Lipid Droplet and Peroxisome Biogenesis: Do They Go Hand-in-Hand?
- Department of Cell Biology and Physiology, The University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
All eukaryotic cells contain membrane bound structures called organelles. Each organelle has specific composition and function. Some of the organelles are generated de novo in a cell. The endoplasmic reticulum (ER) is a major contributor of proteins and membranes for most of the organelles. In this mini review, we discuss de novo biogenesis of two such organelles, peroxisomes and lipid droplets (LDs), that are formed in the ER membrane. LDs and peroxisomes are highly conserved ubiquitously present membrane-bound organelles. Both these organelles play vital roles in lipid metabolism and human health. Here, we discuss the current understanding of de novo biogenesis of LDs and peroxisomes, recent advances on how biogenesis of both the organelles might be linked, physical interaction between LDs and peroxisomes and other organelles, and their physiological importance.
Introduction
Peroxisomes and LDs play important roles in cellular lipid metabolism. These organelles are major metabolic hubs in eukaryotic cells. Both organelles play roles in preventing cell toxicity, albeit in dissimilar ways (Kohlwein et al., 2013). Peroxisomes are sites for beta-oxidation of fatty acids in all eukaryotic cells (Mannaerts and Van Veldhoven, 1996). In yeast and plants, the entire pathway occurs on peroxisomes, whereas in animals it occurs in peroxisomes, and mitochondria (Kunau and Hartig, 1992). Other than oxidation of fatty acids, peroxisomes are essential for detoxification of hydrogen peroxide (Walker et al., 2017). In addition, peroxisomes are sites for synthesis of D-amino acids, plasmalogens, and certain precursors of cholesterol. Understandably, defects in peroxisome function lead to several metabolic disorders (Argyriou et al., 2017). These disorders are caused due to mutations in genes encoding peroxisomal biogenesis proteins (PEX) essential for peroxisome function (Gould and Valle, 2000). Taken as a group, peroxisomal disorders occur in 1 in 5000 individuals (Waterham et al., 2016). Some of the commonly known disorders include X-linked adrenoleukodystrophy and peroxisomal biogenesis disorders (PBD) such as Zellweger Syndrome. For detailed discussion on PBD and other peroxisome related metabolic defects we refer other reviews to the readers (Delille et al., 2006; Wanders, 2014). While peroxisomes are sites of lipid degradation, LDs are organelles that prevent cellular toxicity by sequestering and storing free fatty acids in neutral lipids such as triglycerides (TG) and sterol esters (SE) (Cohen, 2018). Recent studies have elucidated new roles of LDs in protein degradation and protection from ER stress and mitochondrial oxidative stress (Olzmann and Carvalho, 2018). Aberrant LD biogenesis is a hallmark of severe disorders including diabetes, atherosclerosis, lipodystrophy, and neurodegeneration (Krahmer et al., 2013; Onal et al., 2017).
Peroxisome Biogenesis
Peroxisomes are organelles enclosed in a single bilayer membrane. Peroxisomes were observed in early electron micrographs as distinct membrane bound organelles highly associated with the ER (Baudhuin et al., 1965; de Duve and Baudhuin, 1966; Tsukada et al., 1968; de Duve, 1969; Novikoff and Novikoff, 1972). It was later demonstrated that, similar to mitochondria and chloroplasts, peroxisomal matrix proteins are directly imported post-translationally from cytosolic ribosomes. Thus, peroxisomes were known as semi-autonomous organelles that follow the growth and division model where a new organelle is formed from the pre-existing one (Lazarow, 1983; Goldman and Blobel, 2006). Under normal physiological conditions, mature peroxisomes receive membrane lipids and proteins from the ER membrane, which contributes to peroxisome growth prior to division (Motley and Hettema, 2007).
In yeast, growth and division is the major pathway of peroxisome biogenesis. However, peroxisomes can also originate by de novo biogenesis from the ER (Figure 1A; Hoepfner et al., 2005). This pathway was discovered when peroxisome biogenesis mutants (pex) devoid of functional peroxisomes were isolated using genetic screens in yeast (Liu et al., 1992; Van der Leij et al., 1992). These mutants exhibited ghost vesicles which were devoid of many matrix proteins. Depletion of some proteins such as Pex3, a peroxisomal membrane protein (PMP) required for localization and stability of other PMPs, Pex19, a chaperone, and receptor for PMPs, and Pex16, a PMP that can recruit Pex3, resulted in cells that were completely devoid of the ghost vesicles. This suggested that these proteins might play a role in the first steps of peroxisome vesicle formation (Hohfeld et al., 1991; Hettema et al., 2000; Tam et al., 2005; Kim et al., 2006; Fujiki et al., 2014; Erdmann et al., 2015). Interestingly, when the missing protein was re-introduced in these cells, mature peroxisomes reappeared, thus challenging the growth, and division model. The resulting de novo biogenesis model has two main aspects: formation of new pre-peroxisomal vesicles (PPVs) from a mother organelle followed by targeting of PMPs to these vesicles to form functional mature peroxisomes. The mother organelle is usually ER, as many PMPs are targeted to ER membrane in the absence of functional peroxisomes (Geuze, 2003; van der Zand et al., 2010; Agrawal and Subramani, 2013; Tabak et al., 2013; Kim and Hettema, 2015). However, in the absence of peroxisomes, many PMPs are mistargeted to mitochondria in mammalian cells (Kim and Hettema, 2015). The PMPs possibly leave the ER membrane in the newly formed PPVs (Agrawal et al., 2016; Joshi et al., 2016). Two independent in vitro studies reported an essential role for Pex19 in formation of nascent vesicles from the ER membrane. These studies demonstrated that PMPs such as Pex3, Pex15 (a tail-anchored PMP), and Pex11 (required for peroxisome proliferation), are targeted to the ER membrane and traffic to PPVs in a Pex19- and ATP-dependent manner (Agrawal et al., 2011; Lam et al., 2011). The targeting of PMPs to peroxisomes is independent of COPI and COPII proteins (South et al., 2000, 2002; Voorn-Brouwer et al., 2001).
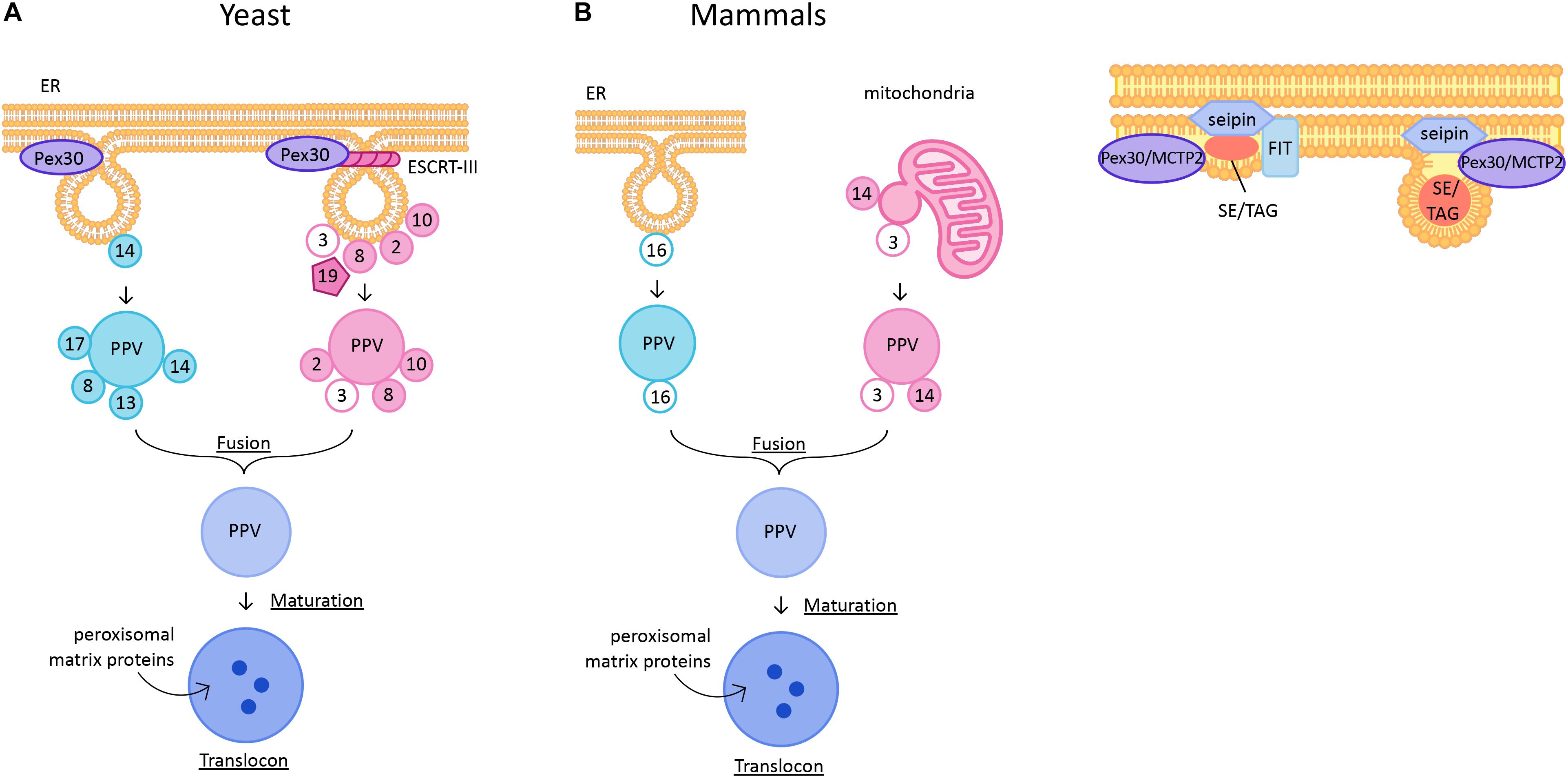
Figure 1. (A) De novo peroxisome biogenesis in yeast and mammalian cells. PPVs containing the docking complex proteins are generated independently of Pex3 and Pex19 at ER subdomains containing Pex30. PPVs containing ring complex proteins are generated in a Pex3- and Pex19-dependent manner. ESCRT-III complex is required for scission of PPVs from the ER membrane. The two pools of PPVs fuse to form an import competent mature peroxisome. In mammals, Pex16-containing PPVs from ER and Pex3 and Pex14-containing PPVs from mitochondrial outer membrane fuse to form a functional peroxisome. (B) Lipid droplet biogenesis. Neutral lipid synthesizing enzymes generate sterol esters (SE) and triglycerides (TG) to form a lens-like structure in the lipid bilayer. The ER regions are enriched with seipin, FIT proteins, and Pex30/MCTP2. As the lens grows the LD buds into the cytoplasm. In yeast, LDs remain attached to the ER subdomains containing Pex30/MCTP2 and seipin but no FIT proteins.
In mammalian cells peroxisomes form through division of preexisting peroxisomes, but can also derive de novo under special conditions. Mammalian peroxisome biogenesis differs from yeast in that mitochondrial outer membrane can be a site for PPV formation (Figure 1A). Artificial targeting of Pex3 to mitochondrial outer membrane in Pex3-deficient cells resulted in de novo peroxisome biogenesis (Rucktäschel et al., 2010). Furthermore, a recent study demonstrated that there are two types of PPVs, one from ER that contains Pex16 and another from mitochondrial outer membrane that contains Pex3 and Pex14. These PPVs fuse to form a functional peroxisome (Sugiura et al., 2017). Existence of more than one type of PPV is controversial in yeast cells. Earlier reports showed that the importomer complex consisting of RING complex (Pex2, Pex10, and Pex12) and docking complex (Pex13, Pex14, and Pex17) sorted in distinct PPVs and fused to form a functional peroxisome (Figure 1A; Van Der Zand et al., 2012). The model for existence of two pools of PPVs is consistent with the report that sorting of RING complex proteins, but not the docking complex proteins, in the ER membrane is dependent on Pex3 (Agrawal et al., 2016). Interestingly, the vesicles consisting of docking complex proteins exist in yeast cells lacking Pex3 or Pex19. Therefore, neither Pex3 nor Pex19 is essential for docking complex-containing PPV formation (Knoops et al., 2014; Wróblewska et al., 2017). Thus, there are two pools of PPVs: Pex3- or Pex19-independent PPVs that contain docking complex proteins, and Pex3- and Pex19- dependent PPVs that contain RING complex proteins. The Pex1-Pex6 AAA ATPase proteins are required for fusion of two pools of PPVs (Figure 1A; Van Der Zand et al., 2012). In Y. lipolytica, the two pools of PPVs were reported to require cytosolic factors, ATP hydrolysis, and functional ATPases (Pex1 and Pex6) for fusion (Titorenko et al., 2000). However, this was challenged by recent findings which suggested that there is only one type of PPV, and that Pex1-Pex6, are mainly involved in import of peroxisomal proteins (Knoops et al., 2015; Motley et al., 2015). Nevertheless, the idea that mature peroxisomes form by fusion of unique pre-peroxisomal structures is appealing, because it provides a mechanism for avoiding the mistargeting of peroxisomal enzymes to parent organelles (mitochondria and/or ER) (Kim, 2017).
Recently, ESCRT-III complex proteins were implicated in scission of PPVs from the ER into the cytosol (Figure 1A; Mast et al., 2018). However, ESCRT proteins are known for budding of vesicles away from the cytosol, for example in mutlivesicular body formation, or virus budding. Therefore, the role of ESCRT-III proteins needs further examination. Also, whether these proteins play a role in scission of all PPVs originating from the ER membrane or only a subset is not known.
We clearly do not completely understand PPV biogenesis from the ER membrane. There is no known mutant background that is completely devoid of PPVs, suggesting that there is redundancy among proteins involved in PPV formation. There are many outstanding questions in the field such as: how are PPVs generated? How do PMPs traffic from the ER or mitochondria to PPVs? Where do the RING and docking complex proteins assemble? We are beginning to understand some of these questions. We will discuss the recent advances in the exit sites for PPV biogenesis in Section “Links Between LD and Peroxisomes Biogenesis.”
LD Biogenesis
Lipid droplets are ubiquitously present unique organelles that are enclosed in a phospholipid monolayer. Unlike other organelles, the LD core is made of neutral lipids such as TG and SE. Embedded in the phospholipid monolayer are more than 100 proteins, including enzymes that synthesize or degrade lipids for storage or energy, respectively (Ducharme and Bickel, 2008; Khor et al., 2013; Bersuker and Olzmann, 2018; Bersuker et al., 2018). In yeast, LDs remain permanently connected to the ER, whereas in mammalian cells at least some mature LDs are released from the ER (Jacquier et al., 2011; Olzmann and Carvalho, 2018). LD biogenesis begins with synthesis of neutrals lipids between the ER bilayer, by ER-associated neutral lipid synthesis enzymes (Jacquier et al., 2011; Kassan et al., 2013; Choudhary et al., 2015; Kimura et al., 2018). How these enzymes are sequestered at the sites of LD biogenesis is not known. Synthesis of neutral lipids leads to formation of a lens-like structure in the bilayer (Figure 1B; Choudhary et al., 2015). After an increase in the concentration of neutral lipids at these sites, they start demixing from the highly charged phospholipid bilayer giving rise to LDs (Choudhary et al., 2018). These LDs are covered with a phospholipid monolayer that eventually acquires several proteins that are required for maturation of LDs (Tan et al., 2014). However, no proteins other than the enzymes involved in synthesizing neutral lipids have been implicated in formation of nascent LDs, suggesting that LD biogenesis is a lipid-driven phenomenon. Indeed, it was demonstrated that the lipid composition of the ER at sites of LD formation regulates the formation of LDs (Zanghellini et al., 2010; Ben M’barek et al., 2017; Deslandes et al., 2017; Choudhary et al., 2018). It was shown that lipids such as lysophospholipids that generate positive intrinsic curvature favor LD budding, whereas lipids such as diacylgycerol (DAG) and phosphatidylethanolamine (PE) that induce negative intrinsic curvature disfavor budding. Lipids and proteins that affect membrane tension also influence the directionality of LD budding (Ben M’barek et al., 2017; Chorlay et al., 2017; Deslandes et al., 2017). There are protein families such as seipin, FIT and Pex30/MCTP2 that play critical roles in LD formation (Figure 1B; Fei et al., 2008; Choudhary et al., 2015, 2018; Grippa et al., 2015; Wang et al., 2016; Joshi et al., 2018). These proteins are at the sites of LD biogenesis and are required for efficient generation of LDs. Here, we discuss the current understanding of these proteins in LD biogenesis.
FIT
The fat inducible transmembrane proteins (FITs) are conserved proteins that play a role in LD biogenesis. There are two homologs of FITs, FIT1, which is muscle specific, and FIT2, which is expressed in most tissues in mammals (Kadereit et al., 2007). In yeast, there are two FIT2 proteins, Scs3 and Yft2, whereas only one FIT protein is present in worms (Choudhary et al., 2015). Depletion of FITs leads to decreased LD biogenesis (Miranda et al., 2014). FIT proteins directly bind to TG (Kadereit et al., 2007). Depletion of FITs in yeast, mammals and worms results in LDs wrapped around with ER membrane suggesting a defect in budding of LDs (Choudhary et al., 2015). Later it was shown that in the absence of FITs, DAG levels in the ER increase, which might affect directionality of LD budding (Choudhary et al., 2018). In the same study, it was demonstrated that supplementation with exogenous lipids such as lyso-phosphatidylcholine and lyso-phosphatidic acid reversed the LD wrapping phenotype observed in the FIT mutants in yeast. Moreover, FIT protein, Yft2, along with DAG, transiently accumulates at sites of LD formation (Figure 1B). Thus, FIT proteins probably regulate DAG levels at the sites where LDs are formed. Maintaining the level of DAG in the ER membrane is vital as its accumulation could be toxic. Deletion of worm FIT and mouse FIT2 is lethal, supporting the importance of the cellular function of FIT proteins (Choudhary et al., 2015, 2018).
Seipin
Seipin is a conserved integral ER membrane protein required for efficient LD biogenesis (Liu et al., 2016). The role of seipin in LD biogenesis was first identified in a screen performed in yeast to identify mutants with aberrant LD morphology. In two independent studies it was shown that in the absence of seipin, yeast cells exhibit multiple smaller, or fewer supersized LDs that are clustered together (Li et al., 2007; Fei et al., 2008). It was later shown that seipin is required for efficient incorporation of both proteins and lipids into the LDs (Salo et al., 2016). Seipin forms discrete foci in the ER membrane where nascent LDs are formed. Seipin along with Ldb16 in yeast is localized at ER-LD contact sites (Wang et al., 2014; Grippa et al., 2015). Seipin is highly mobile on the ER membrane until it encounters a nascent LD. Loss of seipin leads to smaller LDs that eventually fuse to form large and fewer LDs (Wang et al., 2016). Recent cryo-EM studies describe oligomeric structures of human (undecamers) and fly (dodecamers) seipin. Each monomer consists of a hydrophobic helix placed toward the ER bilayer and a β-sandwich domain which is structurally similar to lipid-binding proteins. It was shown that this domain binds to anionic phospholipids such as phosphatidic acid (Sui et al., 2018; Yan et al., 2018). Thus, seipin could regulate the local phospholipid levels at the site of LD budding.
NVJ
Lipid droplet biogenesis occurs at specialized nuclear vacuolar junctions (NVJs) during nutrient stress. Mdm1, a molecular tether at the NVJ, and localizes at the site of LD formation. Overexpression of Mdm1 causes accumulation of LDs at the NVJ, supporting its role in LD biogenesis (Hariri et al., 2018). Other players in LD biogenesis at the NVJ include Ldo16 and Ldo45, which are overlapping genes with shared amino acid sequence. These proteins are required for accumulation of LDs at the NVJ during stationary growth phase. These proteins generate new LDs at NVJ under nutrient stress conditions to facilitate lipophagy, the breakdown of LDs through autophagy (Eisenberg-Bord et al., 2018; Teixeira et al., 2018). Whether spatial compartmentalization of LD biogenesis specific to nutrient stress occurs in higher eukaryotes remains to be investigated.
Links Between LD and Peroxisomes Biogenesis
Several reports have suggested that LDs and peroxisomes are intimately associated (Schrader, 2001; Binns et al., 2006; Valm et al., 2017). Yeast cells grown in the presence of oleic acid exhibit peroxisomes that adhere stably with LDs by forming extended membrane processes called gnarls. These extensions were enriched in fatty acid beta oxidation enzymes, suggesting coupling of lipolysis within the LDs with peroxisomal enzymes (Binns et al., 2006). A recent report suggests that M1 spastin on the LDs physically tethers with ABCD1 on peroxisomal membranes. Moreover, M1 spastin recruits ESCRT III proteins to facilitate fatty acid trafficking from LDs to peroxisomes (Chang et al., 2019). Thus, there is a clear metabolic link between these organelles. In this section we discuss the shared machinery required for the de novo biogenesis and function of peroxisomes and LDs. Both organelles originate from the ER membrane (Joshi et al., 2017). Until recently, the sites in the ER membrane for formation of these organelles was not known. Also, it was unclear whether these sites form stochastically or are pre-determined.
Pex30 Subdomains: Sites for Nascent LD and PPV Generation
Recent studies in yeast demonstrated that Pex30 localizes to discrete regions of the ER called ER subdomains. Pex30 is a resident ER protein, however, it localizes to peroxisomes or PPVs when cells are exposed to oleic acid or when Pex30 is overexpressed (Vizeacoumar et al., 2006; Joshi et al., 2016). Pex30 and Pex30-like proteins contain a reticulon homology domain (RHD), a transmembrane domain that is similar to reticulon proteins. Like reticulons, Pex30 proteins tubulate the ER membrane. However, unlike the reticulons, Pex30 and Pex30-like proteins are low abundance proteins localized to discrete regions in the ER. In cells devoid of peroxisomes, it was demonstrated using fluorescence and electron microscopy that newly expressed Pex14 is targeted to ER regions enriched with Pex30. Pex14 eventually leaves the ER membrane in a newly formed PPV (Joshi et al., 2016). This suggests that Pex30 subdomains are novel exit sites of nascent PPV formation (Figure 1A). Interestingly, these sites do not overlap with ER exit sites for COPII vesicles (Joshi et al., 2016). Whether other types of PPVs also form at these subdomains remains to be investigated. Deletion of Pex30 and the Pex30-like protein, Pex31, generates small clusters of PPVs closely associated with the ER membrane. The rate of formation of new peroxisomes is also decreased in cells devoid of Pex30 and Pex31 (Joshi et al., 2016). Thus, Pex30 and Pex31 are essential for efficient formation of peroxisomes.
The number of Pex30 subdomains per cell is much greater than the number of PPVs per cell. Thus, Pex30 subdomains could have additional roles. Indeed, it was demonstrated that Pex30 subdomains are also the sites for nascent LD formation ((Figure 1B; Joshi et al., 2018). Furthermore, in yeast cells PPVs were associated with LDs at Pex30 subdomains, implying that PPV and LD biogenesis might occur at the same sites within the ER (Figure 2). It is possible that Pex30 also localizes to the site of new LD formation at the NVJ, as Pex30 is enriched at NVJ especially during stationary growth phase (Joshi et al., 2016). Pex30 subdomains exist even in the absence of LDs, suggesting that the sites at which LDs and PPVs form are stable, pre-determined, and not random. Pex30 has a functional homolog, multiple C2 domain containing transmembrane protein 2 (MCTP2), in higher eukaryotes, which also has an RHD (Joshi et al., 2018). Mammalian cells have two MCTP proteins, MCTP1 and MCTP2, whereas flies and worms have only one MCTP (Shin et al., 2005). Similar to Pex30, MCTP2 is also a low abundance protein that localizes to the sites of LD biogenesis (Figure 1B). MCTP2 subdomains also dynamically associate with the peroxisomal vesicles. However, MCTP2 does not localize to peroxisomes (Joshi et al., 2018).
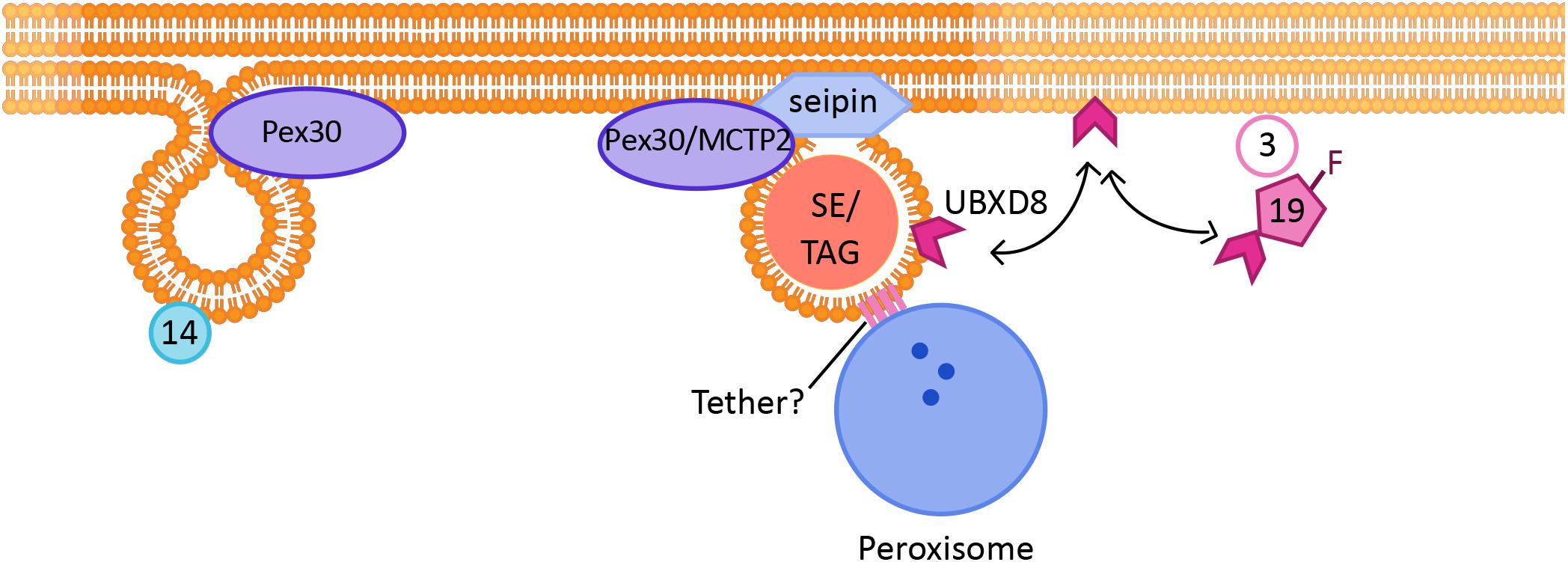
Figure 2. Link between peroxisome and LD biogenesis: The formation of new Pex14-containing PPVs and LDs occur at the same ER subdomains, which are positive for Pex30/MCTP2 proteins. Mature peroxisomes also physically associate with LDs. Tethers identified so far include ABCD1 on peroxisomes, which interacts with Spastin on LDs. Other tethers may also be involved in peroxisome-LD contacts. Pex19 is a cytosolic protein required for peroxisomal protein targeting, which when farnesylated targets a LD protein UBXD8 to LDs.
It is possible that in addition to organelle biogenesis, these ER subdomains play roles in lipid trafficking and signaling. In the absence of Pex30, LDs are small and clustered and there is ER membrane proliferation. An independent study showed that loss of Pex31, a paralog of Pex30, also leads to smaller LDs (Lv et al., 2019). The rate of new LD formation was also significantly decreased in cells devoid of Pex30 (Joshi et al., 2018). It is fascinating that loss of Pex30 has similar morphological effects on both PPVs and LDs, suggesting that the mechanism involved in biogenesis of the organelles is shared (Joshi et al., 2016, 2018). Pex30 also colocalizes with several ER membrane proteins, including seipin, that are known markers for the sites of LD biogenesis. Deletion of seipin affected the distribution of Pex30 in the ER, as there was a significant decrease in the number of Pex30 punctae in the seipin mutant. Cells devoid of both Pex30 and seipin exhibited a severe growth defect, and highly clustered small and large LDs (Joshi et al., 2018; Wang et al., 2018). Seipin also affects peroxisome biogenesis, as deletion of seipin alone decreased the rate of formation of new peroxisomes (Wang et al., 2018). Thus, there is a clear link between LD biogenesis and peroxisome biogenesis.
Shared Protein Machinery for Targeting of Peroxisomal and LD Proteins
Pex19, a well-established cytosolic chaperone protein, and Pex3, a PMP, are required for targeting of UBXD8, an LD protein, in the ER membrane of mammalian cells. UBXD8 is delivered to Pex3-enriched ER subdomains by farnesylated Pex19 ((Figure 2). UBXD8 was mistargeted to the peroxisome if Pex19 was not farnesylated, suggesting that the post-translational modification is vital to gain specificity of protein targeting (Schrul and Kopito, 2016). It is possible that enzymes that are required for LD maturation and growth follow the same route, as cells devoid of Pex3 or Pex19 exhibit smaller LDs (Wang et al., 2013). Another example of a link between peroxisomes and LDs comes from a recent study which demonstrated that in mammalian cells the peroxisomal protein fatty acyl CoA reductase 1 (Far1), is targeted to LDs upon increased triglyceride synthesis. Far1 exhibits two different topologies that differ in orientation of the short hydrophilic C-terminus toward the cytosol or lumen. Out of the two closely spaced hydrophobic domains, one is enough to target the protein to the LDs. Thus, Far1 exhibits dual topologies to peroxisomes and LDs dependent on cellular lipid metabolism (Exner et al., 2019).
Conclusion and Future Perspectives
There are many open questions about the links between peroxisomes, LDs, and other organelles that originate from the ER. What is the common theme in the biogenesis of peroxisomes and LDs? Structurally, these organelles are very different. It is possible that Pex30 and seipin generate specialized ER subdomains for PPV and LD biogenesis. Seipin specifically binds to anionic phospholipids (Yan et al., 2018), whereas studies using sensor proteins such as the DAG-sensing PKD domain from mammalian cells and Dga1 protein demonstrated that Pex30 subdomains have enriched DAG and possibly phosphatidic acid, especially upon induction of LDs with oleic acid (Joshi et al., 2018). In addition, the LD defect in cells devoid of seipin and Pex30 was rescued by manipulating ER phospholipid composition (Wang et al., 2018). Thus, the generation of these organelles could be influenced by lipids. Pex30 could induce curvature with its RHD, which might be conducive for the synthesis of certain lipids to facilitate the generation of LDs and PPVs. It is also possible that the biogenesis of these organelles from a common subdomain allows their formation to be coordinated in response to stimuli that require the proliferation of both, such as a lipid challenge. It has been demonstrated that Pex30 subdomains do not overlap with ER exit sites. However, it remains to be determined whether these domains play a role in the biogenesis of other organelles that originate from the ER, such as autophagosomes, which can also be induced in response to excess lipids (Singh et al., 2009). There are hints that LD and autophagosome biogenesis may be related: both autophagosomes and LDs have been observed cradled within “cups” of ER membrane, and LC3 and Atg2, which are involved in autophagosome biogenesis, also localize to LDs (Robenek et al., 2006; Hayashi-Nishino et al., 2009; Shibata et al., 2009, 2010; Velikkakath et al., 2012). These observations suggest that autophagosome and LD biogenesis may share some common machinery, but whether Pex30 ER subdomains are involved in autophagosome biogenesis is currently unknown.
Finally, the ER subdomains involved in organelle biogenesis may also be membrane contact sites through which peroxisomes and LDs communicate with the ER after their biogenesis. In this case they could play roles in organelle remodeling, or in the trafficking of proteins and/or lipids between the ER, peroxisomes, and LDs. Peroxisomes in mammalian cells are tethered to the ER via ACBD5-VAP proteins. This tether is essential for peroxisomes to receive lipids from the ER required for peroxisomal membrane expansion (Costello et al., 2017; Hua et al., 2017). Several tethers have been proposed for ER-LD contact sites. These include interactions between NRZ-SNARE on the ER and Rab18 on LDs in mammalian cells, and between the lipid synthesis enzymes FATP1 on the ER and DGAT2 on LDs in worms (Xu et al., 2012, 2018). Whether Pex30 ER subdomains colocalize with any of these tethers, or whether they form distinct membrane contact sites, is currently unknown. The precise protein composition of these ER-LD-peroxisome membrane contact sites, their physiological function, and regulation all remain to be determined. The recent development of new biochemical and microscopic tools to study membrane microdomains and membrane contact sites is sure to open up many new areas of research in this exciting field.
Author Contributions
AJ and SC conceived and wrote the manuscript.
Funding
This work was supported by The University of North Carolina at Chapel Hill, by the National Institute on Aging of the National Institutes of Health under award number R00AG052570, and by the Alzheimer’s Association under award number 2018-AARG-590347.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Sarah Monroe for help with the figures.
References
Agrawal, G., Fassas, S. N., Xia, Z. J., and Subramani, S. (2016). Distinct requirements for intra-ER sorting and budding of peroxisomal membrane proteins from the ER. J. Cell Biol. 212, 335–345. doi: 10.1083/jcb.201506141
Agrawal, G., Joshi, S., and Subramani, S. (2011). Cell-free sorting of peroxisomal membrane proteins from the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 108, 9113–9118. doi: 10.1073/pnas.1018749108
Agrawal, G., and Subramani, S. (2013). Emerging role of the endoplasmic reticulum in peroxisome biogenesis. Front. Physiol. 4:286. doi: 10.3389/fphys.2013.00286
Argyriou, C., D’Agostino, M. D., and Braverman, N. (2017). “Peroxisome biogenesis disorders,” in Metabolic Diseases: Foundations of Clinical Management, Genetics, and Pathology, eds E. Gilbert-Barness, L. A. Barness, and P. M. Farrell (Amsterdam: IOS Press), 847–880. doi: 10.3233/978-1-61499-718-4-847
Baudhuin, P., Beaufay, H., and De Duve, C. (1965). Combined biochemical and morphological study of particulate fractions from rat liver. Analysis of preparations enriched in lysosomes or in particles containing urate oxidase, D-amino acid oxidase, and catalase. J. Cell Biol. 26, 219–243. doi: 10.1083/jcb.26.1.219
Ben M’barek, K., Ajjaji, D., Chorlay, A., Vanni, S., Forêt, L., and Thiam, A. R. (2017). ER membrane phospholipids and surface tension control cellular lipid droplet formation. Dev. Cell 41, 591–604.e7. doi: 10.1016/j.devcel.2017.05.012
Bersuker, J. A. (2018). In close proximity: the lipid droplet proteome and crosstalk with the endoplasmic reticulum. Contact 1, 1–3.
Bersuker, K., Peterson, C. W. H., To, M., Sahl, S. J., Savikhin, V., Grossman, E. A., et al. (2018). A proximity labeling strategy provides insights into the composition and dynamics of lipid droplet proteomes. Dev. Cell 44, 97–112.e7. doi: 10.1016/j.devcel.2017.11.020
Binns, D., Januszewski, T., Chen, Y., Hill, J., Markin, V. S., Zhao, Y., et al. (2006). An intimate collaboration between peroxisomes and lipid bodies. J. Cell Biol. 173, 719–731. doi: 10.1083/jcb.200511125
Chang, C.-L., Weigel, A. V., Ioannou, M. S., Pasolli, H. A., Xu, C. S., Peale, D. R., et al. (2019). Spastin tethers lipid droplets to peroxisomes and directs fatty acid trafficking through ESCRT-III. bioRxiv [Preprint]. doi: 10.1101/544023
Chorlay, A., Forêt, L., Vanni, S., Thiam, A. R., Ajjaji, D., and Ben M’barek, K. (2017). ER membrane phospholipids and surface tension control cellular lipid droplet formation. Dev. Cell 41, 591–604.e7. doi: 10.1016/j.devcel.2017.05.012
Choudhary, V., Golani, G., Joshi, A. S., Cottier, S., Schneiter, R., Prinz, W. A., et al. (2018). Architecture of lipid droplets in endoplasmic reticulum is determined by phospholipid intrinsic curvature. Curr. Biol. 28, 915–926.e9. doi: 10.1016/j.cub.2018.02.020
Choudhary, V., Ojha, N., Golden, A., and Prinz, W. A. (2015). A conserved family of proteins facilitates nascent lipid droplet budding from the ER. J. Cell Biol. 211, 261–271. doi: 10.1083/jcb.201505067
Cohen, S. (2018). Lipid droplets as organelles. Int. Rev. Cell Mol. Biol. 337, 83–110. doi: 10.1016/bs.ircmb.2017.12.007
Costello, J. L., Castro, I. G., Hacker, C., Schrader, T. A., Metz, J., Zeuschner, D., et al. (2017). ACBD5 and VAPB mediate membrane associations between peroxisomes and the ER. J. Cell Biol. 216, 331–342. doi: 10.1083/jcb.201607055
de Duve, C. (1969). Evolution of the peroxisome. Ann. N. Y. Acad. Sci. 168, 369–381. doi: 10.1111/j.1749-6632.1969.tb43124.x
de Duve, C., and Baudhuin, P. (1966). Peroxisomes (microbodies and related particles). Physiol. Rev. 46, 323–357. doi: 10.1152/physrev.1966.46.2.323
Delille, H. K., Bonekamp, N. A., and Schrader, M. (2006). Peroxisomes and disease - an overview. Int. J. Biomed. Sci. 2, 308–314.
Deslandes, F., Thiam, A. R., and Forêt, L. (2017). Lipid droplets can spontaneously bud off from a symmetric bilayer. Biophys. J. 113, 15–18. doi: 10.1016/j.bpj.2017.05.045
Ducharme, N. A., and Bickel, P. E. (2008). Minireview: lipid droplets in lipogenesis and lipolysis. Endocrinology 149, 942–949. doi: 10.1210/en.2007-1713
Eisenberg-Bord, M., Mari, M., Weill, U., Rosenfeld-Gur, E., Moldavski, O., Castro, I. G., et al. (2018). Identification of seipin-linked factors that act as determinants of a lipid droplet subpopulation. J. Cell Biol. 217, 269–282. doi: 10.1083/jcb.201704122
Erdmann, R., Baumgart, E., Linkert, M., Kunau, W.-H., Götte, K., Girzalsky, W., et al. (2015). Pex19p, a farnesylated protein essential for peroxisome biogenesis. Mol. Cell. Biol. 18, 616–628. doi: 10.1128/mcb.18.1.616
Exner, T., Romero-Brey, I., Yifrach, E., Rivera-Monroy, J., Schrul, B., Zouboulis, C. C., et al. (2019). An alternative membrane topology permits lipid droplet localization of peroxisomal fatty acyl-CoA reductase 1. J. Cell Sci. 132:223016. doi: 10.1242/jcs.223016
Fei, W., Shui, G., Gaeta, B., Du, X., Kuerschner, L., Li, P., et al. (2008). Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol. 180, 473–482. doi: 10.1083/jcb.200711136
Fujiki, Y., Okumoto, K., Mukai, S., Honsho, M., and Tamura, S. (2014). Peroxisome biogenesis in mammalian cells. Front. Physiol. 5:307. doi: 10.3389/fphys.2014.00307
Geuze, H. J. (2003). Involvement of the endoplasmic reticulum in peroxisome formation. Mol. Biol. Cell 14, 2900–2907. doi: 10.1091/mbc.e02-11-0734
Goldman, B. M., and Blobel, G. (2006). Biogenesis of peroxisomes: intracellular site of synthesis of catalase and uricase. Proc. Natl. Acad. Sci. U.S.A. 75, 5066–5070. doi: 10.1073/pnas.75.10.5066
Gould, S. J., and Valle, D. (2000). Peroxisome biogenesis disorders: genetics and cell biology. Trends Genet. 16, 340–345. doi: 10.1016/S0168-9525(00)02056-4
Grippa, A., Buxó, L., Mora, G., Funaya, C., Idrissi, F. Z., Mancuso, F., et al. (2015). The seipin complex Fld1/Ldb16 stabilizes ER-lipid droplet contact sites. J. Cell Biol. 211, 829–844. doi: 10.1083/jcb.201502070
Hariri, H., Rogers, S., Ugrankar, R., Liu, Y. L., Feathers, J. R., and Henne, M. (2018). Lipid droplet biogenesis is spatially coordinated at ER–vacuole contacts under nutritional stress. EMBO Rep. 19, 57–72. doi: 10.15252/embr
Hayashi-Nishino, M., Fujita, N., Noda, T., Yamaguchi, A., Yoshimori, T., and Yamamoto, A. (2009). A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11, 1433–1437. doi: 10.1038/ncb1991
Hettema, E. H., Girzalsky, W., Van Den Berg, M., Erdmann, R., and Distel, B. (2000). Saccharomyces cerevisiae Pex3p and Pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J. 19, 223–233. doi: 10.1093/emboj/19.2.223
Hoepfner, D., Schildknegt, D., Braakman, I., Philippsen, P., and Tabak, H. F. (2005). Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122, 85–95. doi: 10.1016/j.cell.2005.04.025
Hohfeld, J., Veenhuis, M., and Kunau, W. H. (1991). PAS3, a Saccharomyces cerevisiae gene encoding a peroxisomal integral membrane protein essential for peroxisome biogenesis. J. Cell Biol. 114, 1167–1178. doi: 10.1083/jcb.114.6.1167
Hua, R., Cheng, D., Coyaud,É., Freeman, S., Di Pietro, E., Wang, .Y, et al. (2017). VAPs and ACBD5 tether peroxisomes to the ER for peroxisome maintenance and lipid homeostasis. J. Cell Biol. 216, 367–377. doi: 10.1083/jcb.201608128
Jacquier, N., Choudhary, V., Mari, M., Toulmay, A., Reggiori, F., and Schneiter, R. (2011). Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Sci. 124(Pt 14), 2424–2437. doi: 10.1242/jcs.076836
Joshi, A. S., Huang, X., Choudhary, V., Levine, T. P., Hu, J., and Prinz, W. A. (2016). A family of membrane-shaping proteins at ER subdomains regulates pre-peroxisomal vesicle biogenesis. J. Cell Biol. 215, 515–529. doi: 10.1083/jcb.201602064
Joshi, A. S., Nebenfuehr, B., Choudhary, V., Satpute-Krishnan, P., Levine, T. P., Golden, A., et al. (2018). Lipid droplet and peroxisome biogenesis occur at the same ER subdomains. Nat. Commun. 9:2940. doi: 10.1038/s41467-018-05277-3
Joshi, A. S., Zhang, H., and Prinz, W. A. (2017). Organelle biogenesis in the endoplasmic reticulum. Nat. Cell Biol. 19, 876–882. doi: 10.1038/ncb3579
Kadereit, B., Kumar, P., Torregroza, I., Wang, W.-J., Severina, N., Silver, D. L., et al. (2007). Evolutionarily conserved gene family important for fat storage. Proc. Natl. Acad. Sci. U.S.A. 105, 94–99. doi: 10.1073/pnas.0708579105
Kassan, A., Herms, A., Fernández-Vidal, A., Bosch, M., Schieber, N. L., Reddy, B. J. N., et al. (2013). Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J. Cell Biol. 203, 985–1001. doi: 10.1083/jcb.201305142
Khor, V. K., Shen, W. J., and Kraemer, F. B. (2013). Lipid droplet metabolism. Curr. Opin. Clin. Nutr. Metab. Care 16, 632–637. doi: 10.1097/MCO.0b013e3283651106
Kim, P. (2017). Peroxisome biogenesis: a union between two organelles. Curr. Biol. 27, R271–R274. doi: 10.1016/j.cub.2017.02.052
Kim, P. K., and Hettema, E. H. (2015). Multiple pathways for protein transport to peroxisomes. J. Mol. Biol. 427(6 Pt A), 1176–1190. doi: 10.1016/j.jmb.2015.02.005
Kim, P. K., Mullen, R. T., Schumann, U., and Lippincott-Schwartz, J. (2006). The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J. Cell Biol. 173, 521–532. doi: 10.1083/jcb.200601036
Kimura, H., Arasaki, K., Ohsaki, Y., Fujimoto, T., Ohtomo, T., Yamada, J., et al. (2018). Syntaxin 17 promotes lipid droplet formation by regulating the distribution of acyl-CoA synthetase 3. J. Lipid Res. 59, 805–819. doi: 10.1194/jlr.M081679
Knoops, K., De Boer, R., Kram, A., and Van Der Klei, I. J. (2015). Yeast pex1 cells contain peroxisomal ghosts that import matrix proteins upon reintroduction of Pex1. J. Cell Biol. 211, 955–962. doi: 10.1083/jcb.201506059
Knoops, K., Manivannan, S., Cepińska, M. N., Krikken, A. M., Kram, A. M., Veenhuis, M., et al. (2014). Preperoxisomal vesicles can form in the absence of Pex3. J. Cell Biol. 204, 659–668. doi: 10.1083/jcb.201310148
Kohlwein, S. D., Veenhuis, M., and van der Klei, I. J. (2013). Lipid droplets and peroxisomes: key players in cellular lipid homeostasis or a matter of fat-store ‘em up or burn ’em down. Genetics 193, 1–50. doi: 10.1534/genetics.112.143362
Krahmer, N., Farese, R. V., and Walther, T. C. (2013). Balancing the fat: lipid droplets and human disease. EMBO Mol. Med. 5, 973–983. doi: 10.1002/emmm.201100671
Kunau, W. H., and Hartig, A. (1992). Peroxisome biogenesis in Saccharomyces cerevisiae. Antonie Van Leeuwenhoek 62, 63–78. doi: 10.1007/BF00584463
Lam, S. K., Yoda, N., and Schekman, R. (2011). A vesicle carrier that mediates peroxisome protein traffic from the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 107, 21523–21528. doi: 10.1073/pnas.1103526108
Lazarow, P. B. (1983). Biogenesis of peroxisomal content proteins: in vivo and in vitro studies. Methods Enzymol. 96, 721–728. doi: 10.1016/S0076-6879(83)96061-5
Li, W.-P., Szymanski, K. M., Goodman, J. M., Grishin, N. V., Anderson, R. G. W., Binns, D., et al. (2007). The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. U.S.A. 104, 20890–20895. doi: 10.1073/pnas.0704154104
Liu, H., Tan, X., Veenhuis, M., McCollum, D., and Cregg, J. M. (1992). An efficient screen for peroxisome-deficient mutants of Pichia pastoris. J. Bacteriol. 174, 4943–4951. doi: 10.1128/jb.174.15.4943-4951.1992
Liu, X. N., Lin, Q., Walther, T. C., Upadhyayula, S., Farese, R. V., Housden, B. E., et al. (2016). Seipin is required for converting nascent to mature lipid droplets. eLife 5:e16582. doi: 10.7554/elife.16582
Lv, X., Liu, J., Qin, Y., Liu, Y., Jin, M., Dai, J., et al. (2019). Identification of gene products that control lipid droplet size in yeast using a high-throughput quantitative image analysis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1864, 113–127. doi: 10.1016/j.bbalip.2018.11.001
Mannaerts, G. P., and Van Veldhoven, P. P. (1996). Functions and organization of peroxisomal β-oxidation. Ann. N. Y. Acad. Sci. 804, 99–115. doi: 10.1111/j.1749-6632.1996.tb18611.x
Mast, F. D., Herricks, T., Strehler, K. M., Miller, L. R., Saleem, R. A., Rachubinski, R. A., et al. (2018). ESC RT-III is required for scissioning new peroxisomes from the endoplasmic reticulum. J. Cell Biol. 217, 2087–2102. doi: 10.1083/jcb.201706044
Miranda, D. A., Kim, J. H., Nguyen, L. N., Cheng, W., Tan, B. C., Goh, V. J., et al. (2014). Fat storage-inducing transmembrane protein 2 is required for normal fat storage in adipose tissue. J. Biol. Chem. 289, 9560–9572. doi: 10.1074/jbc.M114.547687
Motley, A. M., Galvin, P. C., Ekal, L., Nuttall, J. M., and Hettema, E. H. (2015). Reevaluation of the role of Pex1 and dynamin-related proteins in peroxisome membrane biogenesis. J. Cell Biol. 211, 1041–1056. doi: 10.1083/jcb.201412066
Motley, A. M., and Hettema, E. H. (2007). Yeast peroxisomes multiply by growth and division. J. Cell Biol. 178, 399–410. doi: 10.1083/jcb.200702167
Novikoff, P. M., and Novikoff, A. B. (1972). Peroxisomes in absorptive cells of mammalian small intestine. J. Cell Biol. 53, 532–560. doi: 10.1083/jcb.53.2.532
Olzmann, J. A., and Carvalho, P. (2018). Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 20, 137–155. doi: 10.1038/s41580-018-0085-z
Onal, G., Kutlu, O., Gozuacik, D., and Dokmeci Emre, S. (2017). Lipid droplets in health and disease. Lipids Health Dis. 16:128. doi: 10.1186/s12944-017-0521-7
Robenek, H., Hofnagel, O., Buers, I., Robenek, M. J., Troyer, D., and Severs, N. J. (2006). Adipophilin-enriched domains in the ER membrane are sites of lipid droplet biogenesis. J. Cell Sci. 119(Pt 20), 4215–4224. doi: 10.1242/jcs.03191
Rucktäschel, R., Halbach, A., Girzalsky, W., Rottensteiner, H., and Erdmann, R. (2010). De novo synthesis of peroxisomes upon mitochondrial targeting of Pex3p. Eur. J. Cell Biol. 89, 947–954. doi: 10.1016/j.ejcb.2010.06.012
Salo, V. T., Belevich, I., Li, S., Karhinen, L., Vihinen, H., Vigouroux, C., et al. (2016). Seipin regulates ER–lipid droplet contacts and cargo delivery. EMBO J. 35, 2699–2716. doi: 10.15252/embj.201695170
Schrader, M. (2001). Tubulo-reticular clusters of peroxisomes in living COS-7 cells: dynamic behavior and association with lipid droplets. J. Histochem. Cytochem. 49, 1421–1429. doi: 10.1177/002215540104901110
Schrul, B., and Kopito, R. R. (2016). Peroxin-dependent targeting of a lipid-droplet-destined membrane protein to ER subdomains. Nat. Cell Biol. 18, 740–751. doi: 10.1038/ncb3373
Shibata, M., Yoshimura, K., Furuya, N., Koike, M., Ueno, T., Komatsu, M., et al. (2009). The MAP1-LC3 conjugation system is involved in lipid droplet formation. Biochem. Biophys. Res. Commun. 382, 419–423. doi: 10.1016/j.bbrc.2009.03.039
Shibata, M., Yoshimura, K., Tamura, H., Ueno, T., Nishimura, T., Inoue, T., et al. (2010). LC3, a microtubule-associated protein1A/B light chain3, is involved in cytoplasmic lipid droplet formation. Biochem. Biophys. Res. Commun. 393, 274–279. doi: 10.1016/j.bbrc.2010.01.121
Shin, O. H., Hau, W., Wang, Y., and Südhof, T. C. (2005). Evolutionarily conserved multiple C2 domain proteins with two transmembrane regions (MCTPs) and unusual Ca2+ binding properties. J. Biol. Chem. 280, 1641–1651. doi: 10.1074/jbc.M407305200
Singh, R., Kaushik, S., Wang, Y., Xiang, Y., Novak, I., Komatsu, M., et al. (2009). Autophagy regulates lipid metabolism. Nature 458, 1131–1135. doi: 10.1038/nature07976
South, S. T., Gould, S. J., Li, X., Sacksteder, K. A., and Liu, Y. (2002). Inhibitors of COPI and COPII do not block PEX3-mediated peroxisome synthesis. J. Cell Biol. 149, 1345–1360. doi: 10.1083/jcb.149.7.1345
South, S. T., Sacksteder, K. A., Li, X., Liu, Y., and Gould, S. J. (2000). Inhibitors of COPI and COPII do not block PEX3-mediated peroxisome synthesis. J. Cell Biol. 149, 1345–1360. doi: 10.1083/jcb.149.7.1345
Sugiura, A., Mattie, S., Prudent, J., and Mcbride, H. M. (2017). Newly born peroxisomes are a hybrid of mitochondrial and ER-derived pre-peroxisomes. Nature 542, 251–254. doi: 10.1038/nature21375
Sui, X., Arlt, H., Brock, K. P., Lai, Z. W., DiMaio, F., Marks, D. S., et al. (2018). Cryo–electron microscopy structure of the lipid droplet–formation protein seipin. J. Cell Biol. 217, 4080–4091. doi: 10.1083/jcb.201809067
Tabak, H. F., Braakman, I., and van der Zand, A. (2013). Peroxisome formation and maintenance are dependent on the endoplasmic reticulum. Annu. Rev. Biochem. 82, 723–744. doi: 10.1146/annurev-biochem-081111-125123
Tam, Y. Y. C., Fagarasanu, A., Fagarasanu, M., and Rachubinski, R. A. (2005). Pex3p initiates the formation of a preperoxisomal compartment from a subdomain of the endoplasmic reticulum in Saccharomyces cerevisiae. J. Biol. Chem. 280, 34933–34939. doi: 10.1074/jbc.M506208200
Tan, J. S. Y., Seow, C. J. P., Goh, V. J., and Silver, D. L. (2014). Recent advances in understanding proteins involved in lipid droplet formation, growth and fusion. J. Genet. Genomics 41, 251–259. doi: 10.1016/j.jgg.2014.03.003
Teixeira, V., Johnsen, L., Martínez-Montañés, F., Grippa, A., Buxó, L., Idrissi, F. Z., et al. (2018). Regulation of lipid droplets by metabolically controlled Ldo isoforms. J. Cell Biol. 217:127. doi: 10.1083/jcb.201704115
Titorenko, V. I., Chan, H., and Rachubinski, R. A. (2000). Fusion of small peroxisomal vesicles in vitro reconstructs an early step in the in vivo multistep peroxisome assembly pathway of Yarrowia lipolytica. J. Cell Biol. 148, 29–44. doi: 10.1083/jcb.148.1.29
Tsukada, H., Mochizuki, Y., and Konishi, T. (1968). Morphogenesis and development of microbodies of hepatocytes of rats during pre- and postnatal growth. J. Cell Biol. 37, 231–243. doi: 10.1083/jcb.37.2.231
Valm, A. M., Cohen, S., Legant, W. R., Melunis, J., Hershberg, U., Wait, E., et al. (2017). Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546, 162–167. doi: 10.1038/nature22369
Van der Leij, I., Van den Berg, M., Boot, R., Franse, M., Distel, B., and Tabak, H. F. (1992). Isolation of peroxisome assembly mutants from Saccharomyces cerevisiae with different morphologies using a novel positive selection procedure. J. Cell Biol. 119, 153–162. doi: 10.1083/jcb.119.1.153
van der Zand, A., Braakman, I., and Tabak, H. F. (2010). Peroxisomal membrane proteins insert into the endoplasmic reticulum. Mol. Biol. Cell 21, 2057–2065. doi: 10.1091/mbc.e10-02-0082
Van Der Zand, A., Gent, J., Braakman, I., and Tabak, H. F. (2012). Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell 149, 397–409. doi: 10.1016/j.cell.2012.01.054
Velikkakath, A. K. G., Nishimura, T., Oita, E., Ishihara, N., and Mizushima, N. (2012). Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol. Biol. Cell 23, 896–909. doi: 10.1091/mbc.e11-09-0785
Vizeacoumar, F. J., Vreden, W. N., Aitchison, J. D., and Rachubinski, R. A. (2006). Pex19p binds Pex30p and Pex32p at regions required for their peroxisomal localization but separate from their peroxisomal targeting signals. J. Biol. Chem. 281, 14805–14812. doi: 10.1074/jbc.M601808200
Voorn-Brouwer, T., Kragt, A., Tabak, H. F., and Distel, B. (2001). Peroxisomal membrane proteins are properly targeted to peroxisomes in the absence of COPI- and COPII-mediated vesicular transport. J. Cell Sci. 114(Pt 11), 2199–2204.
Walker, C. L., Pomatto, L. C. D., Tripathi, D. N., and Davies, K. J. A. (2017). Redox regulation of homeostasis and proteostasis in peroxisomes. Physiol. Rev. 98, 89–115. doi: 10.1152/physrev.00033.2016
Wanders, R. J. A. (2014). Metabolic functions of peroxisomes in health and disease. Biochimie 98, 36–44. doi: 10.1016/j.biochi.2013.08.022
Wang, C.-W., Miao, Y.-H., and Chang, Y.-S. (2014). Control of lipid droplet size in budding yeast requires the collaboration between Fld1 and Ldb16. J. Cell Sci. 127(Pt 6), 1214–1228. doi: 10.1242/jcs.137737
Wang, H., Becuwe, M., Housden, B. E., Chitraju, C., Porras, A. J., Graham, M. M., et al. (2016). Seipin is required for converting nascent to mature lipid droplets. eLife 5:e16582. doi: 10.7554/eLife.16582
Wang, S., Horn, P. J., Liou, L. C., Muggeridge, M. I., Zhang, Z., Chapman, K. D., et al. (2013). A peroxisome biogenesis deficiency prevents the binding of alpha-synuclein to lipid droplets in lipid-loaded yeast. Biochem. Biophys. Res. Commun. 438, 452–456. doi: 10.1016/j.bbrc.2013.07.100
Wang, S., Idrissi, F. Z., Hermansson, M., Grippa, A., Ejsing, C. S., and Carvalho, P. (2018). Seipin and the membrane-shaping protein Pex30 cooperate in organelle budding from the endoplasmic reticulum. Nat. Commun. 9:2939. doi: 10.1038/s41467-018-05278-2
Waterham, H. R., Ferdinandusse, S., and Wanders, R. J. A. (2016). Human disorders of peroxisome metabolism and biogenesis. Biochim. Biophys. Acta Mol. Cell Res. 1863, 922–933. doi: 10.1016/j.bbamcr.2015.11.015
Wróblewska, J. P., Cruz-Zaragoza, L. D., Yuan, W., Schummer, A., Chuartzman, S. G., de Boer, R., et al. (2017). Saccharomyces cerevisiae cells lacking Pex3 contain membrane vesicles that harbor a subset of peroxisomal membrane proteins. Biochim. Biophys. Acta Mol. Cell Res. 1864, 1656–1667. doi: 10.1016/j.bbamcr.2017.05.021
Xu, D., Li, Y., Wu, L., Li, Y., Zhao, D., Yu, J., et al. (2018). Rab18 promotes lipid droplet (LD) growth by tethering the ER to LDs through SNARE and NRZ interactions. J. Cell Biol. 217, 975–995. doi: 10.1083/jcb.201704184
Xu, N., Zhang, S. O., Cole, R. A., McKinney, S. A., Guo, F., Haas, J. T., et al. (2012). The FATP1-DGAT2 complex facilitates lipid droplet expansion at the ER-lipid droplet interface. J. Cell Biol. 198, 895–911. doi: 10.1083/jcb.201201139
Yan, R., Qian, H., Lukmantara, I., Gao, M., Du, X., Yan, N., et al. (2018). Human SEIPIN binds anionic phospholipids. Dev. Cell 47, 248–256.e4. doi: 10.1016/j.devcel.2018.09.010
Keywords: lipid droplet, peroxisome, organelle biogenesis, membrane trafficking, lipid metabolism
Citation: Joshi AS and Cohen S (2019) Lipid Droplet and Peroxisome Biogenesis: Do They Go Hand-in-Hand? Front. Cell Dev. Biol. 7:92. doi: 10.3389/fcell.2019.00092
Received: 12 March 2019; Accepted: 14 May 2019;
Published: 31 May 2019.
Edited by:
Guillaume Thibault, Nanyang Technological University, SingaporeReviewed by:
MIke Henne, UT Southwestern Medical Center, United StatesJoel M. Goodman, UT Southwestern Medical Center, United States
Michael Schrader, University of Exeter, United Kingdom
Copyright © 2019 Joshi and Cohen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amit S. Joshi, amtya21@gmail.com; Sarah Cohen, sarahcoh@med.unc.edu
 Amit S. Joshi
Amit S. Joshi Sarah Cohen
Sarah Cohen