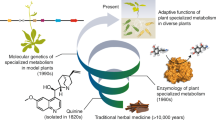
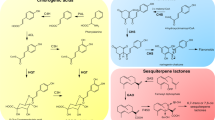
Abstract
Plants biosynthesize a broad range of natural products through specialized and species-specific metabolic pathways that are fuelled by core metabolism, together forming a metabolic network. Specialized metabolites have important roles in development and adaptation to external cues, and they also have invaluable pharmacological properties. A growing body of evidence has highlighted the impact of translational, transcriptional, epigenetic and chromatin-based regulation and evolution of specialized metabolism genes and metabolic networks. Here we review the forefront of this research field and extrapolate to medicinal plants that synthetize rare molecules. We also discuss how this new knowledge could help in improving strategies to produce useful plant-derived pharmaceuticals.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Carrington, Y. et al. Evolution of a secondary metabolic pathway from primary metabolism: shikimate and quinate biosynthesis in plants. Plant J. 95, 823–833 (2018).
Barra, L., Awakawa, T., Shirai, K., Hu, Z. & Bashiri, G. β-NAD as a building block in natural product biosynthesis. Nature 600, 754–758 (2021).
Garagounis, C., Delkis, N. & Papadopoulou, K. K. Unraveling the roles of plant specialized metabolites: using synthetic biology to design molecular biosensors. N. Phytol. 231, 1338–1352 (2021).
Wurtzel, E. T. & Kutchan, T. M. Plant metabolism, the diverse chemistry set of the future. Science 353, 1232–1236 (2016).
Kulagina, N., Méteignier, L., Papon, N., O’Connor, S. E. & Courdavault, V. More than a Catharanthus plant: a multicellular and pluri-organelle alkaloid-producing factory. Curr. Opin. Plant Biol. 67, 102200 (2022).
Ozber, N. & Facchini, P. J. Phloem-specific localization of benzylisoquinoline alkaloid metabolism in opium poppy. J. Plant Physiol. 271, 153641 (2022).
Nielsen, J. Systems biology of metabolism. Annu. Rev. Biochem. 86, 245–275 (2017).
Lacchini, E. & Goossens, A. Combinatorial control of plant specialized metabolism: mechanisms, functions, and consequences. Annu. Rev. Cell Dev. Biol. 36, 291–313 (2020).
Colinas, M. & Goossens, A. Combinatorial transcriptional control of plant specialized metabolism. Trends Plant Sci. 23, 324–336 (2018).
Zhang, Y. & Fernie, A. R. Metabolons, enzyme–enzyme assemblies that mediate substrate channeling, and their roles in plant metabolism. Plant Commun. 2, 100081 (2021).
Zhang, Y. et al. A moonlighting role for enzymes of glycolysis in the co-localization of mitochondria and chloroplasts. Nat. Commun. 11, 4509 (2020).
Mucha, S. et al. The formation of a camalexin-biosynthetic metabolon. Plant Cell 31, 2697–2710 (2019).
Gaudinier, A. et al. Transcriptional regulation of nitrogen-associated metabolism and growth. Nature 563, 259–264 (2018).
Tang, M. et al. A genome‐scale TF–DNA interaction network of transcriptional regulation of Arabidopsis primary and specialized metabolism. Mol. Syst. Biol. 17, e10625 (2021).
Ihmels, J., Levy, R. & Barkai, N. Principles of transcriptional control in the metabolic network of Saccharomyces cerevisiae. Nat. Biotechnol. 22, 86–92 (2004).
Xiong, Y. et al. Glucose–TOR signalling reprograms the transcriptome and activates meristems. Nature 496, 181–186 (2013).
Dong, Y. et al. Sulfur availability regulates plant growth via glucose–TOR signaling. Nat. Commun. 8, 1174 (2017).
Salazar-Díaz, K. et al. TOR senses and regulates spermidine metabolism during seedling establishment and growth in maize and Arabidopsis. iScience 24, 103260 (2021).
Meteignier, L.-V. et al. Translatome analysis of an NB–LRR immune response identifies important contributors to plant immunity in Arabidopsis. J. Exp. Bot. 68, 2333–2344 (2017).
De Vleesschauwer, D. et al. Target of rapamycin signaling orchestrates growth–defense trade-offs in plants. N. Phytol. 217, 305–319 (2017).
Groth, M. et al. MTHFD1 controls DNA methylation in Arabidopsis. Nat. Commun. 7, 11640 (2016).
Samo, N., Ebert, A., Kopka, J. & Mozgová, I. Plant chromatin, metabolism and development—an intricate crosstalk. Curr. Opin. Plant Biol. 61, 102002 (2021).
Escaray, F., Felipo-Benavent, A. & Vera, P. Linking plant metabolism and immunity through methionine biosynthesis. Mol. Plant 15, 6–8 (2022).
González, B. & Vera, P. Folate metabolism interferes with plant immunity through 1C methionine synthase-directed genome-wide DNA methylation enhancement. Mol. Plant 12, 1227–1242 (2019).
Zhai, K. et al. NLRs guard metabolism to coordinate pattern- and effector-triggered immunity. Nature 601, 245–251 (2021).
Méteignier, L.-V. et al. Topoisomerase VI participates in an insulator-like function that prevents H3K9me2 spreading. Proc. Natl Acad. Sci. USA 119, e2001290119 (2022).
Latrasse, D. et al. Dual function of MIPS1 as a metabolic enzyme and transcriptional regulator. Nucleic Acids Res. 41, 2907–2917 (2013).
Meng, J. et al. METHIONINE ADENOSYLTRANSFERASE4 mediates DNA and histone methylation. Plant Physiol. 177, 652–670 (2018).
Venturelli, S. et al. Plants release precursors of histone deacetylase inhibitors to suppress growth of competitors. Plant Cell 27, 3175–3189 (2015).
Zhao, K., Kong, D., Jin, B., Smolke, C. D. & Rhee, S. Y. A novel bivalent chromatin associates with rapid induction of camalexin biosynthesis genes in response to a pathogen signal in Arabidopsis. eLife 10, e69508 (2021).
Catoni, M. & Cortijo, S. in Advances in Botanical Research Vol. 88 (eds Mirouze, M. et al.) 87–116 (Elsevier, 2018).
Kooke, R. et al. Epigenetic mapping of the Arabidopsis metabolome reveals mediators of the epigenotype–phenotype map. Genome Res. 29, 96–106 (2019).
Shirai, K. et al. Positive selective sweeps of epigenetic mutations regulating specialized metabolites in plants. Genome Res. 31, 1060–1068 (2021).
de Bernonville, T. D. et al. Developmental methylome of the medicinal plant Catharanthus roseus unravels the tissue-specific control of the monoterpene indole alkaloid pathway by DNA methylation. Int. J. Mol. Sci. 21, 6028 (2020).
Zhan, C. et al. Plant metabolic gene clusters in the multi-omics era. Trends Plant Sci. 27, 981–1001 (2022).
Zhan, C. et al. Selection of a subspecies-specific diterpene gene cluster implicated in rice disease resistance. Nat. Plants 6, 1447–1454 (2020).
Polturak, G. et al. Pathogen-induced biosynthetic pathways encode defense-related molecules in bread wheat. Proc. Natl Acad. Sci. USA 119, e2123299119 (2022).
Shen, S. et al. An Oryza-specific hydroxycinnamoyl tyramine gene cluster contributes to enhanced disease resistance. Sci. Bull. 66, 2369–2380 (2021).
Chen, W. et al. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat. Genet. 46, 714–721 (2014).
Fang, H. et al. A monocot-specific hydroxycinnamoylputrescine gene cluster contributes to immunity and cell death in rice. Sci. Bull. 66, 2381–2393 (2021).
Grützner, R. et al. High-efficiency genome editing in plants mediated by a Cas9 gene containing multiple introns. Plant Commun. 2, 100135 (2021).
Gardiner, J., Ghoshal, B., Wang, M. & Jacobsen, S. E. CRISPR–Cas-mediated transcriptional control and epi-mutagenesis. Plant Physiol. 188, 1811–1824 (2022).
Smit, S. J. & Lichman, B. R. Plant biosynthetic gene clusters in the context of metabolic evolution. Nat. Prod. Rep. 39, 1465–1482 (2022).
Zhou, X. & Liu, Z. Unlocking plant metabolic diversity: a (pan)-genomic view. Plant Commun. 3, 100300 (2022).
Laverty, K. U. et al. A physical and genetic map of Cannabis sativa identifies extensive rearrangements at the THC/CBD acid synthase loci. Genome Res. 29, 146–156 (2019).
Conart, C. et al. Duplication and specialization of NUDX1 in Rosaceae led to geraniol production in rose petals. Mol. Biol. Evol. 39, msac002 (2022).
Kominek, J. et al. Eukaryotic acquisition of a bacterial operon. Cell 176, 1356–1366.e10 (2019).
Xia, J. et al. Whitefly hijacks a plant detoxification gene that neutralizes plant toxins. Cell 184, 1693–1705.e17 (2021).
Wu, D., Jiang, B., Ye, C.-Y., Timko, M. P. & Fan, L. Horizontal transfer and evolution of the biosynthetic gene cluster for benzoxazinoids in plants. Plant Commun. 3, 100320 (2022).
Barco, B., Kim, Y. & Clay, N. K. Expansion of a core regulon by transposable elements promotes Arabidopsis chemical diversity and pathogen defense. Nat. Commun. 10, 3444 (2019).
Schweizer, F. et al. An engineered combinatorial module of transcription factors boosts production of monoterpenoid indole alkaloids in Catharanthus roseus. Metab. Eng. 48, 150–162 (2018).
Brophy, J. A. N. et al. Synthetic genetic circuits as a means of reprogramming plant roots. Science 377, 747–751 (2022).
Bharadwaj, R., Kumar, S. R., Sharma, A. & Sathishkumar, R. Plant metabolic gene clusters: evolution, organization, and their applications in synthetic biology. Front. Plant Sci. 12, 697318 (2021).
Guo, L. et al. The opium poppy genome and morphinan production. Science 362, 343–347 (2018).
Liu, Z. et al. Formation and diversification of a paradigm biosynthetic gene cluster in plants. Nat. Commun. 11, 5354 (2020).
Zhao, S. et al. Plant HP1 protein ADCP1 links multivalent H3K9 methylation readout to heterochromatin formation. Cell Res. 29, 54–66 (2019).
Zhong, Z. et al. DNA methylation-linked chromatin accessibility affects genomic architecture in Arabidopsis. Proc. Natl Acad. Sci. USA 118, e2023347118 (2021).
Dong, P. et al. 3D chromatin architecture of large plant genomes determined by local A/B compartments. Mol. Plant 10, 1497–1509 (2017).
Yang, X. et al. Three chromosome-scale Papaver genomes reveal punctuated patchwork evolution of the morphinan and noscapine biosynthesis pathway. Nat. Commun. 12, 6030 (2021).
Nützmann, H. W. et al. Active and repressed biosynthetic gene clusters have spatially distinct chromosome states. Proc. Natl Acad. Sci. USA 117, 13800–13809 (2020).
Ariel, F. et al. R-loop mediated trans action of the APOLO long noncoding RNA. Mol. Cell 77, 1055–1065.e4 (2020).
Roulé, T. et al. The lncRNA MARS modulates the epigenetic reprogramming of the marneral cluster in response to ABA. Mol. Plant 15, 840–856 (2022).
Li, Y. et al. Subtelomeric assembly of a multi-gene pathway for antimicrobial defense compounds in cereals. Nat. Commun. 12, 2563 (2021).
Choi, J. Y. et al. Natural variation in plant telomere length is associated with flowering time. Plant Cell 33, 1118–1134 (2021).
Farrell, C. et al. A complex network of interactions governs DNA methylation at telomeric regions. Nucleic Acids Res. 50, 1449–1464 (2022).
Campitelli, B. E. et al. Plasticity, pleiotropy and fitness trade‐offs in Arabidopsis genotypes with different telomere lengths. N. Phytol. 233, 1939–1952 (2022).
Wegel, E., Koumproglou, R., Shaw, P. & Osbourn, A. Cell type-specific chromatin decondensation of a metabolic gene cluster in oats. Plant Cell 21, 3926–3936 (2009).
Wang, J. et al. Manipulation of TAD reorganization by chemical-dependent genome linking. STAR Protoc. 2, 100799 (2021).
Winkler, J. et al. Visualizing protein–protein interactions in plants by rapamycin-dependent delocalization. Plant Cell 33, 1101–1117 (2021).
Lioy, V. S. et al. Dynamics of the compartmentalized Streptomyces chromosome during metabolic differentiation. Nat. Commun. 12, 5221 (2021).
Won, T. H. et al. Copper starvation induces antimicrobial isocyanide integrated into two distinct biosynthetic pathways in fungi. Nat. Commun. 13, 4828 (2022).
Wang, B., Guo, F., Dong, S. H. & Zhao, H. Activation of silent biosynthetic gene clusters using transcription factor decoys. Nat. Chem. Biol. 15, 111–114 (2019).
Park, J., Yim, S. S. & Wang, H. H. High-throughput transcriptional characterization of regulatory sequences from bacterial biosynthetic gene clusters. ACS Synth. Biol. 10, 1859–1873 (2021).
Zhang, M. M. et al. CRISPR–Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nat. Chem. Biol. 13, 607–609 (2017).
Gacek-Matthews, A. et al. KdmB, a Jumonji histone H3 demethylase, regulates genome-wide H3K4 trimethylation and is required for normal induction of secondary metabolism in Aspergillus nidulans. PLoS Genet. 12, e1006222 (2016).
Keller, N. P. Fungal secondary metabolism: regulation, function and drug discovery. Nat. Rev. Microbiol. 17, 167–180 (2019).
Greco, C., Pfannenstiel, B. T., Liu, J. C. & Keller, N. P. Depsipeptide aspergillicins revealed by chromatin reader protein deletion. ACS Chem. Biol. 14, 1121–1128 (2019).
Schüller, A. et al. A novel fungal gene regulation system based on inducible VPR–dCas9 and nucleosome map-guided sgRNA positioning. Appl. Microbiol. Biotechnol. 104, 9801–9822 (2020).
Kumar, S., Singh, B. & Singh, R. Catharanthus roseus (L.) G. Don: a review of its ethnobotany, phytochemistry, ethnopharmacology and toxicities. J. Ethnopharmacol. 284, 114647 (2022).
Rai, A. et al. Chromosome-level genome assembly of Ophiorrhiza pumila reveals the evolution of camptothecin biosynthesis. Nat. Commun. 12, 405 (2021).
Kang, M. et al. A chromosome-level Camptotheca acuminata genome assembly provides insights into the evolutionary origin of camptothecin biosynthesis. Nat. Commun. 12, 3531 (2021).
Zhao, X. et al. Chromosome-level assembly of the Neolamarckia cadamba genome provides insights into the evolution of cadambine biosynthesis. Plant J. 109, 891–908 (2022).
Li, C. et al. Single-cell multi-omics enabled discovery of alkaloid biosynthetic pathway genes in the medical plant Catharanthus roseus. Preprint at bioRxiv https://doi.org/10.1101/2022.07.04.498697 (2022).
Bonetti, A. et al. RADICL-seq identifies general and cell type–specific principles of genome-wide RNA-chromatin interactions. Nat. Commun. 11, 1018 (2020).
Yamamoto, K. et al. The complexity of intercellular localisation of alkaloids revealed by single-cell metabolomics. N. Phytol. 224, 848–859 (2019).
Tian, F., Yang, D. C., Meng, Y. Q., Jin, J. & Gao, G. PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 48, D1104–D1113 (2020).
Gräwe, C., Stelloo, S., van Hout, F. A. H. & Vermeulen, M. RNA-centric methods: toward the interactome of specific RNA transcripts. Trends Biotechnol. 39, 890–900 (2021).
Yaschenko, A. E., Fenech, M., Mazzoni-Putman, S., Alonso, J. M. & Stepanova, A. N. Deciphering the molecular basis of tissue-specific gene expression in plants: can synthetic biology help? Curr. Opin. Plant Biol. 68, 102241 (2022).
Acknowledgements
We acknowledge funding from the ANR MIACYC project (grant no. ANR-20-CE43-0010), the ARD CVL Biopharmaceutical programme of the Région Centre-Val de Loire (ETOPOCentre project) and Le Studium Loire Valley Institute for Advanced Studies (consortium fellowship). A.O. acknowledges funding from the John Innes Foundation and the BBSRC Institute Strategic Programme Grant ‘Molecules from Nature—Products and Pathways’ (grant no. BBS/E/J/000PR9790). H.-W.N. acknowledges support from the University of Bath and the Royal Society (grant no. UF160138).
Author information
Authors and Affiliations
Contributions
L.-V.M., H.-W.N., N.P., A.O. and V.C. conceived the project and wrote the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Alisdair Fernie, Jing-Ke Weng and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Méteignier, LV., Nützmann, HW., Papon, N. et al. Emerging mechanistic insights into the regulation of specialized metabolism in plants. Nat. Plants 9, 22–30 (2023). https://doi.org/10.1038/s41477-022-01288-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-022-01288-7
This article is cited by
-
Intergrative metabolomic and transcriptomic analyses reveal the potential regulatory mechanism of unique dihydroxy fatty acid biosynthesis in the seeds of an industrial oilseed crop Orychophragmus violaceus
BMC Genomics (2024)
-
Pan-evolutionary and regulatory genome architecture delineated by an integrated macro- and microsynteny approach
Nature Protocols (2024)