2 weeks later, the root were harvested and frozen at –80°C until processing. Three replicates were obtained for each growth condition. In all the experiments, the fungal colonization of the roots was confirmed by trypan blue staining.
K+ Uptake Experiments and Ion Content Measurements
High-affinity K+ transport of S. indica was assessed in K+-starved fungal cultures by measuring K+ or Rb+ (used as the K+ analog tracer) disappearance from the external medium. For that, fungal cultures were initially grown for 1 week in AP medium with 1 mM KCl. After that, the mycelium was transferred to AP medium and incubated for another week in the absence of K+. After this week of K+-starvation, cultures were centrifuged, washed twice with Milli Q water and resuspended in 50 ml fresh AP medium without K+. At zero time, 50 μM KCl was added, alone or in the presence of the respective inhibitors assayed and the culture was maintained at 28°C and constant shaking. At intervals, 1 ml samples of the medium were taken, centrifuged and 500 μl of the supernatant were used for K+ determinations.
For K+ transport kinetics carried out with the yeast mutant expressing the S. indica SiHAK1 or Neurospora NcHAK1 cDNAs, the transformants were previously grown overnight in AP medium with 1 mM K+ at 28°C with vigorous shaking. At OD550nm of 0.2 approximately, cells were centrifuged, washed twice with MilliQ water and resuspended in AP medium in the absence of K+. After 4 h of K+ starvation, cells were centrifuged, washed and resuspended in AP medium in the absence of K+. At time point zero, 100 μM KCl was added, and at intervals 1 ml samples of the medium were taken, centrifuged and 500 μl of the supernatant were used for K+ determinations.
High-affinity K+ transport kinetics of Arabidopsis were carried out with previously K+-starved plants obtained as described above. Cut roots were then washed with MES-Ca2+ pH 6.5 and resuspended in 10 ml of 10 mM MES-Ca2+ and they were maintained at RT with shaking. At zero time, 50 μM RbCl was added and 1 ml samples of the buffer were taken at intervals to determine the disappearance of Rb+.
Analysis of the internal cations contents of plant samples was conducted in fractions of biological material, roots and shoots. For roots, they were previously washed in 10 mM MES-Ca2+ pH 6.5 and both, roots and shoots were dried, weighed and extracted with 1 M HNO3. The K+ and Rb+ contents of the supernatants, fungal and plant extracts were determined by atomic emission spectrophotometry. The results are reported as the mean values of at least four experiments.
Combinations of K+ and Pi Starvation of Infected Plants
To study the ability of S. indica to enhance the growth of Arabidopsis at varying K+ and Pi concentrations, seeds of Col 0 and hak5/akt1 mutant, were germinated directly in agar plates with modified Hoagland medium containing the corresponding salt concentration of the respective nutrients. The K+ concentrations tested were zero and 1 mM and the phosphate concentrations were zero and 0.2 mM. In the growth conditions in which the K+ concentration was zero, the double mutant hak5/akt1 did not germinate.
Analysis of Root Colonization of Serendipita indica by Trypan Blue Staining
To check fungal root colonization, five root samples were randomly selected from the plants. Samples were softened in 10% KOH solution for 15 min, acidified with 1 N HCl for 10 min, and finally stained with 0.02% Trypan blue overnight (Jogawat et al., 2016). The samples were de-stained with 50% lacto-phenol for 1–2 h, followed by 3–4 washes with 100% ethanol and suspended in 60% glycerol prior to their inspection under a light microscope.
Visualization of the Serendipita indica GFP Strain by Confocal Laser Scanning Microscopy
To study the level of fungal root colonization at different K+ conditions, a GFP expressing S. indica strain kindly provided by Dr. Alga Zuccaro was used (Hilbert et al., 2012). Sterilized Arabidopsis seeds were sown on cellophane in square plates of modified Hoagland medium in the absence of K+ or in the presence of 1 mM KCl and maintained for 7 days. After that, the roots were inoculated with spores of the S. indica strain that expresses GFP and the plates were incubated for 3 and 7 days more. The green fluorescence corresponding to the fungus was visualized by exciting at 488 nm. Pictures were taken with a Zeiss LSM 880 Airyscan confocal microscope.
Fungal and Root RNA Extraction, Obtaining of Full-cDNAs and qRT-PCR Analysis
For gene expression of the S. indica encoding K+ transporters, fungal samples, with a mixture of mycelia and chlamydospores, or fungal-inoculated plant root samples, were crushed to a fine powder in liquid nitrogen with mortars and pestles. For gene expression analysis of Arabidopsis HAK5, inoculated or non-inoculated plant root samples were also used. Total RNA was extracted with TRIzol reagent (Invitrogen) according to the manufacturer’s instructions, and RNA was purified by passage through Nucleospin RNA Plant columns (Macherey-Nagel). Fungal samples were obtained from S. indica grown for 1 week in liquid AP medium with 1 mM KCl and another additional week in AP medium at the indicated K+ concentration. Fungal-inoculated plant root samples were obtained after 2 weeks of inoculation. The first 4–5 cm of the roots from the seed were excised and immediately frozen at –80°C in liquid nitrogen. qRT-PCR was performed with the One-step TB Green Primescript RT-PCR kit (Perfect Real Time, TAKARA) following the manufacturer’s recommendations. Transcription levels of the Serendipita and Arabidopsis genes were expressed by relating the amount of target transcript to SiTUBULIN (Protein ID in Mycocosm: 76292; mRNA:PIIN_05487) or AtUBIQUITIN10 (At4g05320) as Serendipita and Arabidopsis reference gene, respectively. In order to compare the gene expression in all samples, the ratio cDNA molecules of any transporter encoding gene/cDNA molecules of tubulin has been considered. Standard curves were used to calculate the absolute numbers of each cDNA molecules in each cDNA sample, and these values were then normalized against the corresponding tubulin signals. Standard curves for each gene were obtained from dilution series of known quantities of cDNA fragments used as templates. The normalization factor was calculated assuming that Ct is inversely proportional to the logarithm of the quantity of target cDNA present at the PCR start (Cuéllar et al., 2010). The primers used in this work are listed in Supplementary Table 1. Full-length cDNAs were obtained by using the 5′/3′ RACE Kit (Roche) according to the manufacturer’s instructions.
Statistical Analyses
Significant differences in plant growth or in K+ content was determined by Student’s test. Gene expression data were analyzed by one-way ANOVA test using Graphpad Prism 9 (2021). Tukey test (P < 0.05) was used to pairwise multiple comparisons between the growth conditions.
Results
Growth of Arabidopsis Plants Inoculated With Serendipita indica in K+ Limiting Conditions
In order to know whether the colonization of roots by S. indica has any effect on plant growth under K+ starving conditions, 1-week-old Arabidopsis seedlings were grown in agar plates of modified Hoagland medium in the absence of K+ for 1 week. Thereafter, plant roots of half of the plates were inoculated with a chlamydospore suspension (1.5 × 106 spores ml–1) and the rest of the plates were maintained without inoculation as control. In parallel and for comparison, the same experiment was carried out with plants grown in the presence of 5 mM KCl. To estimate the plant growth under the different conditions in the test, the dry weight of roots and shoots was measured after 1 week of inoculation. The results showed that while under normal K+ conditions, co-cultivation with the fungus significantly improved plant growth, under K+ limiting conditions growth was clearly reduced and the reduction was similar for inoculated and non-inoculated plants (Figure 1A). To find out if in a more K+ restrictive condition of plants is needed to observe a fungal beneficial effect on plant growth, the single and double Arabidopsis mutants hak5, akt1 and hak5/akt1 were used. These mutants are defective in their main high-affinity K+ transporters and their growth at low K+ is seriously compromised. The results confirmed that under K+-starved conditions these mutants grew worse than the wild type and no growth promotion was observed when these mutants were inoculated with the fungus (Supplementary Figure 1). The K+ content of the plants grown under the different conditions was also determined. As it has already been described in the literature, under conditions of K+ deficiency in non-inoculated plants, a clear depletion of the internal K+ was observed, in both roots and shoots, and surprisingly, the co-cultivation with S. indica not only did not increase the K+ of the plant but also produced a significant decreased in the K+ accumulation in both tested plant parts (Figure 1B). When plants were grown under normal K+ conditions, a significant decrease was also observed in the K+ content of plant roots while no changes were detected in the aerial part of the plants. Similar results were obtained when the K+ content was determined in the mutants (Supplementary Figure 1). These results indicated that S. indica neither improves the growth nor the K+ content of the plants when they grow under K+ restrictive conditions, which suggests that the fungus does not participate in the K+ nutrition of the plant.
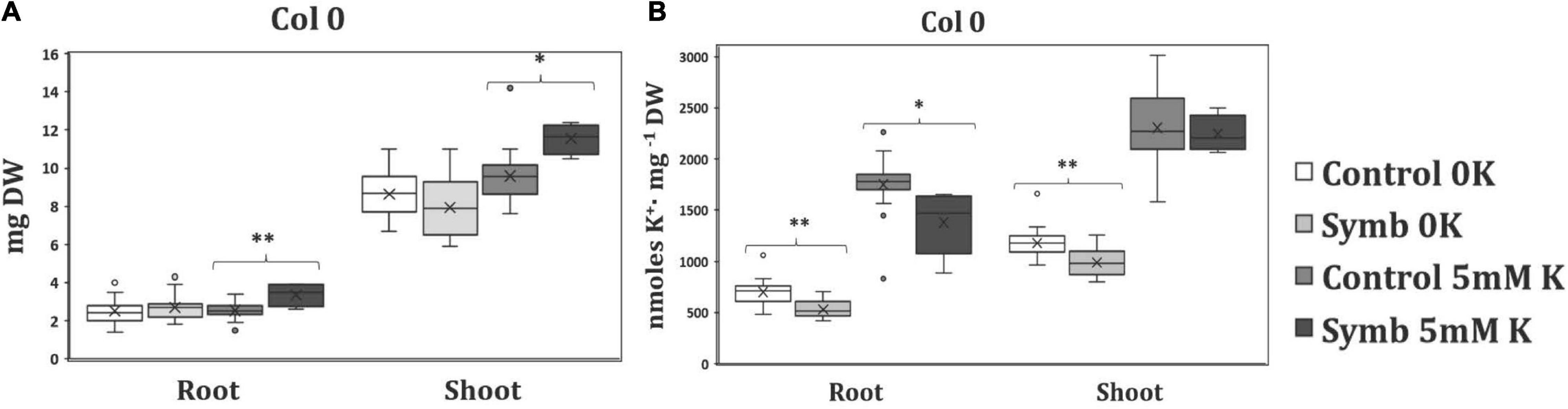
Figure 1. (A). Dry weight measurements of roots and shoots of Arabidopsis plants grown at different K+ concentrations in the presence (Symb) and absence (Control) of Serendipita indica. The plants were grown in agar modified Hoagland medium with 5 mM KCl (5 mM) or under K+ starvation (0K). (B) K+ content of roots and shoots of infected and non-infected plants growing as described in (A). Data represents the means of eighteen values ± SD. Different letter indicates significant differences between conditions (P-value; *P < 0.05, **P < 0.01) based on Student’s t-test.
High-Affinity K+ Transport of Infected Arabidopsis Plants
To study more precisely whether there is any positive effect of the fungus at the plant root level, which is where it colonizes, the kinetics of high-affinity K+ transport of infected and non-infected plants was determined. For this, roots of K+ starved plants (see section “Materials and Methods”) inoculated with the fungus were exposed to 50 μM RbCl, an analogous tracer of K+, and the disappearance of Rb+ from the external medium was determined. As shown in Figure 2, in non-colonized roots a decrease in external Rb+ was observed throughout the time of the experiment and this decrease was significant less in colonized roots. These results indicate that the colonization of roots by S. indica does not have any clear-cut improvement on the K+ absorption from the medium, suggesting that the Rb+ uptake that took place along the experiment would be mainly a consequence of the activity of the plant endogenous high-affinity transport systems expressed in roots (Figure 2).
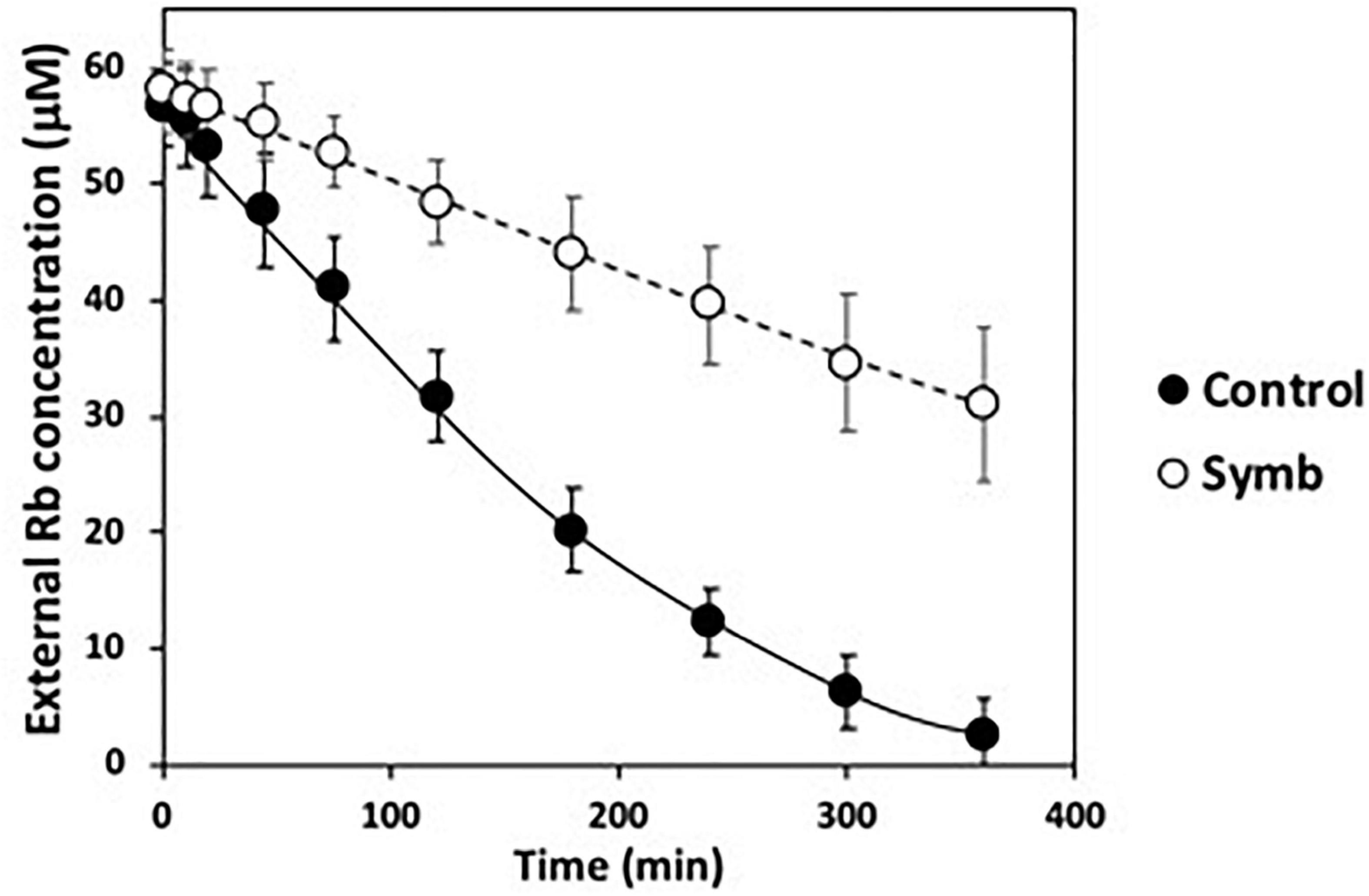
Figure 2. High-affinity Rb+ transport of Arabidopsis roots tested alone (black circles) or in symbiosis with Serendipita indica (white circles). Cut roots of 2-weeks K+ starved plants where resuspended in MES-Ca buffer to which RbCl was added. At intervals, the Rb+ disappearance from the external medium was determined.
Physiological Characterization of High-Affinity K+ Uptake of Serendipita indica in Free Living Conditions
As other endophytic fungi, S. indica also grows under axenic conditions, independent of the plant partner. To investigate how the fungus ensures its own K+ nutrition in free living conditions, it was grown on agar plates with AP minimal medium at different K+ concentrations (Figure 3A). The results showed that a clear fungal growth inhibition took place when exposed to moderate K+ concentrations such as 25 mM KCl, presenting a complete inhibition of growth at concentrations reaching 200 mM KCl (Figure 3A). This was a striking performance because these are K+ concentrations at which other fungi normally grow without restrictions (Jones and Jennings, 1965; Yenush, 2016). On the contrary, S. indica was able to grow at low K+ concentrations such a 50 μM KCl (Figure 3A), and even lower, when no KCl was added to the medium and only traces of K+ were contained (not shown). These latter results suggested that S. indica, as other fungi, may possess a K+-transport system that ensures the K+ uptake when K+ availability is minimal. To study the ability of S. indica to take up K+ at low concentrations, a K+-starved fungal culture was exposed to 50 μM KCl and the K+ disappearance from the external medium was determined. Figure 3B shows that S. indica was able to take up K+ in such a way that, after 30 min almost no K+ was left in the medium indicating that S. indica indeed possesses a high-affinity K+ transport system.
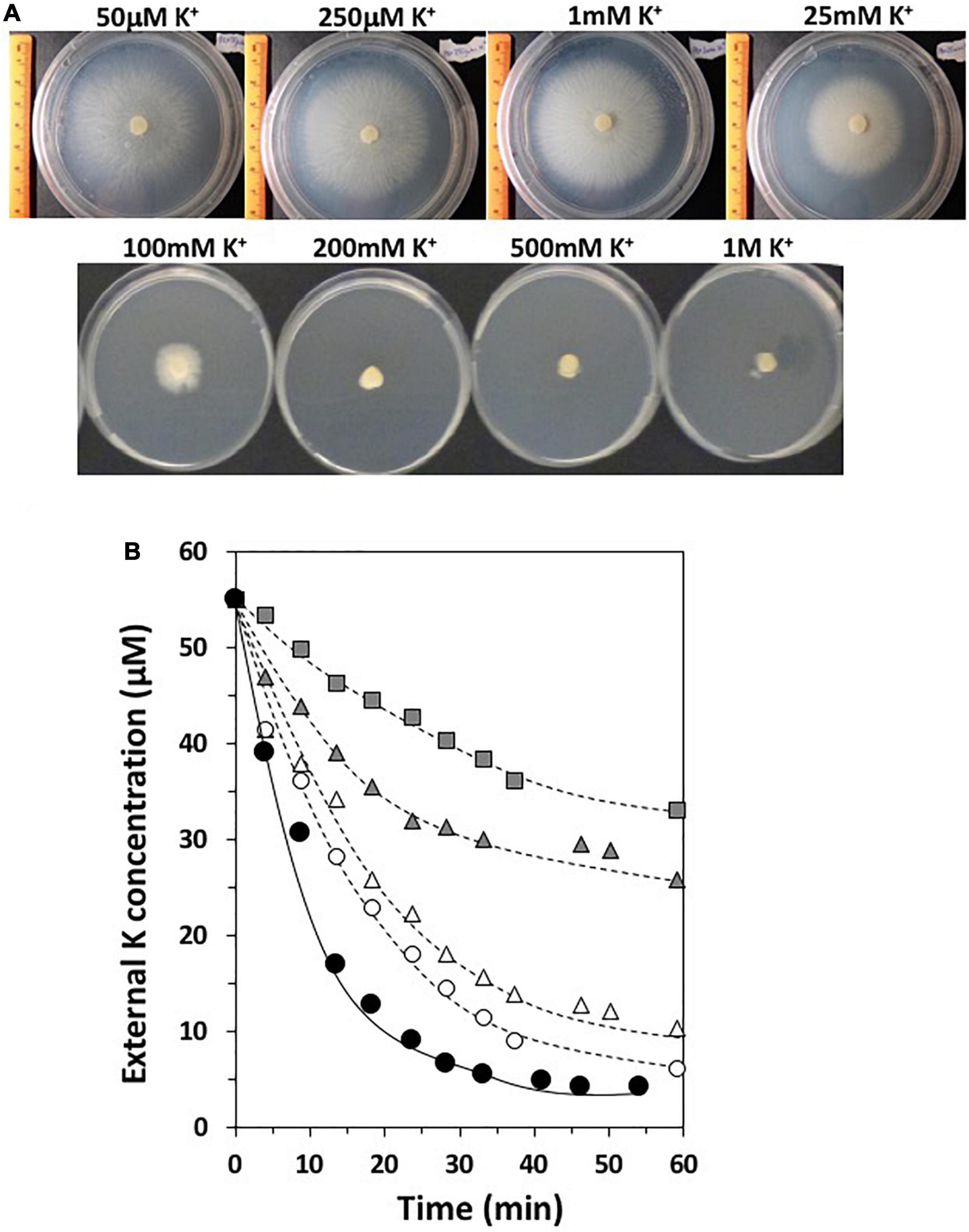
Figure 3. (A) Growth of Serendipita indica in free living conditions in AP medium at different K+ concentrations for 1 week. (B) High-affinity K+ transport of Serendipita indica. The disappearance of external K+ concentration of a fungal culture in the presence or absence of different inhibitors is represented: KCl alone without inhibitors (black circles); 5 mM BaCl2 (white circles); 5 mM NaCl (white triangles); 5 mM NH4Cl (gray triangles); 2 mM CsCl (gray squares).
In order to obtain further evidence regarding putative K+ transport systems which may operate under low K+ conditions, K+ transport was studied in the presence of known inhibitors for K+ transporters and channels described in the literature (Rubio et al., 2010b). Thus, the K+ disappearance at 50 μM was studied in the presence of NH4+ and Cs+, which are known inhibitors for HAK type-K+ transporters, Na+, which inhibits HAK and some TRK transporters, and in the presence of Ba2+, which inhibits K+ channels. The results showed that most of the high-affinity K+ transport tested was primarily inhibited by NH4+ and/or Cs+, suggesting that an HAK transporter may be involved in the high-affinity transport of this fungus (Figure 3B).
Fungal K+ Transport Systems Identified in the Serendipita indica Genome
To elucidate the putative K+ transporters that might be involved in K+ absorption and intracellular accumulation in S. indica, an in silico BLASTP search was performed. By using amino acid sequences of known K+ transporters of other fungi as queries, we identified homologous sequences to all of them in the S. indica genome.2 Genomic analysis revealed that S. indica has one gene encoding a protein homologous to fungal HAK transporters (PIIN_08798, Protein ID: 79610), three genes encoding proteins homologous to fungal TRK transporters (initially named TRK1, PIIN_00119, Protein ID: 70870; TRK2, PIIN_06219, Protein ID: 77027; and TRK3, PIIN_06700, Protein ID: 77504) and one gene encoding a TOK-like K+ channel (SiTOK1, PIIN_03455, Protein ID: 74258). In other fungi, all of these proteins are localized in the plasma membrane so that they may be good candidates to mediate the uptake and/or exchange of K+ from/to the soil and from/to the plant. Apart from these genes, one gene encoding a homologous protein to the basidiomycete SKC channels was also identified (PIIN_07335, Protein ID: 78142). Figure 4A shows an unrooted phylogenetic tree that includes representatives of known fungal K+ transport systems.
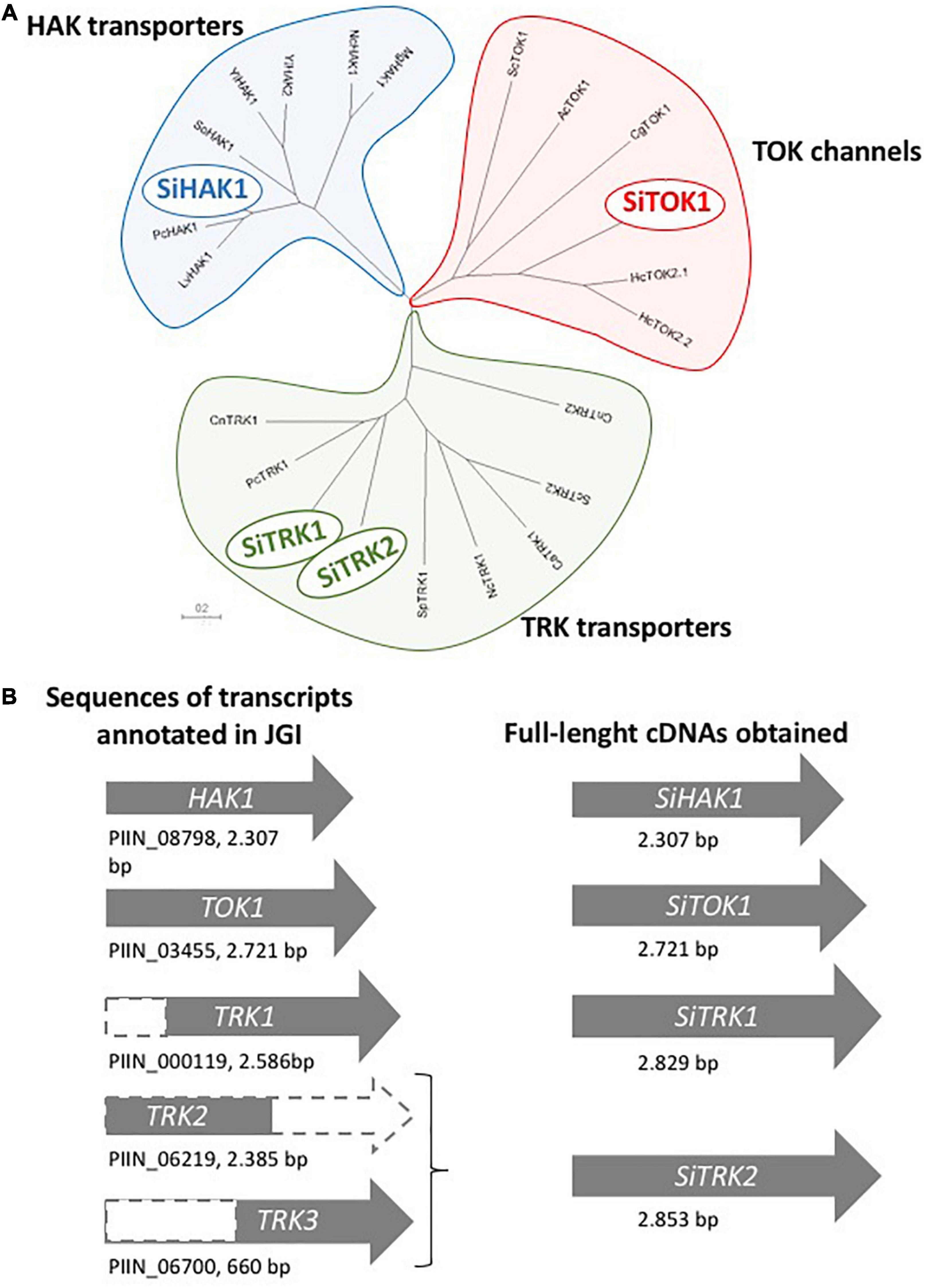
Figure 4. (A) Phylogenetic tree of the amino acid sequence of the K+ transport systems of Serendipita indica. The protein representatives of the different families of fungal K+ transporters and channels included in the tree were (name in the tree: species, protein ID): HAK transporters, LvHAK1: Laccaria bicolor, XP_001876329.1; PcHAK1: Phanerochaete sordida, GJE85382.1; SiHAK1: Serendipita indica, CCA74829.1; SoHAK1: Schwanniomyces occidentalis, P50505.2; YlHAK1: Yarrowia lipolytica, XP_503821.1; YlHAK2: Y. lipolytica, XP_501676.1; NcHAK1: Neurospora crassa, XP_964946.1; MgHAK1: Magnaporthe grisea, XP_003708896.1; TRK transporters, CnTRK1: Cryptococcus neoformans, XP_012047874.1; PcTRK1: P. sordida, GJE91363.1; SiTRK1: S. indica, CAG7854666.1; SiTRK2: S. indica, CAG7854847.1; SpTRK1: Schizosaccharomyces pombe, NP_593934.1; NcTRK1: N. crassa, XP_957340.1; CaTRK1: Candida albicans, AAF72203.1; ScTRK2: Saccharomyces cerevisiae, NP_012976.1; CnTRK2: C. neoformans, XP_012048291.1; TOK proteins, ScTOK1: S. cerevisiae, NP_012442.1; AcTOK1: Aspergillus clavatus, XP_001268834.1; CgTOK1: Cryptococcus gattii, KIR46677.1; SiTOK1: S. indica, CAG7847745.1; HcTOK2.1: Hebeloma cylindrosporum, KIM42222.1; HcTOK2.2: H. cylindrosporum, KIM42108.1. (B) Schematic representation of the transcript sequences annotated in the JGI encoding for the K+ transporters and the products obtained by 5′ RACE and the nested PCR strategy using oligo-dT and specific primers for TRKs.
A thorough comparison of the sequences of the three translated S. indica TRK proteins with other fungal TRK proteins showed that the sequences of the TRK1, TRK2, and TRK3 genes annotated in the genome were truncated versions. The TRK1 and TRK3 genes lack part of the 5′-end, and TRK2 has shorter sequence at the 3′-end (see Figure 4B). To identify the correct full length of the corresponding cDNAs, 5′RACE PCR was performed from RNA of K+ starved cells of S. indica. In the case of SiTRK1, the synthetized full-length cDNA was 243 base pairs longer than the annotated sequence. In the case of TRK2 and TRK3, a single cDNA was obtained whose sequence at the 5′ end corresponded to annotated sequence of TRK2 and at the 3′ end corresponded to that of TRK3. Therefore, this full-length cDNA was renamed and, from now on, is termed SiTRK2. An exhaustive analysis in the genome showed that the TRK3 gene sequence was located at one end of a contig (PIRI_contig_0186:39283-40208) which could explain the incorrect annotation of the SiTRK2 gene since the ends of contigs are sometimes difficult to sequence. In any case, it cannot be ruled out that another SiTRK2 cDNA sequence different from the one obtained in this work may be transcribed under other conditions of growth of the fungus.
Cloning of K+ Transport Systems of Serendipita indica and Functional Characterization by Heterologous Expression in Yeast Mutants
To functionally characterize the S. indica proteins, the respective cDNA sequences of the SiHAK1, SiTRK1, SiTRK2, and SiTOK1 genes were cloned into the pYPGE15 vector which facilitates the expression under the control of the constitutive PGK1 promoter (Brunelli and Pall, 1993) in WΔ6 yeast mutants. This mutant lacks its endogenous TRK1 and TRK2 K+ uptake transport systems, which makes it especially sensitive to grow at low K+ concentrations (Haro and Rodríguez-Navarro, 2003). The drop test analysis showed that the expression of all the cDNAs suppressed the defective growth of the yeast mutant at low concentrations of KCl but to different extents. While the growth of transformants expressing SiTOK1, SiTRK1, and SiTRK2 was very subtle, SiHAK1 expression reverted the defect of mutant growth at a concentration as low as 50 μM KCl (Figure 5A). In fact, a better growth was observed at 50 μM K+ compared to 500 μM suggesting a better activity of this transporter in the high-affinity range. In parallel to the expression of the S. indica transporters, the NcHAK1 cDNA encoding the homologous HAK protein of the soil fungus Neurospora crassa (Haro et al., 1999) was also tested, showing a stronger complementation, which may probably correlate with a better transport activity (Figure 5A).
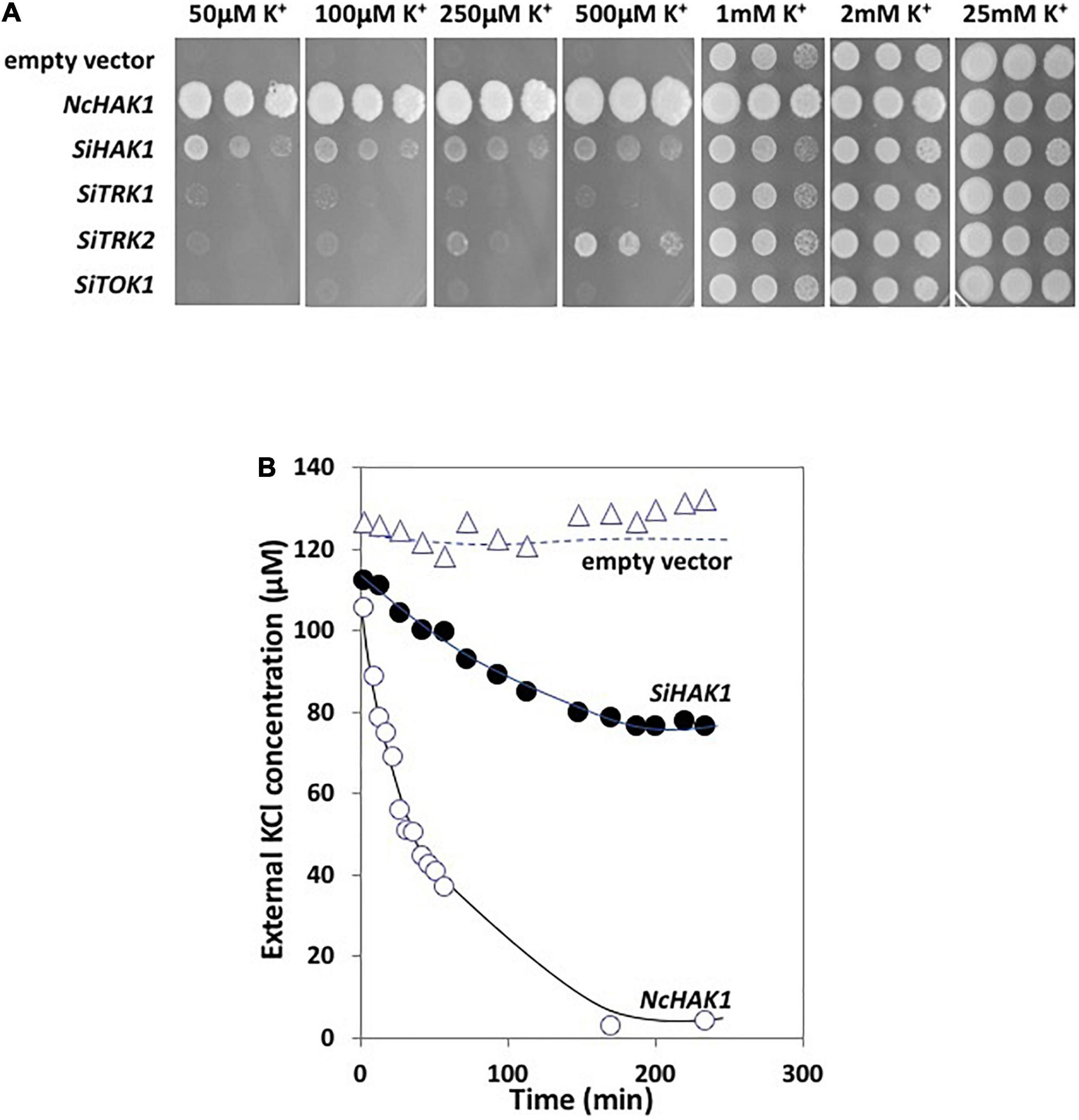
Figure 5. (A) Drop test assay for the complementation of the WΔ6 yeast mutant transformed with the different cDNAs encoding the fungal K+ transport systems. The growth of the transformants were tested in AP medium in the presence of variable KCl concentrations. The four first panels show the fungal growth of plates incubated for 5 days at 28°C to better visualize the drops, the three last panels were incubated for 3 days. (B) High-affinity K+ transport in WΔ6 yeast mutants expressing SiHAK1. As controls, the K+ transport of the transformant expressing NcHAK1 from Neurospora crassa or with the empty vector were also performed.
The kinetic analysis of K+ uptake of yeast transformants expressing the fungal HAK proteins corroborated the differences in growth observed in the drop test since the NcHAK1 transformants showed a very rapid depletion of the external K+ in the medium, while a slower K+ uptake was observable for the SiHAK1 expressing transformants (Figure 5B).
Expression of K+ Transport Systems of Serendipita indica in Free Living Conditions and During Symbiosis
High levels of expression of a certain gene could be indicative of the condition in which the activity of the encoded protein can play a relevant role. To elucidate the contribution of each of the S. indica K+ transporters, an expression analysis of corresponding genes was undertaken, contrasting gene expression in the fungus in the presence of normal K+ conditions (1 mM KCl) and under K+ starvation. In addition, their expression was also studied during the symbiosis with Arabidopsis. The results indicated that SiTRK1 was the gene that showed the highest expression level in any condition. The expression was almost constitutive since no significant differences in expression were observed between free life and symbiosis, or between the two external K+ concentrations tested (Figure 6). In contrast, SiTRK2 and SiTOK1 genes were expressed at very low levels and SiTOK1 did not respond significantly to K+ depletion. On the contrary, SiHAK1 was strongly induced under K+ starvation conditions (Figure 6). In free living conditions at normal K+, SiHAK1 expression was low, but it was highly induced under K+ restriction. When grown in symbiosis under normal K+ conditions, SiHAK1 showed a higher level of expression than at the same K+ concentration in free living conditions, and when plants were subjected to K+ starvation, the induction of SiHAK1 was also observed although it was lower than that observed in free living conditions (Figure 6). This
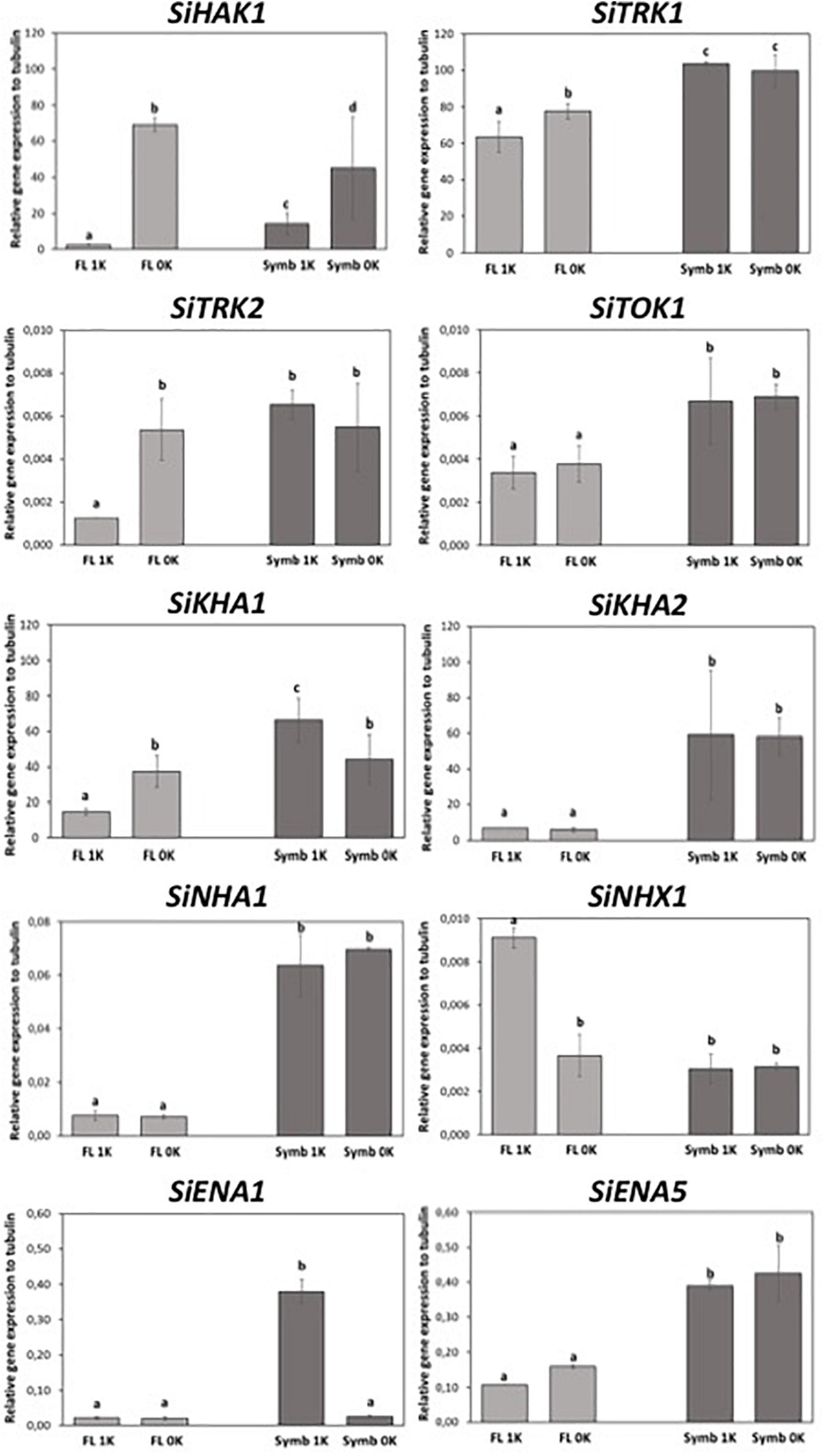
Figure 6. Expression of the genes encoding the different K+ transport systems of Serendipita indica in free living conditions (FL) or during symbiosis (Symb) at different external K+ concentrations, either 1 mM KCl (1K) or under K+ starvation (0K). Data were statistically processed by applying one-way ANOVA test. Different letter indicates statistically significant differences between conditions (P < 0.05) based on Tukey test.
indicates that during symbiosis the fungus does not reach the same level of K+ restriction than when living alone.
Besides the K+ uptake systems studied in this work, all fungi have other transporters that contribute to the K+ efflux, translocation and accumulation across the plasma and internal membranes of fungal cells and to know their gene expression could be useful to unravel their contribution on K+ homeostasis, either in free living conditions or under symbiosis. With the aim of identifying the genes existing in S. indica, a BLASTP search with known fungal proteins in S. indica genome was performed (see Supplementary Figure 2). In order to monitor their expression in our tested conditions, oligonucleotide primers were designed, and qRT-PCR assays were performed. The expression analysis showed that SiKHA1 and SiKHA2, two genes encoding transporters that are generally located in Golgi and internal vesicles (Maresova and Sychrova, 2005), showed a relatively high level of expression in both lifestyles, but were especially induced in symbiosis, regardless of the K+ concentrations of the medium (Figure 6). Other genes whose expression was studied were SiNHA1 and SiNHX1, both encoding Na+/K+-H+ antiporters that are typically located in the plasma membrane and in the tonoplast or in vesicle membranes, respectively (Nass et al., 1997; Bañuelos et al., 1998). Their expression was very low under all tested conditions. However, it was possible to observe an induction of SiNHA1 triggered by symbiosis, in both K+ conditions. In addition, we observed the induction of SiNHX1 in free living conditions at the highest K+ concentration (Figure 6). Finally, two other genes whose expression was studied in these K+ conditions were SiENA1 and SiENA5. These two genes encode for two plasma membrane P-type ATPases involved in Na+ and/or K+ efflux, and specifically SiENA1 has been shown to pump K+ out of the cells when are exposed to high K+ concentrations (Lanza et al., 2019). The expression analysis showed that under free-living conditions, SiENA1 was poorly expressed and non-responsive to K+ changes but under symbiosis, its expression was induced at the highest concentration of K+ tested (Figure 6). SiENA5 was somewhat stronger expressed compared to SiENA1 under free-living conditions, and it was clearly induced in response to symbiosis but showed no response to changes in K+ conditions (Figure 6).
Taken together, the S. indica gene expression results highlight that: (i) there are three genes whose expression is regulated by K+: SiHAK1 and SiTRK2, which are the only genes responsive to K+ restriction, with SiHAK1 being the one that shows the strongest induction; and SiENA1 which is induced at the higher K+ concentration in symbiosis; (ii) we identified a number of genes that were significantly and specifically induced under symbiosis such as SiKHA2, SiENA5, and SiNHA1 (they would be called “symbiosis responsive genes”). The role of these transporters deserves to be addressed in further studies.
HAK5 is known to be the main high-affinity K+ transporter in Arabidopsis whose expression and regulation has been fully characterized (Rubio et al., 2008), so it was interesting to study its expression under the symbiosis with S. indica. As it is already described, in non-infected plants AtHAK5 was induced under the K+ restrictive conditions (0K), but interestingly, the K+-restricted induction response was significantly higher in infected plants (Supplementary Figure 3). These results indicated that the presence of the fungus enhances the K+-starving response from AtHAK5 gene which would suggest that, under symbiosis in the K+ limiting conditions (which is the condition that took place in Figure 2), a stimulation of the K+ uptake should take place. As shown in Figure 2, this was not the case. The reason why higher AtHAK5 expression in infected plants does not translate into more active K+ transport deserves more research.
Serendipita indica Colonization of Arabidopsis Roots Is Enhanced in K+ Limiting Conditions
The results obtained so far indicated that the symbiotic association did not produce any positive effect on the growth or in the K+ reserves of the plant. However, it remained unclear as to whether the fungus would have any growth advantage when associated with the host plant under K+ restrictive conditions. To address this question, we studied the degree of fungal colonization of roots during K+ starvation and under normal K+ conditions. For that, a constitutive GFP-expressing S. indica strain (Hilbert et al., 2012) was used to infect plant roots and confocal microscopy to visualize the level of colonization. Interestingly, in plants grown under K+ limiting conditions, the roots appeared more densely colonized by hyphae, while in plants grown at 1 mM K+, less hyphae were observed surrounding and within plant roots (Figure 7), suggesting that K+ starvation favors root colonization. If, according to the results presented above, an improvement in the fitness of the plant has not been observed, this would suggest that with regard to the nutrition of K+, the fungus would be the only and clear beneficiary of the symbiotic association.
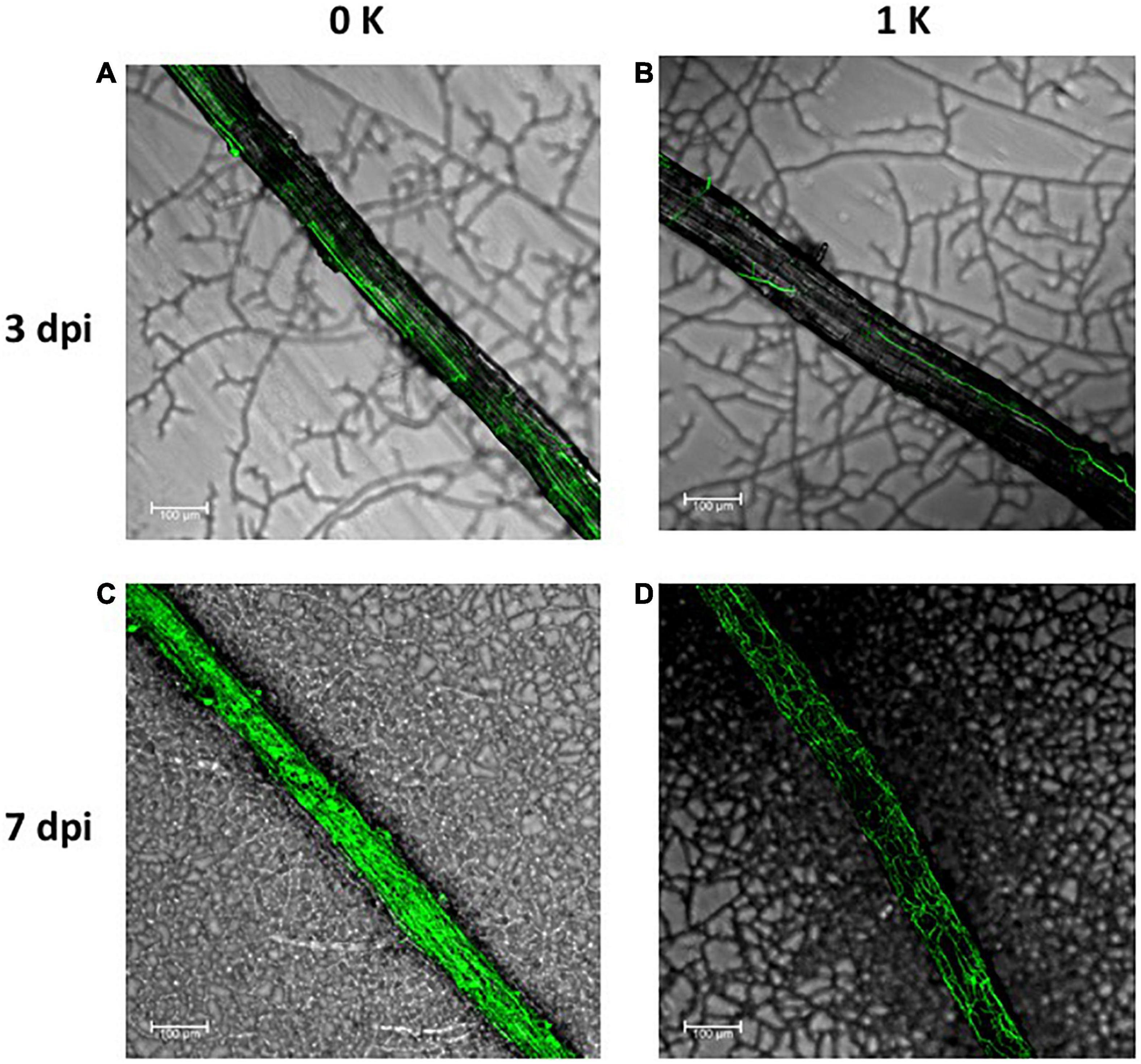
Figure 7. Serendipita indica root colonization of Arabidopsis plants grown in the absence of K+ (0 K) (A,C) or in the presence of 1 mM K+ (1K) (B,D). GFP expressing fungal hyphae were visualized in the confocal microscope after 3 (A,B) and 7 (C,D) days post infection. For comparison, micrographs were taken in the same region of the root with respect to the inoculation site of the fungus. The bars represent 100 μm.
Serendipita indica Improves Plant Growth on P Deprivation but Not on K+ Deprivation
It has been previously described that S. indica improves Arabidopsis growth under low Pi conditions (Bajaj et al., 2018), which would mean that, although Pi and K+ are both essential, the fungus would behave very differently in the nutrition depending on the nutrient. To confirm this claim, we compared the effect of fungus co-cultivation on plant growth under our experimental conditions, either without Pi or K+ added to the medium. As it is shown in Figure 8, the absence of Pi produced detrimental plant growth compared to plants grown in control conditions (i.e., with 1 and 0.2 mM P, which are their respective concentrations in the Hoagland medium used). However, plant growth was promoted by the fungal inoculation. Even when Pi and K+ restriction were applied together, a beneficial effect of fungal co-cultivation was observed on plant growth, although under these conditions the plants were more affected. In contrast, without K+, the plant growth was reduced, and fungal inoculation did not produce any appreciable improvement in plant growth. Similar results were obtained when the double mutant hak5/akt1 mutant was tested (Figure 8). Overall, it may be concluded that S. indica does not exert a general beneficial effect on plant nutrition but it appears to be specific for Pi supply under restrictive nutritional conditions.
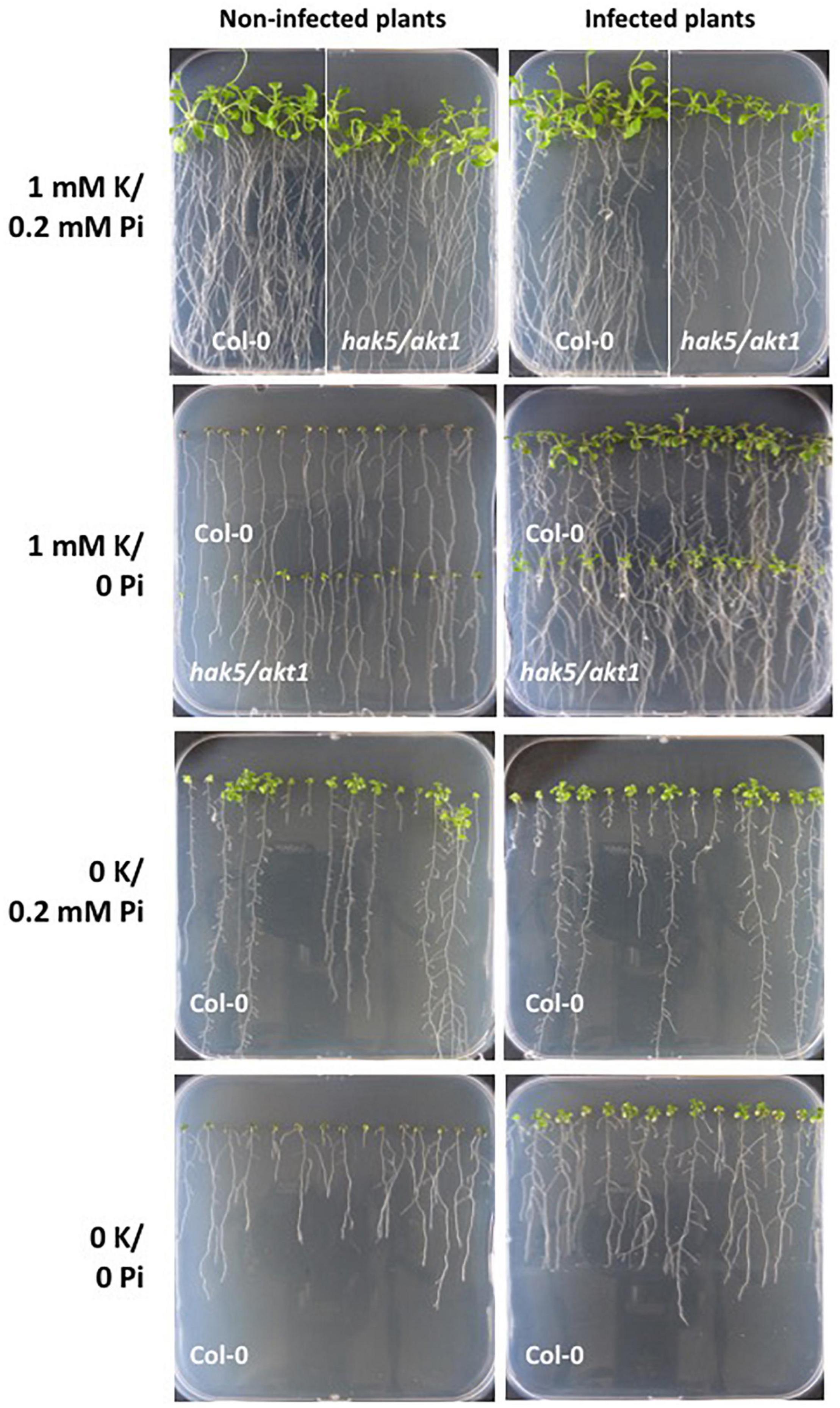
Figure 8. Growth of Arabidopsis plants infected with the fungus under P and/or K+ limiting conditions. Seeds of Col 0 and hak5/akt1 mutant were germinated directly in agar plates containing the modified Hoagland medium with the K+ and Pi concentrations indicated in each panel. Plant growth was followed after 14 days post infection. At 0K the double mutant hak5/akt1 did not germinate (not shown).
Discussion
This work addressed the study of the K+ nutrition of the endosymbiotic fungus Serendipita indica under free-living conditions and during symbiosis, the identification of the fungal K+ transporters involved, and the determination of whether the fungus has any relevant role in the K+ nutrition of the plant. Our results indicate that under free living conditions, the fungus grows better at low K+ concentrations, even at micromolar K+, conditions in which it shows a high-affinity K+ transport capacity (Figures 3A,B). On the contrary, S. indica cannot grow in moderate to high K+ concentrations such as 25–200 mM (Figure 3A), which is striking considering that these concentration ranges are normal in its natural environment where it lives, that is, inside the plant roots. The sensitivity of the fungus to moderate K+ concentrations makes it essential for fungal survival to be equipped with a K+ exclusion system that allows its growth within the plant. In this sense, an excellent candidate would be the K+ efflux pump SiENA1 (Lanza et al., 2019) so it would be interesting to obtain mutants to clarify this point. On the other hand, to decipher the fungal transporters involved in maintaining K+ homeostasis, three transporters and a K+ channel have been identified in the genome, cloned, and functionally characterized in yeast mutants in this work. The comparison of these transporters with the “K+ transportome” of other fungi indicates that S. indica is endowed with typical K+ transporters. It has a HAK transporter that, according to kinetic analysis (Figure 5) and transcriptional expression (Figure 6), is probably the main transporter responsible for the high-affinity K+ transport of the fungus. The discrepancy observed between the good growth of S. indica at low concentrations of K+ (Figure 3) compared to the kinetic analysis of K+ depletion shown in Figure 5B, in which no depletion of external K+ below 80 μM took place, could be explained by the difference in transport activity when studied in vivo or by heterologous expression in yeast. Another possibility is that some other of the K+ transporters of the fungus not yet characterized participates in addition to SiHAK1 in the absorption of K+ in the high affinity range of concentrations. The comparison between the function of Neurospora NcHAK1 and SiHAK1 showed a clear difference in the transport efficiency that is higher in the former (Figures 5A,B). The explanation for this difference could be based on the different habitat where the respective fungi usually live: Neurospora is a soil fungus that lives in an environment where the commonly available K+ concentration is very low, especially compared to the higher K+ concentrations commonly encountered by a fungal plant symbiont. However, it cannot be ruled out that the differences in the kinetics of K+ transport were due, in part, to a different efficiency in the heterologous expression of both cDNAs carried out in yeast.
The other K+ transporters identified in the S. indica genome (two TRK transporters and one TOK1) were also partially characterized in this work. The drop test analysis indicated that they transport K+ although their efficiency was very low (Figure 5A). Their function and whether they have any relevant contribution to the K+ nutrition of the fungus, living alone in the soil or in the host plant, needs to be determined in further studies. Regarding the gene expression results presented here, it is noteworthy that SiTRK1 was the gene that showed the highest expression in all conditions tested, although its expression was not induced by K+ restriction (Figure 6). Its constitutive expression is compatible with being involved in K+ absorption, either under free living conditions or in symbiosis, under starved or normal K+ growth conditions. It is not yet known whether the main function of K+ transport mediated by this protein may be relevant for the maintenance of the membrane potential of fungal cells as other TRK proteins (Madrid et al., 1998), is pending to be studied.
The other aspect that has been pursued in this work was to find out whether the endosymbiotic fungus Serendipita indica has any role in the K+ nutrition of the plant. For that, we have studied Arabidopsis growth in the presence or absence of the fungus under K-limiting conditions, which are nutritional restrictive conditions in which the plant could especially benefit from the presence of a partner “K+ supplier.” The results indicated that S. indica colonization does not improve plant growth during K+ starvation or restore the internal K+ content that falls sharply under this condition (Figures 1A,B, and Supplementary Figure 1). The decrease in the internal K+ content observed in symbiosis could be due to the death and release of the cytoplasmic content suffered by some root cells in the advanced stages of colonization when the fungus is stablished. However, it is worth noting that symbiosis improves plant growth at sufficient K conditions (Figure 1A). The beneficial effect of S. indica on the growth of various plants was already known; the novelty observed in this work is that the improvement of plant growth seems to take place independently of the maintenance of the internal K+ content of the plant since more growth was observed even when there was a significant decrease in the K+ content in roots (Figure 1B).
Despite the evidences provided, it cannot be rule out that the fungus can contribute to improving plant fitness with respect to K+ nutrition in other plant growth conditions not studied here. In nature, for example, plants are exposed to environmental conditions in which changes in K+ homeostasis also occur, such as salinity. Under saline stress, plants are affected by Na+ toxicity, but also suffer from K+ deficiency. In this condition, excessive accumulation of Na+ in the soil can cause disruption of K+ acquisition by the root of plants (Shabala and Cuin, 2008). In line with this idea, it has recently been found that S. indica improves the K+ content in the shoots of maize (Yun et al., 2018), or tomato plants (Ghorbani et al., 2019) exposed to salinity. It could be interesting to find out if any of the fungal K+ transporters identified in this work could perform relevant tasks, such as the uptake or mobilization of K+ that would lead to restoration of the K+ content in this plant stress condition.
Interestingly, the results obtained in this work show a clear difference between the contribution of S. indica to the nutrition of K+ and Pi of plants in symbiosis, confirming that inoculation improves plant growth under Pi limiting conditions, as previously described (Bajaj et al., 2018), while no effect was shown under K+ limiting conditions (Figure 8). In our experiments Pi deprivation was even taken to the extreme than in previous works, because no Pi was added in the medium (instead of, for example, 250 μM Pi which was used in Bajaj et al., 2018), and under this condition, a clear promotion of plant growth was observed. P and K+ (apart from N) are essential macronutrients for all living organisms, and both elements in their freely available forms are found in limiting concentrations in most of the soils of the planet. However, in previous studies, the comparison of individual nutritional deficiencies of plants with dual or triple deficiencies (-N, -P, -K) evidenced that Pi starvation had a dominant behavior over K+ starvation (Hermans et al., 2006; Nieves-Cordones et al., 2019). Our work supports this notion and, according to it, shows that the beneficial role of this endosymbiont in plant growth is clearer in the most extreme nutrient-limiting condition such as Pi deprivation. The P reserves in plant cells are generally much lower than those of K+, so the dependence of taking P from the external environment may be greater and any collaboration in the Pi absorption received from symbionts can be totally decisive. Regarding K+, it can be hypothesized that the evolution of endosymbiotic relationships, such as that established by S. indica with plants, has led to a compromise situation, a “give and take deal” between both organisms: The fungus gives P to the plant and takes K+ (besides sucrose) from the plant. This would not exclude the possibility that there may be other mutualistic fungi that can supply K+ to associated plants having a determinant role in plant K+ nutrition.
A small number of previous studies (for review see: Haro and Benito, 2019) indicated that symbionts can induce changes in the expression of plant genes that may ultimately activate K+ transporters of the host plant, thus improving K+ nutrition. In this sense, another example has recently been published in which the tomato transporter from the HAK family SlHAK10, was specifically induced in mycorrhizal plants (Liu et al., 2019). In our work, the effect of S. indica co-culture on AtHAK5 expression has been studied and showed that symbiosis significantly induced the expression of this plant transporter under K+ starved conditions (Supplementary Figure 3). As no improvement was observed in inoculated plants, neither in K+ transport kinetic analysis (Figure 2) nor in plant K+ content (Figure 1), the transcriptional regulation of AtHAK5 observed does not appear to correlate with the activation of the AtHAK5 protein as previously published in other works (Rubio et al., 2014). Perhaps the observed induction of AtHAK5 is triggered to compensate for the reduction in plant K+ uptake, being more than a cause, the consequence of sensing the K+ content depletion. In fact, it is known that K+ starvation induces changes to only a small number of genes, especially those encoding K+ transporters (Maathuis et al., 2003; Armengaud et al., 2004; Gierth et al., 2005). The Arabidopsis genome contains a total of 13 HAK genes, so it would be interesting to determine whether any of them has a specific role in maintaining plant K+ nutrition during symbiotic interactions.
This report shows that during symbiosis under K+ limiting conditions, Arabidopsis does not benefit from the association with S. indica. But what about the fungus? Does symbiosis benefit K+ nutrition for the fungus? The preliminary results shown here indicate that under K+ limiting conditions, the fungal colonization is stimulated (Figure 7) suggesting that, indeed, symbiosis can benefit the fungus in terms of guaranteeing the K+ supply. It is reasonable to think that when the fungus lives inside the plant, the K+ requirement is met. Either living within the cell, where 100 mM K+ is the existing physiological concentration; or in the apoplast, where although the concentration is lower, probably exceeds that present in the soil, S. indica has ensured enough K+ to support fungal life. In fact, as mentioned above, an active K+ efflux system would be required to withstand the high K+ concentration present in the root cells. Unfortunately, as far as the literature shows, no information has been collected on K+ nutrition in other fungi that share the S. indica lifestyle, so it cannot be known whether the results obtained here can be generalized to other endosymbionts.
In summary, it is generalized accepted that in the symbiotic association between fungi and plants, the contribution of the fungus is in providing water and mineral nutrients to the plant in exchange for sugars synthesized by the plant. In view of these results, this statement should be qualified, in such a way that in symbiosis the contribution of the plant may not only be sugars for the fungus, but also K+ that it needs for its optimal development during the association. While sugars are secreted into the rhizosphere in exudates, K+ is not, and this would explain the stimulation of colonization by the endophyte that is observed in this work under K+ starving conditions.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
BB and RH conceived and designed the research. LC conducted all the experimental work. BB wrote the manuscript. RH designed most of the figures. All authors have made a substantial and intellectual contribution to the work and they have reviewed and approved it for publication.
Funding
This work was supported by grant PID2020-119441RB-I00, AGL2016-80593-R, and BIO2016-81957-REDT funded by MCIN/AEI/10.13039/501100011033. Additional financial support was provided by the DGUI-UPM/CAM Research Group Program. LC was supported by the UPM R+D+I program.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Alga Zuccaro for their gift of the Serendipita indica strain expressing GFP protein used in this work. We also want to thank Stephan Pollmann for the critical reading of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.789371/full#supplementary-material
Footnotes
- ^ https://www.undp.org/content/undp/en/home/sustainable-development-goals.html
- ^ https://genome.jgi.doe.gov/Pirin1/Pirin1.home.html
References
Abdelaziz, M. E., Kim, D., Ali, S., Fedoroff, N. V., and Al-Babili, S. (2017). The endophytic fungus Piriformospora indica enhances Arabidopsis thaliana growth and modulates Na+/K+ homeostasis under salt stress conditions. Plant Sci. 263, 107–115. doi: 10.1016/j.plantsci.2017.07.006
Achatz, B., von Rüden, S., Andrade, D., Neumann, E., Pons-Kühnemann, J., Kogel, K. H., et al. (2010). Root colonization by Piriformospora indica enhances grain yield in barley under diverse nutrient regimes by accelerating plant development. Plant Soil 333, 59–70. doi: 10.1007/s11104-010-0319-0
Álvarez-Aragón, R., Haro, R., Benito, B., and Rodríguez-Navarro, A. (2016). Salt intolerance in Arabidopsis: shoot and root sodium toxicity, and inhibition by sodium-plus-potassium overaccumulation. Planta 243, 97–114. doi: 10.1007/s00425-015-2400-7
Armengaud, P., Breitling, R., and Amtmann, A. (2004). The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiol. 136, 2556–2576. doi: 10.1104/pp.104.046482.2556
Bajaj, R., Huang, Y., Gebrechristos, S., Mikolajczyk, B., Brown, H., Prasad, R., et al. (2018). Transcriptional responses of soybean roots to colonization with the root endophytic fungus Piriformospora indica reveals altered phenylpropanoid and secondary metabolism. Sci. Rep. 8:10227. doi: 10.1038/s41598-018-26809-3
Bañuelos, M. A., Klein, R. D., Alexander-Bowman, S. J., and Rodríguez-Navarro, A. (1995). A potassium transporter of the yeast Schwanniomyces occidentalis homologous to the kup system of Escherichia coli has a high concentrative capacity. EMBO J. 14, 3021–3027. doi: 10.1002/j.1460-2075.1995.tb07304.x
Bañuelos, M. A., Sychrova, H., Bleykasten-Grosshans, C., Souciet, J.-L., and Potier, S. (1998). The Nhal antiporter of Saccharomyces cerevisiae mediates sodium and potassium efflux. Microbiology 144, 2749–2758.
Benito, B., Garciadeblás, B., Fraile-Escanciano, A., and Rodríguez-Navarro, A. (2011). Potassium and sodium uptake systems in Fungi. The transporter diversity of Magnaporthe oryzae. Fungal Genet. Biol. 48, 812–822. doi: 10.1016/j.fgb.2011.03.002
Brunelli, J. P., and Pall, M. L. (1993). A series of yeast- Escherichia coli expression vectors designed for directional cloning of CDNAs and Cre- Lox -mediated plasmid excision. Yeast 9, 1309–1318.
Cabrera, E., Alvarez, M. C., Martín, Y., Siverio, J. M., and Ramos, J. (2012). K+ uptake systems in the yeast Hansenula polymorpha. Transcriptional and post-translational mechanisms involved in high-affinity K+ transporter regulation. Fungal Genet. Biol. 49, 755–763. doi: 10.1016/j.fgb.2012.06.008
Casieri, L., Ait Lahmidi, N., Doidy, J., Veneault-Fourrey, C., Migeon, A., Bonneau, L., et al. (2013). Biotrophic transportome in mutualistic plant-fungal interactions. Mycorrhiza 22, 555–564. doi: 10.1007/s00572-013-0496-9
Corratgé, C., Zimmermann, S., Lambilliotte, R., Plassard, C., Marmeisse, R., Thibaud, J. B., et al. (2007). Molecular and functional characterization of a Na+-K+ transporter from the Trk family in the ectomycorrhizal fungus Hebeloma cylindrosporum. J. Biol. Chem. 282, 26057–26066. doi: 10.1074/jbc.M611613200
Cuéllar, T., Pascaud, F., Verdeil, J. L., Torregrosa, L., Adam-Blondon, A. F., Thibaud, J. B., et al. (2010). A grapevine shaker inward K+ channel activated by the calcineurin B-like calcium sensor 1-protein kinase CIPK23 network is expressed in grape berries under drought stress conditions. Plant J. 61, 58–69. doi: 10.1111/j.1365-313X.2009.04029.x
Dreyer, I., and Michard, E. (2020). High- and low-affinity transport in plants from a thermodynamic point of view. Front. Plant Sci. 10:1797. doi: 10.3389/fpls.2019.01797
Garcia, K., Chasman, D., Roy, S., and Ané, J. M. (2017). Physiological responses and gene co-expression network of mycorrhizal roots under K+ deprivation. Plant Physiol. 173, 1811–1823. doi: 10.1104/pp.16.01959
Garcia, K., Delteil, A., Conéjéro, G., Becquer, A., Plassard, C., Sentenac, H., et al. (2014). Potassium nutrition of ectomycorrhizal pinus pinaster: overexpression of the Hebeloma cylindrosporum HcTrk1 transporter affects the translocation of Both K+ and Phosphorus in the host plant. New Phytol. 201, 951–960. doi: 10.1111/nph.12603
Garcia, K., Doidy, J., Zimmermann, S., Wipf, D., and Courty, P. E. (2016). Take a trip through the plant and fungal transportome of mycorrhiza. Trends Plant Sci. 21, 937–950. doi: 10.1016/j.tplants.2016.07.010
Garcia, K., and Zimmermann, S. (2014). The role of mycorrhizal associations in plant potassium nutrition. Front. Plant Sci. 5:337. doi: 10.3389/fpls.2014.00337
Ghaffari, M. R., Ghabooli, M., Khatabi, B., Hajirezaei, M. R., Schweizer, P., and Salekdeh, G. H. (2016). Metabolic and transcriptional response of central metabolism affected by root endophytic fungus Piriformospora indica under salinity in barley. Plant Mol. Biol. 90, 699–717. doi: 10.1007/s11103-016-0461-z
Ghorbani, A., Omran, V. O. G., Razavi, S. M., Pirdashti, H., and Ranjbar, M. (2019). Piriformospora indica confers salinity tolerance on tomato (Lycopersicon esculentum Mill.) through amelioration of nutrient accumulation, K+/Na+ homeostasis and water status. Plant Cell Rep. 38, 1151–1163. doi: 10.1007/s00299-019-02434-w
Gierth, M., Mäser, P., and Schroeder, J. I. (2005). The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol. 137, 1105–1114. doi: 10.1104/pp.104.057216
Guerrero-Galán, C., Garcia, K., Houdinet, G., and Zimmermann, S. (2018). HcTOK1 participates in the maintenance of K+ homeostasis in the ectomycorrhizal fungus Hebeloma cylindrosporum, which is essential for the symbiotic K+ nutrition of Pinus Pinaster. Plant Signal. Behav. 13:e1480845. doi: 10.1080/15592324.2018.1480845
Haro, R., and Benito, B. (2019). The role of soil fungi in K+ plant nutrition. Int. J. Mol. Sci. 20:3169. doi: 10.3390/ijms20133169
Haro, R., and Rodríguez-Navarro, A. (2003). Functional analysis of the M2D helix of the TRK1 potassium transporter of Saccharomyces cerevisiae. Biochim. Biophys. Acta Biomembr. 1613, 1–6. doi: 10.1016/S0005-2736(03)00132-9
Haro, R., Sainz, L., Rubio, F., and Rodríguez-Navarro, A. (1999). Cloning of two genes encoding potassium transporters in neurospora crassa and expression of the corresponding CDNAs in Saccharomyces cerevisiae. Mol. Microbiol. 31, 511–520. doi: 10.1046/j.1365-2958.1999.01192.x
Hermans, C., Hammond, J. P., White, P. J., and Verbruggen, N. (2006). How do plants respond to nutrient shortage by biomass allocation? Trends Plant Sci. 11, 610–617. doi: 10.1016/j.tplants.2006.10.007
Hilbert, M., Voll, L. M., Ding, Y., Hofmann, J., Sharma, M., and Zuccaro, A. (2012). Indole derivative production by the root endophyte Piriformospora indica is not required for growth promotion but for biotrophic colonization of barley roots. New Phytol. 196, 520–534. doi: 10.1111/j.1469-8137.2012.04275.x
Jogawat, A., Vadassery, J., Verma, N., Oelmüller, R., Dua, M., Nevo, E., et al. (2016). PiHOG1, a stress regulator MAP kinase from the root endophyte fungus Piriformospora indica, confers salinity stress tolerance in rice plants. Sci. Rep. 6:36765. doi: 10.1038/srep36765
Jones, E. B. G., and Jennings, D. H. (1965). The effect of cations on the growth of fungi. New Phytol. 64, 86–100. doi: 10.1111/j.1469-8137.1965.tb05378.x
Lanza, M., Haro, R., Conchillo, L. B., and Benito, B. (2019). The endophyte Serendipita indica reduces the sodium content of Arabidopsis plants exposed to salt stress: fungal ENA ATPases are expressed and regulated at high PH and during plant co-cultivation in salinity. Environ. Microbiol. 21, 3364–3378. doi: 10.1111/1462-2920.14619
Leigh, R. A., and Wyn Jones, R. G. (1984). A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant cell. New Phytol. 97, 1–13. doi: 10.1111/j.1469-8137.1984.tb04103.x
Li, L., Li, L., Wang, X., Zhu, P., Wu, H., and Qi, S. (2017). Plant growth-promoting endophyte Piriformospora indica alleviates salinity stress in Medicago truncatula. Plant Physiol. Biochem. 119, 211–223. doi: 10.1016/j.plaphy.2017.08.029
Liu, J., Liu, J., Liu, J., Cui, M., Huang, Y., Tian, Y., et al. (2019). The potassium transporter SlHAK10 is involved in mycorrhizal potassium uptake. Plant Physiol. 180, 465–479. doi: 10.1104/pp.18.01533
Maathuis, F. J. M., Filatov, V., Herzyk, P., Krijger, G. C., Axelsen, K. B., Chen, S., et al. (2003). Transcriptome analysis of root transporters reveals participation of multiple gene families in the response to cation stress. Plant J. 35, 675–692. doi: 10.1046/j.1365-313X.2003.01839.x
Madrid, R., Gómez, M. J., Ramos, J., and Rodríguez-Navarro, A. (1998). Ectopic potassium uptake in Trk1 Trk2 mutants of Saccharomyces cerevisiae correlates with a highly hyperpolarized membrane potential. J. Biol. Chem. 273, 14838–14844. doi: 10.1074/jbc.273.24.14838
Maresova, L., and Sychrova, H. (2005). Physiological characterization of Saccharomyces cerevisiae Kha1 deletion mutants. Mol. Microbiol. 55, 588–600. doi: 10.1111/j.1365-2958.2004.04410.x
Marschner H. (ed.) (1995). “Functions of mineral nutrients: macronutrients,” in Mineral Nutrition of Higher Plants, (London: Academic Press), 229–312.
Narayan, O. P., Verma, N., Jogawat, A., Dua, M., and Johri, A. K. (2021). Sulfur transfer from the endophytic fungus Serendipita indica improves maize growth and requires the sulfate transporter SiSulT. Plant Cell 33, 1268–1285. doi: 10.1093/plcell/koab006
Nass, R., Cunningham, K. W., and Rao, R. (1997). Intracellular sequestration of sodium by a novel Na+/H+ exchanger in yeast is enhanced by mutations in the plasma membrane H+-ATPase. J. Biol. Chem. 272, 26145–26152. doi: 10.1074/JBC.272.42.26145
Nieves-Cordones, M., Martínez, V., Benito, B., and Rubio, F. (2016). Comparison between Arabidopsis and rice for main pathways of K+ and Na+ uptake by roots. Front. Plant Sci. 7:992. doi: 10.3389/fpls.2016.00992
Nieves-Cordones, M., Ródenas, R., Lara, A., Martínez, V., and Rubio, F. (2019). The combination of K+ deficiency with other environmental stresses: what is the outcome? Physiol. Plant. 165, 264–276. doi: 10.1111/ppl.12827
Prasad, D., Verma, N., Bakshi, M., Narayan, O. P., Singh, A. K., Dua, M., et al. (2019). Functional characterization of a magnesium transporter of root endophytic fungus Piriformospora indica. Front. Microbiol. 10:3231. doi: 10.3389/fmicb.2018.03231
Rivetta, A., Allen, K. E., Slayman, C. W., and Slayman, C. L. (2013). Coordination of K+ transporters in Neurospora: TRK1 is scarce and constitutive, while HAK1 is abundant and highly regulated. Eukaryotic Cell 12, 684–696. doi: 10.1128/ec.00017-13
Rodríguez-Navarro, A. (2000). Potassium transport in fungi and plants. Biochim. Biophys. Acta Rev. Biomembr. 1469, 1–30. doi: 10.1016/S0304-4157(99)00013-1
Rodriguez-Navarro, A., and Ramos, J. (1984). Dual system for potassium transport in Saccharomyces cerevisiae. J. Bacteriol. 159, 940–945.
Rubio, F., Alemán, F., Nieves-Cordones, M., and Martínez, V. (2010a). Studies on Arabidopsis Athak5, Atakt1 double mutants disclose the range of concentrations at which AtHAK5, AtAKT1 and unknown systems mediate K+ uptake. Physiol. Plant. 139, 220–228. doi: 10.1111/j.1399-3054.2010.01354.x
Rubio, F., Arévalo, L., Caballero, F., Botella, M. A., Rubio, J. S., García-Sánchez, F., et al. (2010b). Systems involved in K+ uptake from diluted solutions in pepper plants as revealed by the use of specific inhibitors. J. Plant Physiol. 167, 1494–1499. doi: 10.1016/j.jplph.2010.05.022
Rubio, F., Fon, M., Ródenas, R., Nieves-Cordones, M., Alemán, F., Rivero, R. M., et al. (2014). A low K+ signal is required for functional high-affinity K+ uptake through HAK5 transporters. Physiol. Plant. 152, 558–570. doi: 10.1111/ppl.12205
Rubio, F., Nieves-Cordones, M., Alemán, F., and Martínez, V. (2008). Relative contribution of AtHAK5 and AtAKT1 to K+ uptake in the high-affinity range of concentrations. Physiol. Plant. 134, 598–608.
Shabala, S., and Cuin, T. A. (2008). Potassium transport and plant salt tolerance. Physiol. Plant. 133, 651–669. doi: 10.1111/j.1399-3054.2007.01008.x
Verma, N., Narayan, O. P., Prasad, D., Jogawat, A., Panwar, S. L., Dua, M., et al. (2021). Functional characterization of a high-affinity iron transporter (PiFTR) from the endophytic fungus Piriformospora indica and its role in plant growth and development. Environ. Microbiol. [Epub ahead of print]. doi: 10.1111/1462-2920.15659
Waller, F., Achatz, B., Baltruschat, H., Fodor, J., Becker, K., Fischer, M., et al. (2005). The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc. Natl. Acad. Sci. U.S.A. 102, 13386–13391. doi: 10.1073/pnas.0504423102
Yadav, V., Kumar, M., Deep, A. K., Kumar, H., Sharma, R., Tripathi, T., et al. (2010). A phosphate transporter from the root endophytic fungus Piriformospora indica plays a role in phosphate transport to the host plant. J. Biol. Chem. 285, 26532–26544. doi: 10.1074/jbc.M110.111021
Yun, P., Xu, L., Wang, S.-S., Shabala, L., Shabala, S., and Zhang, W.-Y. (2018). Piriformospora indica improves salinity stress tolerance in Zea mays L. plants by regulating Na+ and K+ loading in root and allocating K+ in shoot. Plant Growth Regul. 86, 1–9. doi: 10.1007/s10725-018-0431-3
Keywords: Serendipita indica, endophyte, symbiosis, plant nutrition, potassium, K+ transporters, abiotic stress
Citation: Conchillo LB, Haro R and Benito B (2021) K+ Nutrition Exchange in the Serendipita-Arabidopsis Symbiosis: Study of the Fungal K+ Transporters Involved. Front. Ecol. Evol. 9:789371. doi: 10.3389/fevo.2021.789371
Received: 04 October 2021; Accepted: 11 November 2021;
Published: 16 December 2021.
Edited by:
Pedro Emilio Gundel, University of Talca, ChileReviewed by:
Francisco Rubio, Center for Edaphology and Applied Biology of Segura, Spanish National Research Council (CSIC), SpainNidhi Verma, Jawaharlal Nehru University, India
Copyright © 2021 Conchillo, Haro and Benito. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Begoña Benito, begona.benito@upm.es
 Lorena B. Conchillo
Lorena B. Conchillo Rosario Haro
Rosario Haro Begoña Benito
Begoña Benito