Abstract
The forthcoming massive genome data generated by the Earth BioGenome Project will open up a new era of comparative genomics, for which genome synteny analysis provides an important framework. Profiling genome synteny represents an essential step in elucidating genome architecture, regulatory blocks/elements and their evolutionary history. Here we describe PanSyn, (https://github.com/yhw320/PanSyn), the most comprehensive and up-to-date genome synteny pipeline, providing step-by-step instructions and application examples to demonstrate its usage. PanSyn inherits both basic and advanced functions from existing popular tools, offering a user-friendly, highly customized approach for genome macrosynteny analysis and integrated pan-evolutionary and regulatory analysis of genome architecture, which are not yet available in public synteny software or tools. The advantages of PanSyn include: (i) advanced microsynteny analysis by functional profiling of microsynteny genes and associated regulatory elements; (ii) comprehensive macrosynteny analysis, including the inference of karyotype evolution from ancestors to extant species; and (iii) functional integration of microsynteny and macrosynteny for pan-evolutionary profiling of genome architecture and regulatory blocks, as well as integration with external functional genomics datasets from three- or four-dimensional genome and ENCODE projects. PanSyn requires basic knowledge of the Linux environment and Perl programming language and the ability to access a computer cluster, especially for large-scale genomic comparisons. Our protocol can be easily implemented by a competent graduate student or postdoc and takes several days to weeks to execute for dozens to hundreds of genomes. PanSyn provides yet the most comprehensive and powerful tool for integrated evolutionary and functional genomics.
Key points
-
PanSyn is a user-friendly pipeline that integrates popular and customized micro- and macrosynteny tools and provides access to external functional genomics datasets for comparative genomic studies.
-
Compared with alternative methods, PanSyn allows advanced microsynteny analysis of regulatory blocks, comprehensive macrosynteny analysis of karyotype evolution and integrated analysis of micro- and macrosynteny for the pan-evolutionary and functional investigation of genome architecture.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data analyzed within this protocol are publicly available. Demo datasets used in the procedure section and expected results are included in the PanSyn package, which are accessible at Zenodo (https://zenodo.org/records/10115240). The accession numbers for the demo datasets used in the PanSyn procedure are listed in Supplementary Table 1. Source data are provided with this paper.
Code availability
All PanSyn source codes are publicly available at the GitHub website (https://github.com/yhw320/PanSyn/tree/main/scripts) and are provided in the Supplementary Code.
References
Lewin, H. A. et al. The Earth BioGenome Project 2020: starting the clock. Proc. Natl Acad. Sci. USA 119, e2115635118 (2022).
ENCODE Project Consortium. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020).
Dekker, J. et al. The 4D nucleome project. Nature 549, 219–226 (2017).
Darwin Tree of Life Project Consortium. Sequence locally, think globally: the Darwin tree of life project. Proc. Natl Acad. Sci. USA 119, e2115642118 (2022).
Meyer, A. & Schartl, M. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr. Opin. Cell Biol. 11, 699–704 (1999).
Simakov, O. et al. Deeply conserved synteny resolves early events in vertebrate evolution. Nat. Ecol. Evol. 4, 820–830 (2020).
Putnam, N. H. et al. The amphioxus genome and the evolution of the chordate karyotype. Nature 453, 1064–1071 (2008).
Simakov, O. et al. Deeply conserved synteny and the evolution of metazoan chromosomes. Sci. Adv. 8, eabi5884 (2022).
Nguyen, N. T. T., Vincens, P., Dufayard, J. F., Roest Crollius, H. & Louis, A. Genomicus in 2022: comparative tools for thousands of genomes and reconstructed ancestors. Nucleic Acids Res. 50, D1025–D1031 (2022).
Lemons, D. & McGinnis, W. Genomic evolution of Hox gene clusters. Science 313, 1918–1922 (2006).
Wang, S. et al. Scallop genome provides insights into evolution of bilaterian karyotype and development. Nat. Ecol. Evol. 1, 120 (2017).
Wilson, M. A. & Makova, K. D. Genomic analyses of sex chromosome evolution. Annu. Rev. Genomics Hum. Genet. 10, 333–354 (2009).
Guo, L. et al. Island-specific evolution of a sex-primed autosome in a sexual planarian. Nature 606, 329–334 (2022).
Han, W. et al. Ancient homomorphy of molluscan sex chromosomes sustained by reversible sex-biased genes and sex determiner translocation. Nat. Ecol. Evol. 6, 1891–1906 (2022).
Dunning, L. T. et al. Lateral transfers of large DNA fragments spread functional genes among grasses. Proc. Natl Acad. Sci. USA 116, 4416–4425 (2019).
Simion, P. et al. Chromosome-level genome assembly reveals homologous chromosomes and recombination in asexual rotifer Adineta vaga. Sci. Adv. 7, eabg4216 (2021).
Harmston, N. et al. Topologically associating domains are ancient features that coincide with Metazoan clusters of extreme noncoding conservation. Nat. Commun. 8, 441 (2017).
Schmidbaur, H. et al. Emergence of novel cephalopod gene regulation and expression through large-scale genome reorganization. Nat. Commun. 13, 2172 (2022).
Méteignier, L. V., Nützmann, H. W., Papon, N., Osbourn, A. & Courdavault, V. Emerging mechanistic insights into the regulation of specialized metabolism in plants. Nat. Plants 9, 22–30 (2023).
Zimmermann, B., Robert, N. S. M., Technau, U. & Simakov, O. Ancient animal genome architecture reflects cell type identities. Nat. Ecol. Evol. 3, 1289–1293 (2019).
Wong, E. S. et al. Deep conservation of the enhancer regulatory code in animals. Science 370, eaax8137 (2020).
Wang, Y. et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49 (2012).
Haas, B. J. et al. DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics 20, 3643–3646 (2004).
Soderlund, C., Bomhoff, M. & Nelson, W. M. SyMAP v3.4: a turnkey synteny system with application to plant genomes. Nucleic Acids Res. 39, e68 (2011).
Wei, J. et al. EDomics: a comprehensive and comparative multi-omics database for animal evo–devo. Nucleic Acids Res. 51, D913–D923 (2023).
Xiao, Z. & Lam, H. M. ShinySyn: a Shiny/R application for the interactive visualization and integration of macro- and micro-synteny data. Bioinformatics 38, 4406–4408 (2022).
Robert, N. S. M., Sarigol, F., Zieger, E. & Simakov, O. SYNPHONI: scale-free and phylogeny-aware reconstruction of synteny conservation and transformation across animal genomes. Bioinformatics 38, 5434–5436 (2022).
Bao, Y. et al. Genomic insights into the origin and evolution of molluscan red-bloodedness in the blood clam Tegillarca granosa. Mol. Biol. Evol. 38, 2351–2365 (2021).
Li, Y. et al. Adaptive bird-like genome miniaturization during the evolution of scallop swimming lifestyle. Genomics Proteom. Bioinforma. 20, 1066–1077 (2022).
Wang, S. et al. Construction of a high-resolution genetic linkage map and comparative genome analysis for the reef-building coral Acropora millepora. Genome Biol. 10, R126 (2009).
Liu, Z. et al. The channel catfish genome sequence provides insights into the evolution of scale formation in teleosts. Nat. Commun. 7, 11757 (2016).
Li, Y. et al. Scallop genome reveals molecular adaptations to semi-sessile life and neurotoxins. Nat. Commun. 8, 1721 (2017).
Li, Y. et al. Sea cucumber genome provides insights into saponin biosynthesis and aestivation regulation. Cell Discov. 4, 29 (2018).
Liu, F. et al. MolluscDB: an integrated functional and evolutionary genomics database for the hyper-diverse animal phylum Mollusca. Nucleic Acids Res. 49, D988–D997 (2021).
Zeng, Q. et al. High-quality reannotation of the king scallop genome reveals no ‘gene-rich’ feature and evolution of toxin resistance. Comput. Struct. Biotechnol. J. 19, 4954–4960 (2021).
Ye, N. et al. The role of zinc in the adaptive evolution of polar phytoplankton. Nat. Ecol. Evol. 6, 965–978 (2022).
Bao, L., Zhong, X., Yang, Y. & Yang, L. Starfish infers signatures of complex genomic rearrangements across human cancers. Nat. Cancer 3, 1247–1259 (2022).
Jiao, Y. & Paterson, A. H. Polyploidy-associated genome modifications during land plant evolution. Philos. Trans. R. Soc. Lond. B 369, 20130355 (2014).
Wu, S. et al. Genome sequences of two diploid wild relatives of cultivated sweetpotato reveal targets for genetic improvement. Nat. Commun. 9, 4580 (2018).
Wu, H. et al. A high-quality Actinidia chinensis (kiwifruit) genome. Hortic. Res. 6, 117 (2019).
Ma, D. et al. Chromosome-level reference genome assembly provides insights into aroma biosynthesis in passion fruit (Passiflora edulis). Mol. Ecol. Resour. 21, 955–968 (2021).
Yin, Y. et al. The chromosome-scale genome of Magnolia officinalis provides insight into the evolutionary position of magnoliids. iScience 24, 102997 (2021).
Xu, Q. et al. Ancestral flowering plant chromosomes and gene orders based on generalized adjacencies and chromosomal gene co-occurrences. J. Comput. Biol. 28, 1156–1179 (2021).
Hong, S. et al. Genome-wide comparative analysis of flowering-time genes: insights on the gene family expansion and evolutionary perspective. Front. Plant Sci. 12, 702243 (2021).
Peng, R. et al. Evolutionary divergence of duplicated genomes in newly described allotetraploid cottons. Proc. Natl Acad. Sci. USA 119, e2208496119 (2022).
Hoopes, G. et al. Phased, chromosome-scale genome assemblies of tetraploid potato reveal a complex genome, transcriptome, and predicted proteome landscape underpinning genetic diversity. Mol. Plant 15, 520–536 (2022).
Luo, J. et al. From asymmetrical to balanced genomic diversification during rediploidization subgenomic evolution in allotetraploid fish. Sci. Adv. 6, eaaz7677 (2020).
Blanc-Mathieu, R. et al. Hybridization and polyploidy enable genomic plasticity without sex in the most devastating plant-parasitic nematodes. PLoS Genet. 13, e1006777 (2017).
Zhao, T. & Schranz, M. E. Network-based microsynteny analysis identifies major differences and genomic outliers in mammalian and angiosperm genomes. Proc. Natl Acad. Sci. USA 116, 2165–2174 (2019).
Gamboa-Tuz, S. D., Pereira-Santana, A., Zhao, T. & Schranz, M. E. Applying synteny networks (SynNet) to study genomic arrangements of protein–coding genes in plants. Methods Mol. Biol. 2512, 199–215 (2022).
Almeida-Silva, F., Zhao, T., Ullrich, K. K., Schranz, M. E. & Van de Peer, Y. Syntenet: an R/Bioconductor package for the inference and analysis of synteny networks. Bioinformatics 39, btac806 (2022).
Sun, P. et al. WGDI: a user-friendly toolkit for evolutionary analyses of whole-genome duplications and ancestral karyotypes. Mol. Plant. 15, 1841–1851 (2022).
Conover, J. et al. pSONIC: ploidy-aware syntenic orthologous networks identified via collinearity. G3 11, jkab170 (2021).
Luo, X. et al. 3D genome of macaque fetal brain reveals evolutionary innovations during primate corticogenesis. Cell 184, 723–740 (2021).
Lu, J., Huang, P., Sun, J. & Liu, J. DupScan: predicting and visualizing vertebrate genome duplication database. Nucleic Acids Res. 51, D906–D912 (2023).
Wang, Y. et al. Comparative genome anatomy reveals evolutionary insights into a unique amphitriploid fish. Nat. Ecol. Evol. 6, 1354–1366 (2022).
Kikuta, H. et al. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res. 17, 545–555 (2007).
Lazar, N. H. et al. Epigenetic maintenance of topological domains in the highly rearranged gibbon genome. Genome Res. 28, 983–997 (2021).
Zhao, T. et al. Whole-genome microsynteny-based phylogeny of angiosperms. Nat. Commun. 12, 3498 (2021).
Zhou, Z. W. et al. GenomeSyn: a bioinformatics tool for visualizing genome synteny and structural variations. J. Genet. Genomics. 49, 1174–1176 (2022).
Shtolz, N. & Mishmar, D. The metazoan landscape of mitochondrial DNA gene order and content is shaped by selection and affects mitochondrial transcription. Commun. Biol. 6, 93 (2023).
Stein, J. C. et al. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat. Genet. 50, 285–296 (2018).
Grueber, C. E. Comparative genomics for biodiversity conservation. Comput. Struct. Biotechnol. J. 13, 370–375 (2015).
Wallace, H. A. et al. Manipulating the mouse genome to engineer precise functional syntenic replacements with human sequence. Cell 128, 197–209 (2007).
Nutzmann, H. W. et al. Plant metabolic clusters—from genetics to genomics. N. Phytol. 211, 771–789 (2016).
Graham, L. A. et al. Horizontal gene transfer in vertebrates: a fishy tale. Trends Genet. 37, 501–503 (2020).
Waterhouse, R. M. et al. Evolutionary superscaffolding and chromosome anchoring to improve Anopheles genome assemblies. BMC Biol. 18, 1 (2020).
Meyer, A. et al. Giant lungfish genome elucidates the conquest of land by vertebrates. Nature 590, 284–289 (2021).
Albertin, C. B. et al. Genome and transcriptome mechanisms driving cephalopod evolution. Nat. Commun. 13, 2427 (2022).
Rhie, A. et al. Towards complete and error-free genome assemblies of all vertebrate species. Nature 592, 737–746 (2021).
Nurk, S. et al. The complete sequence of a human genome. Science 376, 44–53 (2022).
Nakatani, Y. et al. Reconstruction of proto-vertebrate, proto-cyclostome and proto-gnathostome genomes provides new insights into early vertebrate evolution. Nat. Commun. 12, 4489 (2021).
Ip, J. C. et al. Host–endosymbiont genome integration in a deep-sea chemosymbiotic clam. Mol. Biol. Evol. 38, 502–518 (2021).
Kim, J. et al. Reconstruction and evolutionary history of eutherian chromosomes. Proc. Natl Acad. Sci. USA 114, E5379–E5388 (2017).
Simakov, O. et al. Insights into bilaterian evolution from three spiralian genomes. Nature 493, 526–531 (2013).
Li, Y. et al. Contrasting modes of macro and microsynteny evolution in a eukaryotic subphylum. Curr. Biol. 32, 1–9 (2022).
Fernández, R. & Gabaldón, T. Gene gain and loss across the metazoan tree of life. Nat. Ecol. Evol. 4, 524–533 (2020).
Ocaña–Pallarès, E. et al. Divergent genomic trajectories predate the origin of animals and fungi. Nature 609, 747–753 (2022).
Irimia, M. et al. Extensive conservation of ancient microsynteny across metazoans due to cis-regulatory constraints. Genome Res. 22, 2356–2367 (2012).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Buchfink, B., Reuter, K. & Drost, H. G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 18, 366–368 (2021).
Bryson, A. E. et al. Uncovering a miltiradiene biosynthetic gene cluster in the Lamiaceae reveals a dynamic evolutionary trajectory. Nat. Commun. 14, 343 (2023).
Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009).
Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019).
Lopez-Delisle, L. et al. pyGenomeTracks: reproducible plots for multivariate genomic datasets. Bioinformatics 37, 422–423 (2021).
Ayad, L. A. K., Pissis, S. P. & Polychronopoulos, D. CNEFinder: finding conserved non-coding elements in genomes. Bioinformatics 34, i743–i747 (2018).
Tan, G., Polychronopoulos, D. & Lenhard, B. CNEr: a toolkit for exploring extreme noncoding conservation. PLoS Comput. Biol. 15, e1006940 (2019).
Kumar, S., Tamura, K. & Nei, M. MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput. Appl. Biosci. 10, 189–191 (1994).
de Hoon, M. J., Imoto, S., Nolan, J. & Miyano, S. Open source clustering software. Bioinformatics 20, 1453–1454 (2004).
Anand, L. & Rodriguez Lopez, C. M. ChromoMap: an R package for interactive visualization of multi-omics data and annotation of chromosomes. BMC Bioinforma. 23, 33 (2022).
Quigley, S., Damas, J., Larkin, D. M. & Farré, M. syntenyPlotteR: a user-friendly R package to visualize genome synteny, ideal for both experienced and novice bioinformaticians. Bioinforma. Adv. 3, vbad161 (2023).
Servant, N. et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16, 259 (2015).
Adhikari, B., Trieu, T. & Cheng, J. Chromosome3D: reconstructing three-dimensional chromosomal structures from Hi-C interaction frequency data using distance geometry simulated annealing. BMC Genomics 17, 886 (2016).
DeLano, W. L. PyMOL: an open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 40, 82–92 (2002).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Csurös, M. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics 26, 1910–1912 (2010).
Cantalapiedra, C. P., Hernández-Plaza, A., Letunic, I., Bork, P. & Huerta-Cepas, J. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38, 5825–5829 (2021).
Stevens, T. J. et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544, 59–64 (2017).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Engström, P. G., Ho Sui, S. J., Drivenes, O., Becker, T. S. & Lenhard, B. Genomic regulatory blocks underlie extensive microsynteny conservation in insects. Genome Res. 17, 1898–1908 (2007).
Dong, X., Fredman, D. & Lenhard, B. Synorth: exploring the evolution of synteny and long-range regulatory interactions in vertebrate genomes. Genome Biol. 10, R86 (2009).
Muffato, M. et al. Reconstruction of hundreds of reference ancestral genomes across the eukaryotic kingdom. Nat. Ecol. Evol. 7, 355–366 (2023).
Damas, J. et al. Evolution of the ancestral mammalian karyotype and syntenic regions. Proc. Natl Acad. Sci. USA 119, e2209139119 (2022).
Van de Peer, Y., Maere, S. & Meyer, A. The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 10, 725–732 (2009).
Van de Peer, Y., Mizrachi, E. & Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411–424 (2017).
Lee, T. H., Tang, H., Wang, X. & Paterson, A. H. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 41, D1152–D1158 (2013).
Zhao, T. & Schranz, M. E. Network approaches for plant phylogenomic synteny analysis. Curr. Opin. Plant Biol. 36, 129–134 (2017).
Acknowledgements
We thank all developers of useful genome comparison algorithms and tools that have been integrated in the PanSyn pipeline. We also wish to thank J. Zhang (Novogene Bioinformatics Institute) and X. Dai (University of Michigan) for assisting in the early development of macrosynteny pipeline and PanSyn protocol testing, respectively. This research is part of the ongoing M10K+ genome project that is proposed by M10K+ Consortium and targets sequencing of 10,000 molluscan genomes. We acknowledge the grant support from the Science & Technology Innovation Project of Laoshan Laboratory (LSKJ202203001, LSKJ202202804), National Natural Science Foundation of China (32130107, 32222085), National Key Research and Development Program of China (2022YFD2400301), Key R&D Project of Shandong Province (2021ZLGX03, 2022ZLGX01), the Fundamental Research Funds for the Central Universities (842341005) and Taishan Scholar Project Fund of Shandong Province of China.
Author information
Authors and Affiliations
Contributions
S.W. and Y.L. conceived and designed the protocol. H.Y., Y.L., W.H., L.B., F.L., Y.M. and Z.P. developed, optimized and tested the protocol. Q.Z., L.Z. and Z.B. participated in discussions and provided suggestions for protocol improvement. S.W., Y.L. and H.Y. wrote the protocol with the input from other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Steven Cannon, Xiyin Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Wang, S. et al. Nat. Ecol. Evol. 1, 120 (2017): https://doi.org/10.1038/s41559-017-0120
Han, W. et al. Nat. Ecol. Evol. 6, 1891–1906 (2022): https://doi.org/10.1038/s41559-022-01898-6
Wei, J. et al. Nucleic Acids Res. 51, D913–D923 (2023): https://doi.org/10.1093/nar/gkac944
Extended data
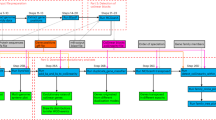
Extended Data Fig. 1 Extended demonstrations of microsynteny analyses.
(a) Dot plot or Circos plot visualization of polyploid plant genomes (left) and microbial genomes (right). In the dot plot, homologous gene pairs are shown as dots, and syntenic gene pairs are aligned together. In the Circos plot, lines linking two chromosomes indicate the location of microsynteny genes. (b) Microsynteny analysis of heteromorphic and homomorphic sex chromosomes in X/Y and Z/W sexual systems, respectively. Lines linking two sex chromosomes indicate the location of microsynteny genes. The location of sex-determining gene is indicated by a green line. (c) Genomic organization of plant gene clusters, which are tandemly connected in metabolic pathways. Homologous genes are represented with rectangles of the same color. Microsynteny between two species is shown with grey curves. (d) Integrative analysis of microsynteny with genomic structural variations. Oryza sativa L. indica rice varieties Minghui63 (MH63) and Zhenshan97 (ZS97) genomes are used for displaying the association of genome synteny and different structural variations (insertions/deletions or inversions).
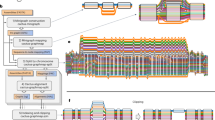
Extended Data Fig. 2 Computational procedure and visualization of network-based microsynteny analysis.
(a, b) Schematic overview of network-based approaches developed for microsynteny network detection and macroevolutionary history inference (see Zhao et al.108 and Robert et al.27 for detailed algorithm descriptions). (c) Network-based microsynteny analysis in 18 animal genomes. The heatmap in the top panel shows the pairwise comparisons for microsynteny conservation between any two species. The adjacent network shows several example clusters after microsynteny network clustering. The middle panel shows a binary matrix constructed by the phylogenomic profiling of all clusters, where rows represent clusters and columns represent species. The bottom panel shows the network representation of one conserved (left) and one Eutheria-specific (right) microsynteny genes.
Extended Data Fig. 3 Functional characterization and regulatory analysis of microsynteny gene clusters.
(a) Association of microsynteny cluster with single-cell transcriptome data of Amphimedon queenslandica (left) and Trichoplax adhaerens (right), with microsynteny genes associated with cell type (top), cell lineage (middle) and co-expression pattern (bottom). (b) Identification of the conserved regulatory CNEs for the well-known pharyngeal gene cluster across four placental mammals. Blue and orange rectangles represent the positions of CNEs on chromosomes that are presented in each species (blue) or conserved across all species (orange). Pink rectangles represent the position of conserved gene cluster on the chromosome of the reference species (human Chr14). (c) Distribution and comparison of TADs around the conserved pharyngeal gene cluster in human and mouse. The chromatin interaction heatmap was generated using the 3D Genome Browser (http://3dgenome.fsm.northwestern.edu/).
Extended Data Fig. 4 Ancestral genome reconstruction and macrosynteny analysis.
(a) Schematic overview of ancestral genome reconstruction approaches for macrosynteny analysis, which are suitable for a wide range of evolutionary distance (see Kim et al.74 and Simakov et al.75 for detailed algorithm descriptions). (b) Various visualizations of genome macrosynteny for human, chimpanzee and mouse in comparison with the deduced karyotype of the eutherian ancestor, including profiling of karyotype evolution and conservation (CI values), identification of chromosome breakage and fusion events, and genome-wide profiling of macrosynteny landscapes for both genomic DNA-based and protein-based analyses.
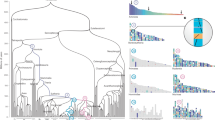
Extended Data Fig. 5 Macrosynteny analysis of 34 representative species across the animal kingdom.
Macrosynteny analysis using the ancestral linkage groups represented by the ancestral genome of Nematostella vectensis is presented, with orange and blue dots representing chromosome-level and scaffold-level genomes, respectively. In the dot plots, dots represent homologous genes distributed in the chromosomes of compared species (x-axis: extant species, y-axis: bilaterian ancestor). Conserved macrosynteny blocks (with statistical significance) are indicated by red dots.
Extended Data Fig. 6 Visualization of karyotype evolution and integration with functional genomics data.
(a) Visualization of karyotype comparison of the bilaterian ancestor with human (top) or mouse (bottom). Each color represents one of the 17 chromosomes of bilaterian ancestor. (b) Integrative analysis of karyotype and regulatory evolution in humans (top) and mice (bottom). The color density in the heatmap represents the relative number of contacts observed within chromosomes. Various epigenetic and regulatory data are collectively shown for the chromosomes under investigation. The chromatin interaction heatmap was generated using the 3D Genome Browser (http://3dgenome.fsm.northwestern.edu/).
Extended Data Fig. 7 Schematic overview of pan-evolutionary analysis of microsynteny and macrosynteny.
(a) Recovering macrosyntenic ancient blocks from microsyntenic gene clusters in extant species. Coloured circles correspond to different OGs. The lengths of the edges in the networks are proportional to the intergenic distance. The schematic overview is adapted from the SYNPHONI pipeline27. (b) Tracing evolutionary trajectories and dynamics of gene contents and gene orders from ancestors to extant species in the aspects of gene gain and loss events and conservative transitions from macrosynteny to microsynteny.
Extended Data Fig. 8 Whole-genome distribution and three-dimensional exhibition of ancient/novel gene clusters.
(a) Distribution of Eutheria-conserved (green), Boreoeutheria-conserved (pink) and Simian-conserved (blue) microsynteny genes along the human chromosomes. (b) Three-dimensional chromosome model of human (top) and mouse (bottom), with color labeling Eutheria-conserved (left), Euarchontoglires-conserved (middle) and Simian/Glires-specific (right) microsynteny locations. Gray thick threads represent the 3D structure of the entire chromosome. 3D genome structures are visualized using the Nucleome Browser (http://www.nucleome.org).
Extended Data Fig. 9 Integrated pan-evolutionary and regulatory analysis of genome microsynteny and macrosynteny.
(a) Chromosomal distribution of macrosynteny, microsynteny and various regulatory information derived from the ENCODE project in human (left) and mouse (right). Rectangles represent conserved synteny genes (blue: macrosynteny, green: microsynteny, red: microsynteny & macrosynteny). Visualization of various associated regulatory data types is shown below. (b) Functional enrichment analysis of macrosynteny/microsynteny genes based on KEGG (top) and GO (bottom) annotation. In the bubble diagrams, the color and size of the bubbles are utilized to convey statistical information, such as the P-value and the number of overlapping genes with the pathway. In the bar charts, the enriched annotated GO terms are shown, with statistical significance indicated by the height of the bars. (c) Detailed presentation of CNEs and other regulatory information from ENCODE for the HOXA gene cluster in human and mouse. Blue and red rectangles represent the position of the HOXA gene cluster and identified CNEs on chromosomes, respectively. Various ENCODE data types are collectively shown for the chromosomal regions under investigation.
Supplementary information
Supplementary Table 1
The sources of all the input datasets used in demonstration.
Supplementary Code 1
All PanSyn source codes and user guidance.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yu, H., Li, Y., Han, W. et al. Pan-evolutionary and regulatory genome architecture delineated by an integrated macro- and microsynteny approach. Nat Protoc (2024). https://doi.org/10.1038/s41596-024-00966-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41596-024-00966-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.