The ATP-Dependent Protease ClpP Inhibits Biofilm Formation by Regulating Agr and Cell Wall Hydrolase Sle1 in Staphylococcus aureus
- 1Department of Laboratory Medicine, School of Medicine, Ren Ji Hospital, Shanghai Jiao Tong University, Shanghai, China
- 2Department of Laboratory Medicine, Shanghai Children's Medical Center, School of Medicine, Shanghai Jiao Tong University, Shanghai, China
- 3Institute of Analytical Chemistry and Synthetic and Functional Biomolecules Center, College of Chemistry and Molecular Engineering, Peking University, Beijing, China
- 4Department of Microbiology and Immunology, Indiana University School of Medicine-Northwest, Gary, IN, USA
Biofilm causes hospital-associated infections on indwelling medical devices. In Staphylococcus aureus, Biofilm formation is controlled by intricately coordinated network of regulating systems, of which the ATP-dependent protease ClpP shows an inhibitory effect. Here, we demonstrate that the inhibitory effect of ClpP on biofilm formation is through Agr and the cell wall hydrolase Sle1. Biofilm formed by clpP mutant consists of proteins and extracellular DNA (eDNA). The increase of the protein was, at least in part, due to the reduced protease activity of the mutant, which was caused by the decreased activity of agr. On the other hand, the increase of eDNA was due to increased cell lysis caused by the higher level of Sle1. Indeed, as compared with wild type, the clpP mutant excreted an increased level of eDNA, and showed higher sensitivity to Triton-induced autolysis. The deletion of sle1 in the clpP mutant decreased the biofilm formation, the level of eDNA, and the Triton-induced autolysis to wild-type levels. Despite the increased biofilm formation capability, however, the clpP mutant showed significantly reduced virulence in a murine model of subcutaneous foreign body infection, indicating that the increased biofilm formation capability cannot compensate for the intrinsic functions of ClpP during infection.
Introduction
Bacteria biofilms are complex communities which have intrinsic resistance to host immune defenses and antibiotic treatment (Paharik and Horswill, 2016). In the important human pathogen Staphylococcus aureus, biofilm enables the bacterium to persist during infections on indwelling medical devices, endocarditis, or chronic wound infections (Otto, 2013). Depending on its constituents, staphylococcal biofilm can be polysaccharide intercellular adhesion (PIA)-dependent or proteins/ extracellular DNA (eDNA)-dependent (McCarthy et al., 2015). PIA is synthesized by the enzyme encoded by the icaADBC operon, and can be an important component of the biofilm matrix (Cramton et al., 1999; Gotz, 2002; Jefferson et al., 2004). However, several studies have shown that S. aureus can form a biofilm by proteins and eDNA without PIA-involvement (Toledo-Arana et al., 2005; Rohde et al., 2007). For proteins, protein A (Spa), and the fibronectin-binding proteins FnbpA and FnbpB, are known to play an important role in the biofilm matrix formation (O'Neill et al., 2008; Merino et al., 2009). In the PIA-independent biofilm, extracellular DNA (eDNA) is also an important component of biofilm matrix (Kiedrowski et al., 2011; Okshevsky and Meyer, 2015). For example, secreted protease Esp inhibits biofilm formation by cleaving murein hydrolase autolysin (Atl) and preventing release of eDNA (Chen et al., 2013).
Biofilm formation in S. aureus is controlled by an intricate network of regulatory systems. Rot (repressor of toxin) contributes to biofilm formation by down-regulating secreted proteases (Mootz et al., 2015). The accessory gene regulator (Agr) is known to inhibit biofilm formation by up-regulating extracellular proteases, an important contributor to the dispersal of established biofilm (Boles and Horswill, 2008). Sigma factor B (sigB) contributes to biofilm formation by suppressing the expression of RNAIII, the effector molecule of Agr, which, in turn, leads to decreased levels of extracellular proteases (Lauderdale et al., 2009; Marti et al., 2010). Recently, AraC-type transcriptional regulator, Rsp, was shown to inhibit biofilm formation by repressing the expression of biofilm-associated genes such as ica operon (Li et al., 2015).
Another important biofilm regulatory system is the ATP-dependent protease ClpP, the proteolytic subunit of Clp proteases (Arnold and Langer, 2002; Dalbey et al., 2012). In Clp proteases, the proteolytic chamber is formed by two hexameric rings of ClpP subunits, whereas the ATPase function is provided by Clp ATPases such as ClpB, ClpC, ClpL, and ClpX. The ClpP proteolytic activity is reported to play a critical role on virulence, stress response, and physiology in S. aureus (Frees et al., 2004). In the S. aureus 8325-4 strain, while ATPases ClpX and ClpC promote biofilm formation, ClpP represses it (Frees et al., 2004). However, it is not known whether the biofilm inhibitory effect of ClpP is universal, and, if so, how ClpP does it.
In this study, we found that the biofilm inhibitory effect of ClpP is conserved among different S. aureus strains. Moreover, we further demonstrate that the inhibitory effect is, at least in part, due to ClpP's effect on the quorum sensing system Agr, and the cell wall hydrolyzing enzyme Sle1.
Materials and Methods
Ethics Statement
All animal experiments protocols were performed following the Guide for the Care and Use of Laboratory Animals of the Chinese Association for Laboratory Animal Sciences (CALAS) and were approved by the ethics committee of Renji Hospital, School of medicine, Shanghai Jiaotong University, Shanghai, China.
Bacterial Strains, Plasmids and Culture Conditions
All experiments were performed with S. aureus Newman (NM) or USA300 as the wild-type strains. The bacterial strains and plasmids used in this study are listed in Supplementary Table 1. Escherichia coli and S. aureus were grown in Luria-Bertani (LB) broth and tryptic soy broth (TSB), respectively. However, for transduction of plasmids, heart infusion broth (HIB) supplemented with 5 mM CaCl2 was used. When necessary, antibiotics were added to the growth media at the following concentrations: ampicillin, 100 μg/ml; erythromycin, 10 μg/ml; and chloramphenicol, 5 μg/ml.
Construction of Plasmids
To construct the plasmid for deleting the lytM, sle1, or agr genes in Newman and USA300 strains, we used a ligation independent cloning (LIC) method (Aslanidis and De Jong, 1990). First, vector DNA was PCR-amplified from pKOR1 using the primers P236/237 (Supplementary Table 2). One- kb DNA fragments, upstream and downstream of lytM, sle1, or agr, were amplified by PCR from the chromosomal DNA with the primer pairs P596/597 and P598/599 (for lytM), P632/633, and P634/637 (for sle1) or P19/20 and P21/22 (for agr) (Supplementary Table 2) and PrimSTAR (Takara), a PCR enzyme with high fidelity. The PCR products were treated with T4 DNA polymerase in the presence of dCTP (vector) or dGTP (insert DNA) and mixed together. The DNA mixture was used to transform E. coli DH5α. The resulting plasmids, pKOR1ΔlytM, pKOR1Δsle1, or pKOR1Δagr were electroporated into S. aureus strain RN4220 and subsequently transduced into NM or USA300 with Φ85. The deletion was carried out as described previously (Bae and Schneewind, 2006).
To generate complement plasmids for clpP mutant strain, First, vector DNA was PCR-amplified from pCL55 using the primers P35/80, the clpP gene full length with its own promoter was amplified with the primer pairs P2525/2526 (Supplementary Table 2). The plasmid was constructed with LIC and the resulting plasmid, pclpP, was electroporated into S. aureus strain RN4220 and then into NMΔclpP with Φ85.
To express Sle1 proteins in S. aureus, the gene sle1 was PCR-amplified with the following primer pairs: PL96/97 (Supplementary Table 2). The amplified fragments were digested with either BamHI/SalI. Then they were inserted into the multi-copy plasmid pOS1 under the control of its own promoter, resulting in pOS1-Sle1-his. The plasmids were transformed into E. coli DH5a, and then into S. aureus RN4220. Finally, the plasmids were transduced by Φ85 into wild type and clpP transposon mutants of S. aureus. The test strains in TSB containing chloramphicol (10 μg/ml) were incubated until early stationary growth phase (OD600 is around 2). The proteins were detected by Western blot analysis with anti-His-tag antibody.
DNA Manipulation
Unless stated otherwise, all restriction enzymes and DNA modification enzymes were purchased from New England Biolabs. Plasmids and genomic DNA were extracted with plasmid miniprep kit (Omega) according to the manufacturer's instruction. Plasmid DNA was introduced into E. coli by the method of Hanahan (Hanahan, 1983) and into S. aureus RN4220 by electroporation with a gene pulser (Bio-Rad). Subsequent transduction of the plasmids into target strains of S. aureus was carried out with Φ 85.
Real-Time Quantitative Reverse Transcription-PCR (RT-PCR)
Cells in 3 ml TSB cultures were harvested at the stationary phase (OD600 value of 2). Cells were disrupted by shaking with a Mini-Beadbeater (Biospec Products) at maximum speed for 30 s. Tubes were then incubated on ice for 5 min. Then, the suspension was centrifuged. Total RNA isolation from the supernatant was performed according to the manufacturer's instructions (Qiagen). After treatment using a TURBO DNA-freeTM kit (Ambion), ~1 μg of total RNA was reverse-transcribed with a HieffTM first Strand cDNA Synthesis Super Mix for RT-PCR kit (Yeasen Bio). The cDNA was used as a template for real-time PCR using SYBR-green PCR reagents (Roche). Reactions were performed in a MicroAmp Optical 96-well reaction plate using a 7,500 Sequence Detector (Applied Biosystems). Primers used are listed in Supplementary Table 2. All RT-PCR experiments were performed in triplicate, with gyrase B (gyrB) used as an internal control.
Western Blot Hybridization
Western blot analysis of proteins was carried out as described previously (Sun et al., 2011). The AgrA antibody was generated by GLbiochem, China. The His-tag antibody was bought from Yeasen Bio, China. All other antibodies were generated by GenScript.
Semi-Quantitative Biofilm Assay
Semi-quantitative biofilm assays were performed as described in our previous work (Liu et al., 2011). Briefly, overnight cultures of S. aureus strains were diluted 1:100 into fresh TSB with 0.5% Glucose. The diluted cultures were pipetted into sterile 96-well flat-bottom tissue culture plates and incubated at 37°C for 24 h. Culture supernatants were gently removed, and wells were washed with phosphate-buffered saline (PBS). The adherent organisms at the bottom of the wells were fixed by Bouin fixative over 1 h. Then the fixative was gently removed, wells were washed with PBS and stained with 0.4% (wt/vol) crystal violet. Biofilm formation was measured with a MicroELISA autoreader.
Biofilm Formation Using a BioFlux Microfluidic Flow Cell System
The BioFlux 1000z microfluidic system (Fluxion Biosciences, CA) was used to assess biofilm formation under flow conditions as described (Benoit et al., 2010). To grow biofilms, the microfluidic channels were primed with TSB supplemented with 0.5% glucose at 2 dynes/cm2 for 10 min. Each channel of a 48-well plate was coated with 20% platelet-poor human plasma in 50 mM carbonate buffer, PH 9.6 and incubated for 24 h at 4°C before biofilm assays were set-up. The exponential cultures were diluted 1:100 in TSB supplemented with 0.5% glucose. Bacterial suspensions were seeded at 0.2 dyn/cm2 for 5 s in all channels. The plate was then incubated at room temperature for 1 h to allow cells to adhere. Excess inoculums were removed and 0.8 ml of TSB supplemented with 0.5% glucose was added to the input wells. Biofilms were grown at 37°C with a flow of fresh media at a constant shear of 0.15 dyn/cm2. Images were taken every 10 min for 24 h at 10 × magnification under brightfield.
Extracellular (e) DNA in Culture Supernatants
Cell cultures were grown to exponential phase. Culture supernatants from strain wild-type and its derivatives were filter sterilized, extracted with phenol-chloroform, and ethanol precipitated. Precipitates were suspended in H2O. The volumes of water were varied to compensate for slight differences in OD600 of the initial cultures. The eDNAs were quantified using Nanodrop 2000 (Thermo Scientific) and visualized by agarose gel.
Triton X-100 Induced Autolysis Assays
Bacterial cells were grown in TSB containing 1 M NaCl to the exponential growth phase at 37°C with shaking and then washed and suspended in 0.05 M Tris-HCl buffer (pH 7.5) containing 0.1% Triton X-100 and were incubated at 30°C with shaking. The optical density was measured in intervals. Results were normalized to an OD600 at time zero.
Zymographic Analysis
Cell cultures were grown to exponential phase, and normalized by OD600. The normalized supernatants were filtered through 0.22 μM filter and concentrated with a Centricon 3 concentrator (Millipore) at 4°C according to the manufacturer's instructions. Concentrated supernatants were mixed with an equal volume of Laemmli sample buffer, and 20 μl was separated by 12% SDS-PAGE supplemented with 0.2% autoclaved S. aureus RN4220 cell (wt/vol) as a substrate. After electrophoresis, the gels were washed four times with distilled water for 30 min at room temperature, incubated in 25 mM Tris-HCl containing 1.25% Triton X-100 (pH 8.0) at 37°C for 4 h, and then stained with methylene blue in 0.01% KOH(wt/vol), followed by destaining in deionized water.
Animal Infection Model
A murine subcutaneous foreign body infection model was performed as described by Pozzi et al. (2012). Briefly, 20 male BALB/c mice were randomly and evenly divided into two groups and infected with wild type or clpP mutant bacteria. One-centimeter long silicon catheters (14 gauge) were implanted subcutaneously on each side at the flank of each mouse. Bacterial cells were grown in TSB to the exponential growth phase and then washed with and suspended in phosphate buffered saline (PBS) to OD600 = 1.0. S. aureus (108 CFU) were injected into the catheter bed. After 7 days, the mice were sacrificed. Catheters were aseptically removed and manipulated with the method described previously except with a sonication time of 15 min (Liu et al., 2011). Furthermore, peri-catheter tissue, liver, spleen, kidneys were dissected, weighed, and homogenized in 1 ml PBS. Then serial dilutions of the wash fluid were plated on TSA blood plates, and the recovered S. aureus colonies were counted. Bacteria present in blood were also quantitatively cultured on TSA blood plates for CFU counting.
Statistics
Statistical analysis was performed using Graph-Pad Prism, version 5. Error bars in all graphs show the standard deviation (± SD).
Results
ClpP, but Not Its ATPases, Inhibits Biofilm Formation in Staphylococcus aureus
ClpP has been reported to inhibit biofilm formation in the S. aureus 8325-4 strain (Frees et al., 2004). To examine whether the biofilm inhibitory effect is universal, we analyzed biofilm formation of clpP mutant in the following three strains: S. aureus Newman, a methicillin-sensitive strain (MSSA), USA300, the predominant community-associated methicillin-resistant strain (CA-MRSA) in the USA and ST59 11-775, the most frequent lineage of community-associated infections in China and adjacent Asian countries (Li et al., 2016). In all three strains, clpP mutants showed enhanced biofilm formation (Figure 1), indicating that the biofilm inhibitory effect of ClpP is likely to be universal among S. aureus strains. When biofilm formation was analyzed in the presence of shear stress (0.15 dynes/cm2), which imitates blood flow in veins, the biofilm inhibitory effect of ClpP was still observed (Figure 1B). On the other hand, the mutants of Clp ATPases showed a wild-type level of biofilm formation (Figure 1C), indicating that either the Clp ATPases are not involved in the regulation of the biofilm formation or they have redundant roles.
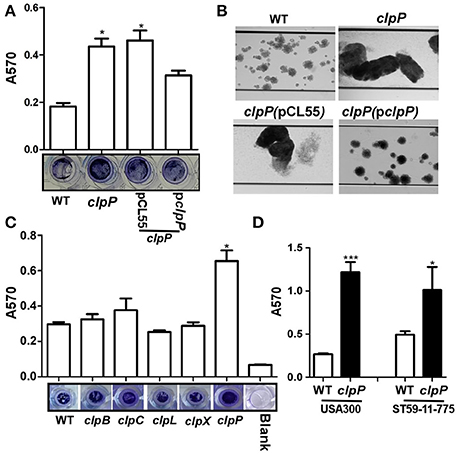
Figure 1. ClpP, but not its ATPases, inhibits biofilm formation in S. aureus. (A) Biofilm formation was determined in clpP mutant and complement strains in comparison to Newman (WT) in vitro by semi-quantitative biofilm assay. The density of crystal violet-stained biofilms was measured at A570. Data were derived from three independent experiments, with four replicates of each experiment. *p < 0.05 (vs. WT). The bottom showed crystal staining for biofilm formation. (B) Newman (WT), clpP mutant and complement strains were grown in TSB supplemented with 0.5% glucose in flow cells pre-coated with human plasma at 0.15 dynes/cm2 shear using a Biofulx 1000z instrument. Brightfield images depicting biofilm accumulation after 24 h were captured at 10 × magnification and are representative of at least three independent experiments. (C) ClpP chaperons are not involved in biofilm formation individually. Biofilm formation was determined in clpB, clpC, clpL, clpX, and clpP mutant in comparison to Newman (WT) in vitro by semi-quantitative biofilm assay. The density of crystal violet-stained biofilms was measured at A570. Data were derived from three independent experiments, with four replicates of each experiment. *p < 0.05 (vs. WT). The bottom showed crystal staining for biofilm formation. (D) Biofilm formation was determined in clpP mutant in comparison to USA300 and ST59-11-775 WT strains in vitro by semi-quantitative biofilm assay. The density of crystal violet-stained biofilms was measured at A570. Data were derived from three independent experiments, with four replicates in each experiment. *P < 0.05; ***P < 0.001 (vs. WT).
Proteins and eDNA Are the Main Components of the Biofilm Formed by clpP Mutant
In the staphylococcal biofilm, the three main components are proteins, extracellular DNA (eDNA) and PIA (Paharik and Horswill, 2016). To determine the main constituents of the biofilm formed by the clpP mutant, we first added Proteinase K or nuclease to the biofilm formed by the Newman clpP mutant. As shown in Figures 2A,B, the biofilm was reduced by either proteinase K or nuclease, indicating proteins and eDNA are the main components of the biofilm formed by the clpP mutant.
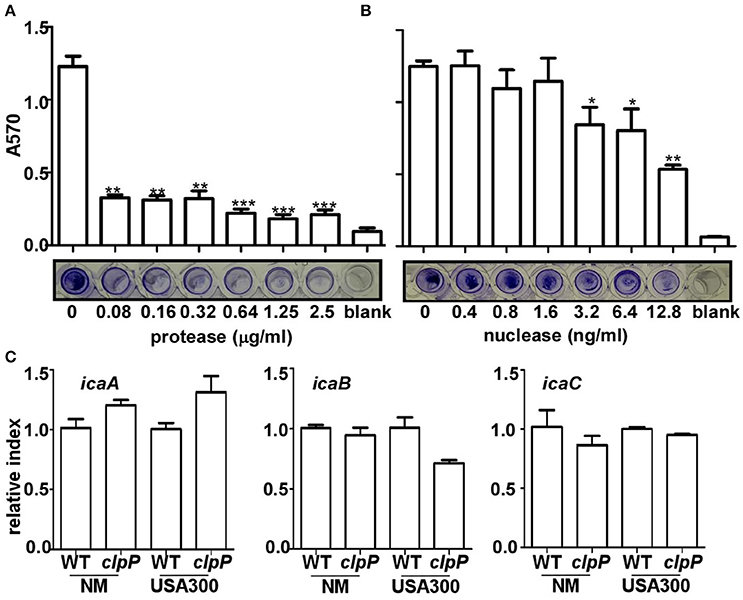
Figure 2. Proteins and eDNA, but not PIA, are involved in biofilm formation by clpP mutant. Degradation of clpP mutant biofilms by various concentrations of protease (A) and nuclease (B). Biofilm formation by clpP mutant was determined in vitro by semi-quantitative biofilm assay. Data were from three independent experiments, with four replicates of each experiment. *P < 0.05; **P < 0.01; ***P < 0.001 (vs. WT). The bottom showed crystal staining for biofilm formation. (C) The expression of the icaA/icaB/icaC genes were determined in samples prepared from stationary-phase (OD600 = 2) cells grown in TSB, using quantitative RT-PCR. The wild-type (WT) and clpP mutant (clpP) strains were used for the assay. Data were derived from three biological repeats.
The involvement of PIA in the biofilm formation by the clpP mutant was assessed indirectly by comparing the transcriptional levels of icaABC, the genes encoding PIA-synthesis enzymes in wild type and clpP mutant. As shown in Figure 2C, the transcript levels of icaABC were not significantly affected by the clpP mutation, suggesting that PIA probably does not contribute to the increased biofilm formation by the clpP mutant.
In Newman and USA300 Strain Backgrounds, the clpP Mutation Lowers the Activity of Agr, Resulting in Reduced Protease Activity
In S. aureus, the quorum sensing two component system Agr is known to suppress biofilm formation by up-regulating the production of extracellular proteases (Boles and Horswill, 2008). In the strain 8325-4, clpP mutation is known to reduce the activity of Agr (Frees et al., 2003; Michel et al., 2006). Indeed, the transcript levels of agrA and agrC were greatly reduced in the clpP mutants of both NM and USA300 strains (Figure 3A). The expression level of the response regulator AgrA was also greatly reduced in the clpP mutants (Figure 3B), further confirming the lower Agr activity in the clpP mutant. Finally, we observed that the protease activity was significantly decreased by mutation in either clpP or agr (Figure 3C), indicating that protease activity was reduced due to low Agr activity in clpP mutant strain.
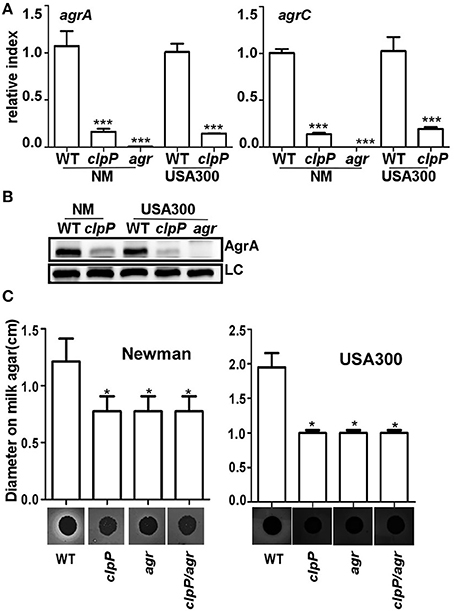
Figure 3. In clpP mutant strain, protease activity is reduced due to the low Agr activity. The transcription of the agrA/agrC genes (A) and the expression of AgrA (B) were determined in samples prepared from stationary-phase (OD600 = 2) cells grown in TSB using quantitative RT-PCR and Western blot. The wild-type (WT) and clpP mutant strains were used for the assay. agr mutant strain was used as a negative control. Data were derived from three biological repeats. ***P < 0.001 (vs. WT). LC, loading control. (C) Relative protease levels detected in wild type, clpP, agr, and clpP/agr double mutant strains grown in broth culture. Images show bacterial colonies and zones of clearing caused by protease activity on milk agar plates (20 g/L non-fat milk). *P < 0.05 (vs. WT).
The Overproduction of the Cell Wall Hydrolase Sle1 Is Responsible for the Increased Release of eDNA
Next we decided to determine the origin of the eDNA in the biofilm of the clpP mutant (Figure 2B). Since the release of eDNA is likely due to autolysis of S. aureus cells, we focused on staphylococcal cell wall hydrolases. Among either proven or putative cell wall hydrolases in S. aureus, LytM and Sle1 are known to be affected by ClpP. The transcription of lytM was up-regulated in the clpP mutant of S. aureus 8325 strain (Michel et al., 2006), whereas, in the S. aureus strains 8325-4 and Newman, Sle1 was shown to bind to ClpPtrap, the protease-deficient mutant of ClpP (Feng et al., 2013). Therefore, we examined whether either LytM or Sle1 play a role in the biofilm formation by the clpP mutant. As can be seen, while the deletion of lytM in the clpP mutant did not decrease the biofilm formation (clpP/lytM in Figure 4A), the sle1 deletion did (clpP/sle1 in Figure 4A), indicating that sle1 is required for the biofilm formation by the clpP mutant. Indeed, a higher level of Sle1 was detected in the clpP mutant as compared with the wild type (Figure 4B). However, the transcription of sle1 was either not significantly affected or reduced in the clpP mutants (Figure 4C). These results are consistent with the idea that the precursor of Sle1 is a ClpP substrate (Feng et al., 2013). Finally, to investigate whether the increased level of Sle1 leads to the release of more eDNA, we compared the amount of eDNA in culture supernatants of the test strains. Indeed, the culture supernatant of the clpP mutant contained a higher level of eDNA, as compared with that of wild type strain (clpP in Figure 4D). However, the level of eDNA was restored to that of wild type by further deletion of sle1 (clpP/sle1 in Figure 4D), strongly suggesting that the increased level of Sle1 is responsible for the heightened release of eDNA, leading to more robust biofilm formation of the clpP mutant.
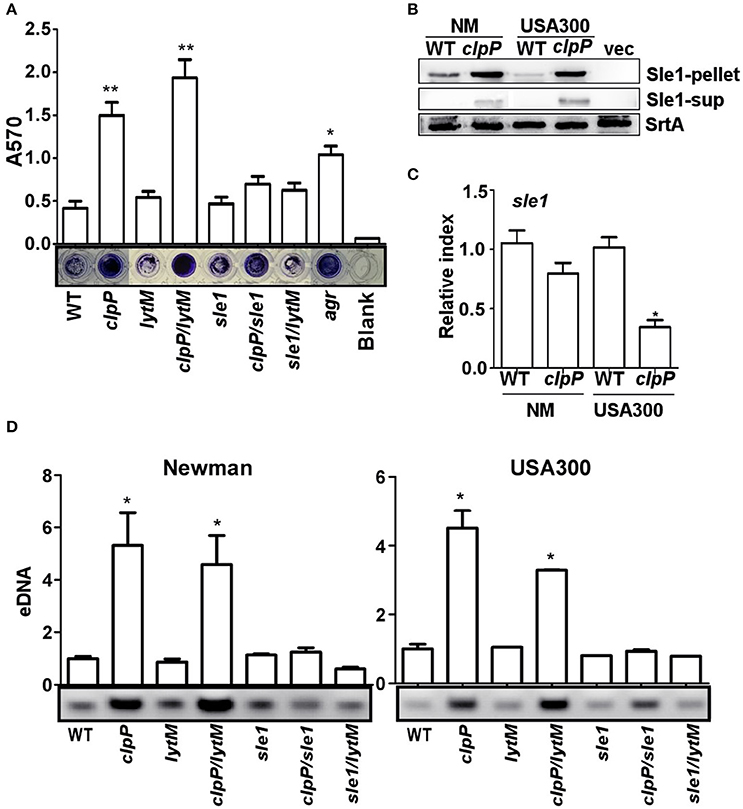
Figure 4. The overproduction of the cell wall hydrolase Sle1 is responsible for the increased release of eDNA. (A) Biofilm formation was determined in sle1, lytM, clpP/sle1, clpP/lytM double mutant strains in comparison to NM strain (WT) in vitro by semi-quantitative biofilm assay. agr mutant strain was used as a positive control. The density of crystal violet-stained biofilms was measured at 570 nm. Data were derived from three independent experiments, with four replicates of each experiment. *P < 0.05; **P < 0.01 (vs. WT). (B) The expression of the Sle1 protein was determined in samples prepared from stationary-phase (OD600 = 2) cells grown in TSB by Western blot. The wild-type (WT) and clpP mutant strains were used for the assay. Sle1-pellet, Sle1 protein detected in the cell pellets; Sle1-sup, Sle1 protein detected in the supernatant by TCA precipitation; SrtA, Sortase A was used as a loading control. (C) The expression of the sle1 gene was determined in samples prepared from stationary-phase (OD600 = 2) cells grown in TSB, using quantitative RT-PCR. The wild-type (WT) and clpP mutant strains were used for the assay. Data were derived from three biological repeats. *P < 0.05 (vs. WT). (D) Agarose gel of isolated eDNAs. Culture supernatants from the wild-type (WT) and its derivatives were filter sterilized, extracted with phenol-chloroform and ethanol precipitated. Precipitates were suspended in H2O normalized according to OD600 of the initial cultures. Data were derived from three biological repeats. *P < 0.05(vs. WT).
Sle1 Is Responsible for the Increased Autolysis of the clpP Mutant
Sle1 is a peptidoglycan hydrolase, preferentially cleaving the N-acetylmuramyl-L-Ala bonds in dimeric cross-bridges that interlink the two murein strands in the peptidoglycan (Kajimura et al., 2005). Therefore, it is likely that the increased amount of eDNA in the culture supernatant of clpP mutant is due to the heightened cell lysis. To examine this notion, we measured autolysis activity of the test strains in the presence of Triton X-100. Indeed, the clpP mutation increased the autolysis activity of S. aureus Newman and USA300 (clpP in Figure 5A). Although the further deletion of lytM did not affect the autolysis activity of the clpP mutant (clpP vs. clpP/lytM in Figure 5A), the deletion of sle1 greatly reduced the autolysis activity of the clpP mutant (clpP vs. clpP/sle1 in Figure 5A). Zymography analysis showed that, in the test conditions, the cell wall hydrolase activities are mostly provided by the major autolysin Atl and Sle1, while the clpP mutant produced a higher level of Atl compared to WT only in USA300 strain. However, the clpP mutation specifically increases the overall activity of Sle1 (clpP vs. clpP/sle1 in Figure 5B).
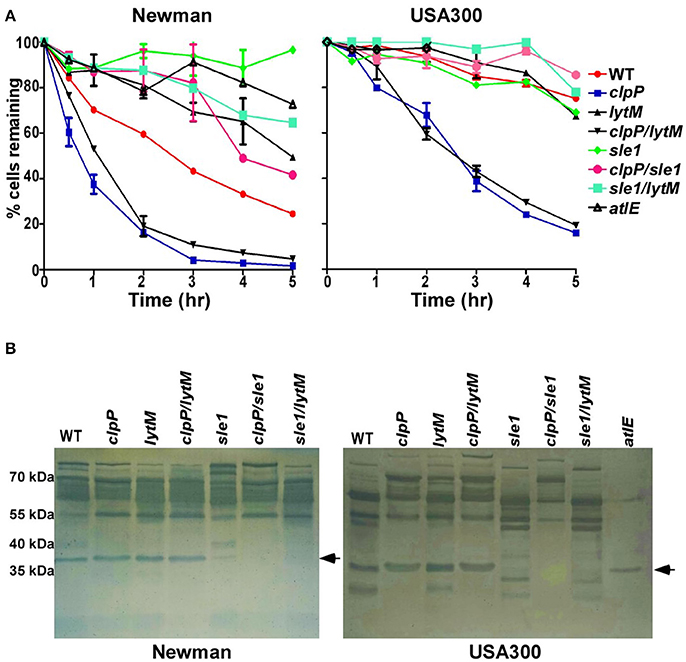
Figure 5. Sle1 is responsible for the increase autolysis of the clpP mutant. (A) Triton X-100 induced autolysis. The average of two independent experiments were shown. (B) Zymogram of S. aureus murein hydrolase activity. Concentrated culture supernatants from the wild-type (WT) and its derivatives were run on 12% acrylamide-SDS gels containing heat killed S. aureus RN4220 cells. Numbers to the left indicate the molecular weights of size standards. Dark bands indicate regions of murein hydrolase activity. The arrows showed the activity of murein hydrolase Sle1.
The Role of clpP in the Pathogenesis of S. aureus Biofilm-Associated Infection
ClpP is required for pathogenesis in a systemic staphylococcal infection as well as skin abscess infection (Frees et al., 2003; Farrand et al., 2013), indicating that ClpP plays important roles in S. aureus pathogenesis. Although the clpP mutant showed a slight growth defect especially during early exponential phase, the final cell density was similar to that of wild-type at lag phase (Supplementary Figure 1). Because clpP mutant showed an increased biofilm formation, we asked whether the increased biofilm formation can compensate the important virulence contributions of ClpP. A murine model of subcutaneous foreign body infection was used to investigate the impact of clpP on biofilm-associated infection in vivo. Progression of disease was measured by determining bacterial loads on implanted catheters while dissemination of the bacteria was examined by detecting bacterial loads on peri-tissues and organs. Wild type and the clpP mutant showed similar bacterial loads in the infected animals (Figure 6A). However, significantly higher number of bacteria were found in peri-catheter tissue as well as blood, liver, spleen, and kidneys in the animals infected by the wild-type strain, as compared with the animals infected by the clpP mutant strain (Figures 6B–F), indicating that ClpP plays a critical role in the dissemination of S. aureus, and that the increased biofilm formation capability cannot compensate the critical role played by ClpP.
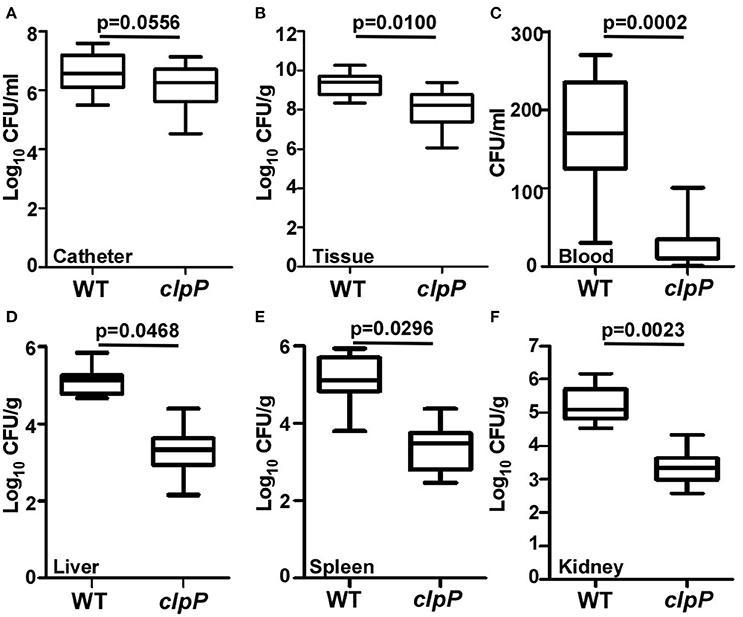
Figure 6. Catheter colonization and dissemination of NM and clpP mutant strains in a mouse device-related infection model. Implanted catheter segments were injected with 1 × 108 bacteria and mice were sacrificed after 7 days. Colony forming units (CFU) per catheter (A), per g peri-catheter tissue (B), per ml blood (C), per g liver (D), per g spleen (E), per g kidney (F) recovered from animals infected with wild type (NM) and clpP mutant strain. Statistical significance (p-values) is indicated in the figure.
Discussion
In S. aureus, biofilm is an important virulence determinant in hospital-associated infections on indwelling medical devices (Joo and Otto, 2012). Staphylococcal biofilm formation is known to be controlled by multiple global regulators including ClpP, the proteolytic component of the Clp proteolysis system. However, it has not been elucidated how ClpP controls biofilm formation (Frees et al., 2004). In this study, we provided evidence that ClpP controls staphylococcal biofilm formation by activating the Agr quorum sensing system, a negative regulator of biofilm formation, and suppressing the expression of the murein hydrolase Sle1 at a post-transcriptional level. Moreover, using a murine model of subcutaneous foreign body infection, we also demonstrated that, in the clpP mutant, the increased biofilm formation cannot compensate the other critical ClpP functions missing in the mutant.
The Agr quorum sensing system plays important roles in staphylococcal virulence by suppressing the expression of surface protein expression and activating the expression of secreted proteins such as hemolysin and enzymes (Le and Otto, 2015). In particular, the Agr system is known to promote the detachment of biofilm by inducing the expression of the metalloprotease Aur and the serine proteases SplABCDEF (Boles and Horswill, 2008). Therefore, it is likely that the reduced expression of those proteins contributed to the decreased protease activity (Figure 3C) and the increased biofilm formation (Figure 1). Then how does ClpP activate the activity of Agr? ClpP is a protease, and the degradation of either AgrA or AgrC by ClpP would result in reduced, not increased, activity of Agr. Therefore, it is likely that ClpP controls Agr indirectly, possibly by degradation of a negative regulator of Agr. For negative regulators of agr transcription, the DNA binding protein SarX and the two component systems ArlSR and SrrAB are known (Fournier et al., 2001; Yarwood et al., 2001; Manna and Cheung, 2006). It is possible that one of those negative regulators is degraded by ClpP. Further, work will need to address whether any of those or hitherto unknown regulators are involved in the ClpP-mediated Agr activation.
Sle1 is a N-acetyl-muramyl-L-alanine amidase and plays an important role in splitting daughter cells during cell division (Kajimura et al., 2005). However, Sle1 does not appear to affect autolysis of staphylococcal cells (Kajimura et al., 2005). Indeed, in our study, deletion of sle1 did not significantly affect autolysis, eDNA release, or biofilm formation (WT vs. sle1 in Figures 4, 5A). In contrast, in clpP mutant background, the deletion of sle1 abolished autolysis, eDNA release, and biofilm formation (clpP vs. clpP/sle1 in Figures 4, 5A). It is likely that, in the wild type cells, ClpP-mediated control of Sle1 limits the Sle1 function only to daughter cell cleavage (Figure 4B). However, in the clpP mutant, due to increased concentration (Figure 4B), Sle1 seems to play not only in daughter cell separation, but also autolysis of the cells (Figure 5A). Unlike Sle1, LytM did not play any role in autolysis, eDNA release, or biofilm formation regardless of the mutation in clpP (lytM and clpP/lytM in Figures 4, 5A). LytM was initially suggested to be a glycylglycine endopeptidase (Ramadurai and Jayaswal, 1997). However, purified LytM did not show any lytic activity against S. aureus cells, and lytM deletion did not affect cell wall lysis (Singh et al., 2010). In our study, although the transcription of lytM was increased in the clpP mutant (Supplementary Figure 2), further deletion of lytM in the clpP mutant did not reduce biofilm formation or autolysis of the clpP mutant (Figures 4A, 5A), suggesting that LytM does not play a significant role in cell wall hydrolysis of S. aureus.
In a previous study, ClpX, an ATP-binding partner for ClpP, was shown to bind Sle1 (Feng et al., 2013). The inactivation of the ClpXP protease increased the β-lactam resistance by controlling cell wall metabolism through regulating Sle1 and Atl in JE2 strain (Baek et al., 2014). However, mutation in clpX did not significantly increase biofilm formation (Figure 1C), indicating that either Sle1 is delivered to ClpP by other ATPase partners or Sle1 overexpression alone is not sufficient to increase biofilm formation in the presence of functional Agr. On the other hand, although the mutation in agr greatly reduced protease production (Figure 3C), its effect on biofilm formation is not as significant as clpP mutation (Figure 4A), demonstrating the critical contribution of Sle1 to the biofilm formation. Since Sle1 is a secreted protein, it is likely that the precursor of Sle1 (i.e., Sle1 with signal peptide) binds to either ClpX or other Clp ATPase and is processed by ClpP.
Figure 7 illustrates our current model of how ClpP represses biofilm formation in S. aureus. In a wild type condition, the Clp protease system is expected to degrade an unidentified negative regulator of Agr and Sle1, resulting in Agr activation and suppression of cell autolysis. The activated Agr and suppression of autolysis will limit the presence of proteins and eDNA and repress the formation of staphylococcal biofilm.
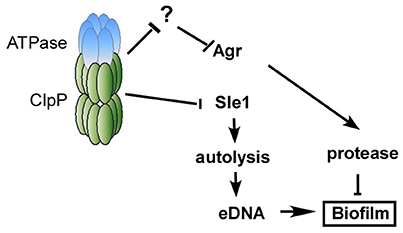
Figure 7. Working model of the mechanism of how ClpP inhibits biofilm formation. In a wild type condition, the Clp protease system is expected to degrade an unidentified negative regulator of Agr, resulting in Agr activation, which will limit the presence of proteins and repress biofilm formation. On the other hand, the Clp protease system degrades the precursor of cell wall hydrolase Sle1, resulting in the suppression of cell autolysis. The suppression of autolysis will decrease the release of eDNA and repress the formation of staphylococcal biofilm.
Author Contributions
TB, QL, ML, and XL conceived the study. QL, XW, JQ, SC, W-SY, LH, and XM performed experiments. QL, XW, TB, ML, and XL analyzed the data. TB, QL, ML, and XL drafted the manuscript. QL, XW, JQ, XL, ML, and TB revised and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by Indiana University (Research Enhancement grant, TB), the NIH (AI121664, TB), the National Natural Science Foundation of China (grant 81501803, QL), the Shanghai Committee of Science and Technology, China (grant 14ZR1424600, QL), the National Natural Science Foundation of China (grants 81671975 and 81371875, ML), and the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (grant 81421001, ML).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fcimb.2017.00181/full#supplementary-material
References
Arnold, I., and Langer, T. (2002). Membrane protein degradation by AAA proteases in mitochondria. Biochim. Biophys. Acta 1592, 89–96. doi: 10.1016/S0167-4889(02)00267-7
Aslanidis, C., and De Jong, P. J. (1990). Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 18, 6069–6074. doi: 10.1093/nar/18.20.6069
Bae, T., and Schneewind, O. (2006). Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55, 58–63. doi: 10.1016/j.plasmid.2005.05.005
Baek, K. T., Grundling, A., Mogensen, R. G., Thogersen, L., Petersen, A., Paulander, W., et al. (2014). β-lactam resistance in methicillin-resistant Staphylococcus aureus USA300 is increased by inactivation of the ClpXP protease. Antimicrob. Agents Chemother. 58, 4593–4603. doi: 10.1128/AAC.02802-14
Benoit, M. R., Conant, C. G., Ionescu-Zanetti, C., Schwartz, M., and Matin, A. (2010). New device for high-throughput viability screening of flow biofilms. Appl. Environ. Microbiol. 76, 4136–4142. doi: 10.1128/AEM.03065-09
Boles, B. R., and Horswill, A. R. (2008). Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4:e1000052. doi: 10.1371/journal.ppat.1000052
Chen, C., Krishnan, V., Macon, K., Manne, K., Narayana, S. V., and Schneewind, O. (2013). Secreted proteases control autolysin-mediated biofilm growth of Staphylococcus aureus. J. Biol. Chem. 288, 29440–29452. doi: 10.1074/jbc.M113.502039
Cramton, S. E., Gerke, C., Schnell, N. F., Nichols, W. W., and Gotz, F. (1999). The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67, 5427–5433.
Dalbey, R. E., Wang, P., and Van Dijl, J. M. (2012). Membrane proteases in the bacterial protein secretion and quality control pathway. Microbiol. Mol. Biol. Rev. 76, 311–330. doi: 10.1128/MMBR.05019-11
Farrand, A. J., Reniere, M. L., Ingmer, H., Frees, D., and Skaar, E. P. (2013). Regulation of host hemoglobin binding by the Staphylococcus aureus Clp proteolytic system. J. Bacteriol. 195, 5041–5050. doi: 10.1128/JB.00505-13
Feng, J., Michalik, S., Varming, A. N., Andersen, J. H., Albrecht, D., Jelsbak, L., et al. (2013). Trapping and proteomic identification of cellular substrates of the ClpP protease in Staphylococcus aureus. J. Proteome Res. 12, 547–558. doi: 10.1021/pr300394r
Fournier, B., Klier, A., and Rapoport, G. (2001). The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41, 247–261. doi: 10.1046/j.1365-2958.2001.02515.x
Frees, D., Chastanet, A., Qazi, S., Sorensen, K., Hill, P., Msadek, T., et al. (2004). Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 54, 1445–1462. doi: 10.1111/j.1365-2958.2004.04368.x
Frees, D., Qazi, S. N., Hill, P. J., and Ingmer, H. (2003). Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol. Microbiol. 48, 1565–1578. doi: 10.1046/j.1365-2958.2003.03524.x
Gotz, F. (2002). Staphylococcus and biofilms. Mol. Microbiol. 43, 1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x
Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. doi: 10.1016/S0022-2836(83)80284-8
Jefferson, K. K., Pier, D. B., Goldmann, D. A., and Pier, G. B. (2004). The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 186, 2449–2456. doi: 10.1128/JB.186.8.2449-2456.2004
Joo, H. S., and Otto, M. (2012). Molecular basis of in vivo biofilm formation by bacterial pathogens. Chem. Biol. 19, 1503–1513. doi: 10.1016/j.chembiol.2012.10.022
Kajimura, J., Fujiwara, T., Yamada, S., Suzawa, Y., Nishida, T., Oyamada, Y., et al. (2005). Identification and molecular characterization of an N-acetylmuramyl-L-alanine amidase Sle1 involved in cell separation of Staphylococcus aureus. Mol. Microbiol. 58, 1087–1101. doi: 10.1111/j.1365-2958.2005.04881.x
Kiedrowski, M. R., Kavanaugh, J. S., Malone, C. L., Mootz, J. M., Voyich, J. M., Smeltzer, M. S., et al. (2011). Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus. PLoS ONE 6:e26714. doi: 10.1371/journal.pone.0026714
Lauderdale, K. J., Boles, B. R., Cheung, A. L., and Horswill, A. R. (2009). Interconnections between Sigma, B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect. Immun. 77, 1623–1635. doi: 10.1128/IAI.01036-08
Le, K. Y., and Otto, M. (2015). Quorum-sensing regulation in staphylococci-an overview. Front. Microbiol. 6:1174. doi: 10.3389/fmicb.2015.01174
Li, M., Wang, Y., Zhu, Y., Dai, Y., Hong, X., Liu, Q., et al. (2016). Increased community-associated infections caused by panton-valentine leukocidin-negative MRSA, Shanghai, 2005-2014. Emerg. Infect. Dis. 22, 1988–1991. doi: 10.3201/eid2211.160587
Li, T., He, L., Song, Y., Villaruz, A. E., Joo, H. S., Liu, Q., et al. (2015). AraC-type regulator Rsp adapts Staphylococcus aureus gene expression to acute infection. Infect. Immun. 84, 723–734. doi: 10.1128/IAI.01088-15
Liu, Q., Fan, J., Niu, C., Wang, D., Wang, J., Wang, X., et al. (2011). The eukaryotic-type serine/threonine protein kinase Stk is required for biofilm formation and virulence in Staphylococcus epidermidis. PLoS ONE 6:e25380. doi: 10.1371/journal.pone.0025380
Manna, A. C., and Cheung, A. L. (2006). Expression of SarX, a negative regulator of agr and exoprotein synthesis, is activated by MgrA in Staphylococcus aureus. J. Bacteriol. 188, 4288–4299. doi: 10.1128/JB.00297-06
Marti, M., Trotonda, M. P., Tormo-Mas, M. A., Vergara-Irigaray, M., Cheung, A. L., Lasa, I., et al. (2010). Extracellular proteases inhibit protein-dependent biofilm formation in Staphylococcus aureus. Microbes Infect. 12, 55–64. doi: 10.1016/j.micinf.2009.10.005
McCarthy, H., Rudkin, J. K., Black, N. S., Gallagher, L., O'Neill, E., and O'gara, J. P. (2015). Methicillin resistance and the biofilm phenotype in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 5:1. doi: 10.3389/fcimb.2015.00001
Merino, N., Toledo-Arana, A., Vergara-Irigaray, M., Valle, J., Solano, C., Calvo, E., et al. (2009). Protein A-mediated multicellular behavior in Staphylococcus aureus. J. Bacteriol. 191, 832–843. doi: 10.1128/JB.01222-08
Michel, A., Agerer, F., Hauck, C. R., Herrmann, M., Ullrich, J., Hacker, J., et al. (2006). Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 188, 5783–5796. doi: 10.1128/JB.00074-06
Mootz, J. M., Benson, M. A., Heim, C. E., Crosby, H. A., Kavanaugh, J. S., Dunman, P. M., et al. (2015). Rot is a key regulator of Staphylococcus aureus biofilm formation. Mol. Microbiol. 96, 388–404. doi: 10.1111/mmi.12943
Okshevsky, M., and Meyer, R. L. (2015). The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit. Rev. Microbiol. 41, 341–352. doi: 10.3109/1040841X.2013.841639
O'Neill, E., Pozzi, C., Houston, P., Humphreys, H., Robinson, D. A., Loughman, A., et al. (2008). A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 190, 3835–3850. doi: 10.1128/JB.00167-08
Otto, M. (2013). Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu. Rev. Med. 64, 175–188. doi: 10.1146/annurev-med-042711-140023
Paharik, A. E., and Horswill, A. R. (2016). The Staphylococcal biofilm: adhesins, regulation, and host response. Microbiol. Spectr. 4:VMBF-0022-2015. doi: 10.1128/microbiolspec.VMBF-0022-2015
Pozzi, C., Waters, E. M., Rudkin, J. K., Schaeffer, C. R., Lohan, A. J., Tong, P., et al. (2012). Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog. 8:e1002626. doi: 10.1371/journal.ppat.1002626
Ramadurai, L., and Jayaswal, R. K. (1997). Molecular cloning, sequencing, and expression of lytM, a unique autolytic gene of Staphylococcus aureus. J. Bacteriol. 179, 3625–3631. doi: 10.1128/jb.179.11.3625-3631.1997
Rohde, H., Burandt, E. C., Siemssen, N., Frommelt, L., Burdelski, C., Wurster, S., et al. (2007). Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials 28, 1711–1720. doi: 10.1016/j.biomaterials.2006.11.046
Singh, V. K., Carlos, M. R., and Singh, K. (2010). Physiological significance of the peptidoglycan hydrolase, LytM, in Staphylococcus aureus. FEMS Microbiol. Lett. 311, 167–175. doi: 10.1111/j.1574-6968.2010.02087.x
Sun, F., Cho, H., Jeong, D. W., Li, C., He, C., and Bae, T. (2011). Aureusimines in Staphylococcus aureus are not involved in virulence. PLoS ONE 5:e15703. doi: 10.1371/journal.pone.0015703
Toledo-Arana, A., Merino, N., Vergara-Irigaray, M., Debarbouille, M., Penades, J. R., and Lasa, I. (2005). Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J. Bacteriol. 187, 5318–5329. doi: 10.1128/JB.187.15.5318-5329.2005
Keywords: Staphylococcus aureus, biofilm, proteolysis system, Agr, cell wall hydrolysis
Citation: Liu Q, Wang X, Qin J, Cheng S, Yeo W-S, He L, Ma X, Liu X, Li M and Bae T (2017) The ATP-Dependent Protease ClpP Inhibits Biofilm Formation by Regulating Agr and Cell Wall Hydrolase Sle1 in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 7:181. doi: 10.3389/fcimb.2017.00181
Received: 06 April 2017; Accepted: 27 April 2017;
Published: 15 May 2017.
Edited by:
Joan A. Geoghegan, Trinity College, Dublin, IrelandReviewed by:
Brian Patrick Conlon, University of North Carolina at Chapel Hill, USAChia Y. Lee, University of Arkansas for Medical Sciences, USA
Copyright © 2017 Liu, Wang, Qin, Cheng, Yeo, He, Ma, Liu, Li and Bae. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian Liu, qq2005011@163.com
Min Li, ruth_limin@126.com
Taeok Bae, tbae@iun.edu
†These authors have contributed equally to this work.
 Qian Liu
Qian Liu Xing Wang
Xing Wang Juanxiu Qin
Juanxiu Qin Sen Cheng
Sen Cheng Won-Sik Yeo4
Won-Sik Yeo4  Lei He
Lei He Xiaoyun Liu
Xiaoyun Liu Min Li
Min Li Taeok Bae
Taeok Bae