Abstract
Mutations in genes encoding cardiac ion channels and their subunits are responsible for several genetic cardiac disorders. We characterised the human gene KCNA7, encoding the voltage-gated potassium channel Kv1.7 and compared its coding sequence with that of the mouse orthologue, kcna7. Both genes are encoded by two exons separated by a conserved intron, unlike all the other Kv1-family genes that contain intronless coding regions. KCNA7 and kcna7 encode proteins of 456 amino acid residues that share >95% sequence identity, and the mouse channel has biophysical and pharmacological properties closely resembling the ultra-rapidly activating delayed rectifier (IKur) in cardiac tissue. Using reverse transcriptase-PCR, KCNA7 mRNA was detected in adult human heart. We determined that KCNA7 resides on chromosome 19q13.3 in a region that also contains the progressive familial heart block I (PFHBI) locus. Direct sequencing of KCNA7's coding sequence in PFHB1-affected individuals revealed no pathogenic sequence changes, but two single nucleotide polymorphisms detected in exon 2 result in amino acid substitutions. These results provide evidence for the exclusion of this candidate as the PFHB1-causative gene, although mutations in regulatory and non-coding regions cannot be excluded. As ion channel-encoding genes have been implicated in a growing number of genetic conditions, the data presented may facilitate further analysis of the role of KCNA7 and its product in the heart.
Similar content being viewed by others
Introduction
Ion channel defects are implicated in a number of inherited cardiac diseases, many of which are characterised by sudden death and associated with conduction abnormalities. The Long QT syndromes (LQTS) are associated with mutations in at least five genes, four of which encode potassium (K+) channels. These K+ channel genes act as two paired sets, namely KCNQ1/KCNE1 and KCNH2/KCNE1, with the products of each pair co-assembling to form the ion channels responsible for the slowly activating potassium current (IKs) and the rapidly activating delayed rectifier potassium current IKr, respectively.1 Interestingly, the fifth LQTS-causative gene, SCN5A1 has also been implicated in three distinct cardiac diseases: Brugada syndrome, in which patients have marked abnormalities in ventricular conduction,1 progressive cardiac conduction defect (PCCD);2 and an isolated cardiac conduction disease.3
Progressive familial heart block type I (PFHBI) is an autosomal dominantly inherited cardiac conduction disorder, which has been mapped to chromosome 19q13.3 in a large South African Afrikaner family.4 Clinically, PFHBI is characterised by right bundle branch block, left anterior hemiblock, or complete heart block with broad QRS complexes.5 Evidence of genetic anticipation in this disorder, with an increase in severity of the disease's presentation in succeeding generations, has been reported.5 A second cardiac conduction disorder, designated isolated cardiac conduction disorder (ICCD), showing similar clinical features to PFHBI, has been mapped to the same region of chromosome 19q13.3 in a Lebanese family.6 Although PFHBI and ICCD exhibit phenotypic similarities and are linked to the same microsatellite markers on chromosome 19q13.3, it is not known if they are the same clinical entity. Recent studies from our group using a number of microsatellite markers have refined the PFHB1 locus to a 4 cM region on 19q13.3 between markers D19S606 and D19S866 (unpublished data).
A plausible candidate gene for PFHBI and ICCD by mapped position, expression profile and function is the voltage-gated K+ channel gene7 designated hKv1.7 in the functional classification and KCNA7 in the HUGO classification (by which it will be referred to in the text). This member of a multigene family8 has been mapped by fluorescent in situ hybridisation (FISH) to human chromosome 19q13.3, 7 placing it within the PFHBI locus, where it is flanked by the muscle glycogen synthase gene (GSY1) and histidine-rich calcium binding protein (HRC).4 The mouse homologue, kcna7, a member of the Shaker-related Kv1-subfamily of K+ channels, is abundantly expressed in mouse heart,7 strengthening the candidacy of the human homologue KCNA7 as the PFHBI-causative gene.
Consequently, we determined the complete coding sequence of KCNA7, defined its genomic organisation, demonstrated its presence in human heart, refined its chromosomal location relative to the PFHBI locus and screened PFHBI-affected family members for mutations. Moreover, we conducted an in silico search in the 5′ and 3′-untranslated regions of KCNA7 for the presence of triplet repeat motifs which might be pathologically expanded in PFHB1-affected family members displaying genetic anticipation.
Materials and methods
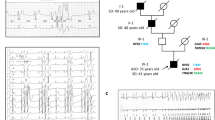
All procedures used were reviewed and approved by the University of Stellenbosch's Ethics Committee (reference 86/085). The clinical details and pedigrees of the Afrikaner families in which PFHBI segregates were described previously.4,5
Characterisation of KCNA7
Determining coding sequence
A BLAST search of the Genoscope database (http://www.genoscope.org) revealed several 19q13.3 human genomic sequences (eg, R0AA003ZF08G1) that aligned with the published mouse kcna7 cDNA sequence (AF032099). Subsequent BLAST searches of GenBank with this inferred human KCNA7 sequence identified significant matches with human BAC clone BC52309 (synonym: CITB-60B18; GenBank accession no. AC008687), mouse genomic contig (AC073711), as well as three mouse placental expressed sequence tags, ESTs (AA021711, AI322534, AI324179). The coding region of KCNA7 was deduced by aligning the mouse cDNA sequence with the human BAC clone and the three mouse EST sequences retrieved from the databases. Based on this determined coding sequence of KCNA7, three overlapping sets of PCR primers were designed to span exon 1 and four sets were designed to span the larger exon 2 (Table 1). These seven primer sets were used in direct DNA sequencing to confirm the sequence deduced by the in silico techniques.
Electrophysiology of kcna7
Since the published mouse cDNA sequence was shown to contain an error (see Results), the corrected mouse kcna7 sequence was generated by PCR from the original published construct and then cloned in-frame downstream to enhanced green fluorescent protein in the eGFP-C3 vector. COS-7 cells were transiently transfected with this construct using FuGeneTM 6 (Roche) according to the manufacturer's protocol and kcna7 currents were recorded 6–12 hours after transfection. COS-7 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% heat-inactivated foetal calf serum (Summit Biotechnology, Fort Collins, CO, USA), 4 mM L-glutamine and 1 mM Na+ pyruvate. All experiments were carried out in the whole-cell configuration of the patch-clamp technique with a holding potential of −80 mV. Currents were recorded in normal Ringer solution (160 mM NaCl, 4.5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, pH 7.4, 290–310 mOsm) with an internal pipette solution containing 134 mM KF, 2 mM MgCl2, 10 mM HEPES, 10 mM EGTA (pH 7.2, 290–310 mOsm). 200-ms depolarising pulses to 40 mV were applied every 30 s and Kd-values were determined by fitting the Hill equation to the reduction of peak current; for verapamil the reduction in the area under the current trace was used to determine the Kd value. Series resistance compensation (60–80%) was used if currents exceeded 2 nA. 4-Aminopyridine (4-AP), amiodarone, flecainide, quinidine, and tetraethylammonium chloride (TEA) were purchased from Sigma. Sotalol was from ICN, charybdotoxin, margatoxin, tertiapin and ShK toxin were from Bachem Biosciences (King of Prussia, PA, USA).
RT–PCR analysis
Reverse transcriptase–PCR (RT–PCR) was performed to determine whether KCNA7 is expressed in human heart tissue. Total RNA was extracted from adult human heart by a standard method.9 First strand cDNA synthesis was performed using MMLV Expand Reverse Transcriptase (Roche, Germany) and oligo dT as a primer following the manufacturer's instructions. RT–PCR was performed using the KCNA7-specific primers in Table 1 and the products were separated by electrophoresis on a 12% polyacrylamide gel and subsequently silver-stained.
In silico searches
Bioinformatic-based in silico mapping using the EST2GENOME software (http://ftp.sanger.ac.uk/pub/EMBOSS) was used to position KCNA7 relative to microsatellite markers and to other genes of interest on chromosome 19q. These included the genes encoding the K+-intermediate/small conductance calcium-activated, subfamily N, member 4 (KCNN4), glycogen synthase 1 (GYS1), histidine-rich calcium-binding protein C (HRC) and K+-voltage-gated channel, Shaw-related subfamily, member 3 (KCCN3). Chromosome 19 is jointly sequenced by the Lawrence Livermore National Laboratory (LLNL; http://www-bio.llnl.gov/) and the Joint Genome Institute (JGI; http://www.jgi.doe.gov/). Draft DNA sequences from 32 BAC and cosmid clones, which span the PFHBIlocus on 19q13.3, were downloaded from the JGI ftp site on 04.18.2000 and used for the alignments. This region extends from BAC clone CITB-E1_3191M6 (BC894691) to cosmid clone LLNL-R_249H9 (R28901). As the sequence was still in draft phase, Escherichia coli sequences were masked out using the program Repeatmasker prior to further analysis http://ftp.genome.washington.edu/cgi-bin/RepeatMasker).
Investigation as a candidate for PFHB1?
PCR–SSCP analysis and direct DNA sequencing of KCNA7
The primers described in Table 1 were used to screen for putative pathogenic mutations in PFHB1-affected individuals in the two exons of KCNA7 and approximately 50 bp of the exon-intron junctions. The seven overlapping fragments were PCR-amplified in a panel of PFHBI family members, following standard SSCP techniques.10 PCR products were size-separated by electrophoresis on 8% and 10% acrylamide SSCP gels containing 5% glycerol at 25 W overnight, after which the gels were silver-stained and analysed for mobility shifts. The alleles of the two polymorphisms identified within KCNA7 were genotyped by PCR–SSCP analysis, using primer sets D and G (Table 1) and their frequencies calculated in 52 unrelated South African Afrikaner individuals.
In addition, all seven overlapping PCR-amplified products of exons 1 and 2 of KCNA7 generated from an affected and an unaffected PFHBI family member were directly sequenced and compared to identify possible pathogenic mutations. To this end, the products were purified in QIAquick spin columns (Qiagen) and sequenced using an ABI 377 DNA Sequencing System following the manufacturer's recommended protocols (Perkin Elmer).
Screening for triplet repeats
The KCNA7-specific primers were used in PCR amplification to verify that BAC clone BC52309 harboured the entire coding sequence for KCNA7. Alignment of the exonic sequence of KCNA7 to BC52309, using the EST2GENOME software facilitated the identification of approximately 2 kb of the putative 5′ and 3′ untranslated regions (UTRs) of this gene (http://ftp.sanger.ac.uk/pub/EMBOSS). These regions were then screened for the presence of a putative pathogenic triplet repeat with the program FindMarker (personal communication; Alan Christoffels).
Results and discussion
The KCNA7 cDNA sequence has been deposited in GenBank under accession no. AF315818. The coding region encodes a protein of 456 amino acid residues containing cytoplasmic N- and C-termini, a central core domain composed of six transmembrane segments (S1–S6) and the characteristic pore-loop, P-region (Figure 1a and Figure 1b). A KCNA7 cDNA sequence has been recently deposited in GenBank (AJ310479; 12 Feb 2001) and the sequence of this entry matches exactly with cDNA sequence reported here, except for two substitutions. We determined that a single intron interrupts the coding region of KCNA7 (indicated by an inverted arrow in Figure 1a). The entire N-terminus, the S1 transmembrane segment and the proximal part of the S1–S2 external loop is encoded by the upstream exon, while the downstream exon encodes the region extending from the S1–S2 loop to the C-terminus. The sequence of the intron-exon junctions of KCNA7 and kcna7 is shown in Figure 1b. The human intron is 1153 bp in length and smaller than that of mouse (1929 bp). Thus, both KCNA7 and kcna7 contain a single conserved intron in the coding region, a feature that differentiates these genes from all other members of the mammalian Kv1-Shaker-related sub-family that have intronless coding regions.8,11 Since this intron is not present in the Drosophila Shaker gene,12 it probably originated after the divergence of flies and mammals.
Comparison of the nucleotide and deduced amino acid sequences of human KCNA7 and mouse kcna7. The six putative membrane-spanning domains (S1 through S6) and the pore-forming region (P-region) are indicated. The potential sites for phosphorylation by tyrosine kinase (TY-K) and protein kinase (PKC) are shown. The arrowhead (▾) indicates the location of the single intron that lies between S1 and S2. Differences between the human and mouse amino acid sequences are in bold and underlined. Two polymorphisms in the human gene are indicated (*) at nucleotide positions 566 C : G and 1253 T : C of the KCNA7 coding sequence. The mouse kcna7 sequence shown has been corrected at several positions based on alignments between the published cDNA sequence (AF032099), the mouse genomic sequence (AC073711) and mouse EST sequences. The missing ‘G’ at position 29 in AF032099 is indicated in bold and underlined. Exon-intron junctions of human KCNA7 and mouse kcna7. Top. Comparison of human KCNA7 genomic sequence (AA008687) with mouse kcna7 genomic sequence (AC073711). The splice donor and acceptor sites which form the boundaries of the human 1153 bp intron and the mouse 1929 bp intron are shown, with the consensus gt/ag sequences in bold and underlined. Bottom. Part of the extended KCNA7 intronic sequence with the positions of the intronic primers C reverse (Crev) and D forward (Dfor) indicated.
Interestingly, the deduced N-terminus encoded by KCNA7 is different from that of the published kcna7 cDNA sequence (AF032099), the first 10 residues of the K+-channel (KCNA7) (MEPRCPPPCG) being replaced by 88 residues in the mouse K+-channel (kcna7).7 Upon investigation, we detected an error in the published kcna7 cDNA sequence; the ‘G’ (GCCCTGCGGCTGCTGCG) at position 29 in the coding region (Figure 1a) is absent in the published sequence (position 264) resulting in a frame-shift. This nucleotide is present in the human and mouse genomic sequences, as well as in three mouse ESTs (AA021711, AI322534, AI324179), which supports the observation that the published AF032099 sequence is incorrect. Based on alignments with the genomic kcna7 sequence and the mouse ESTs, other errors were detected in AF032099 and in Figure 1a, we display the alignment of KCNA7 with the corrected kcna7 sequence. Using the deduced amino acid sequences, the K+-channels of the two species are highly conserved (>95%), there are 22 substitutions, and the mouse also has an additional residue (proline) in the external loop linking the S1–S2 segments.
In order to determine the biophysical and pharmacological properties of the corrected kcna7 channel, we heterologously expressed the construct in COS-7 cells and studied the currents by the patch clamp method (Figure 2). The channel is voltage-dependent (V1/2=−8 mV), activates rapidly (τ=5.8±0.3 ms at 30 mV) and inactivates slowly, with kinetics resembling the ultra-rapidly activating delayed rectifier K+ channel, IKur, in human atrial myocytes.13,14 Like IKur, kcna7 is blocked by the classical K+ channel blockers 4-AP (Kd=150±20 μM) and TEA (Kd=80±5 mM), and by the anti-arrhythmic drugs amiodarone (Kd=35±μM), flecainide (Kd=8±μM) and quinidine (Kd=15±2 μM). The L-type calcium channel antagonist verapamil produced a time-dependent block (Kd=16±1.5 μM), suggesting open-channel block as has been reported for Kv1.5/KCNA5,15 the other cloned channel reported to contribute to the IKur current.16 Sotalol, tertiapine, charybdotoxin, margatoxin and ShK toxin had no effect on the channel. Given the >95% amino acid sequence identity between the mouse and human homologues, especially in the hydrophobic domain, it is highly likely that the properties of KCNA7 will be identical to that of kcna7. Since the macroscopic cardiac IKur current is thought to be composed of kcna5/KCNA5 and another K+ channel,14 we propose that kcna7/KCNA7 might constitute at least part of this current either as a homotetramer or as a heteromultimer with kcna5/KCNA5.
Biophysical and pharmacological properties of kcna7. (a) Family of kcna7 currents. The test potential was changed from −60 to 60 mV in 10 mV increments every 10 s. (b) Peak K+ conductance-voltage relationship for currents shown in (a). The line through the points was fitted with the Boltzmann equation (V1/2 = −8 mV). (c) Slow inactivation in a 2000 ms pulse (depolarising pulse to 40 mV every 10 s). (d) Shift in reversial potential. Currents in a 200 ms voltage-ramp from −120 mV to 40 mV applied every 10 s in normal and in potassium Ringer solution. (e–h) Block of the current by 4-aminopyridine (4-AP), flecainide, verapamil and amiodarone (200 ms depolarising pulsed to 40 mV every 10 s).
To confirm the presence of KCNA7 mRNA in human adult heart, RT–PCR studies were performed using the KCNA7-specific primers. PCR products were obtained with primers A, B, E, F and G that lie within the coding regions (see examples with primers B and F in Figure 3), and, as expected, no products were obtained for the two primer sets (C and D) spanning exon-intron junctions (data not shown). These results provide evidence that KCNA7 is expressed in human adult heart and importantly, also confirm the gene organisation deduced by the in silico methods.
Bioinformatic in silico mapping data placed KCNA7 on BAC BC52309, with the relative order of the five genes on 19q being: (cen)-KCNN4-KCNA7-GYS1-HRC-KCNC3-(tel) (Figure 4). Interestingly, this data places KCNA7 between markers D19S596 and D19S604, within the PFHB1 4 cM locus defined by D19S606 and D19S866, thereby implicating it as a positional candidate gene for this disease (Figure 4).
Ideogram of genetic and physical maps of chromosome 19q depicting the position of KCNA7 relative to microsatellite markers spanning the PFHB1 locus and other genes in that region (not drawn to scale). The electronic placement of genes or genetic markers on clones is indicated by a horizontal line. LLNL clone names are used and the genetic distances were taken from NCBI's Genemap99 (http://www.ncbi.nlm.nih.gov/genemap99/). D19S879, KCNA7 and GYSI were shown to reside on the same BAC, BC52309 (GenBank accession no. AC008687). The appropriate mRNA sequences were retrieved from GenBank using the following accession numbers (in brackets): K+ intermediate/small conductance calcium-activated channel, subfamily N, member 4 (KCNN4; NM_002250), glycogen synthase 1 (GYS1; NM_002103), histidine-rich calcium-binding protein C (HRC; M91083), K+-voltage-gated channel, Shaw-related subfamily, member 3 (KCNC3; NM_004977). cM = centiMorgans.
The chromosomal location of KCNA7 within the PFHB1 locus, its expression in human heart and its functional resemblance to the IKur current in atrial myocytes, strengthens its candidature as the PFHBI-causative gene. Using the primer sets described in Table 1, the coding region of KCNA7 was screened for putative pathogenic mutations in selected PFHBI-affected family members. PCR–SSCP analysis and direct DNA sequencing did not reveal any pathogenic mutations in the coding regions of the affected individuals tested. However, two single nucleotide polymorphisms (SNPs) were detected in exon 2, both of which resulted in amino acid changes (Figure 1a). These polymorphisms were deposited in NCBI's dbSNP under accession nos. AF315818-566 and AF315818-1253. The first of these (C : G at nucleotide 566 of KCNA7) in the external S1–S2 loop changes proline to arginine at codon 189, while the second (T : C at nucleotide 1253 of KCNA7) in the C-terminus changes methionine to threonine at codon 418. The allele frequencies for these SNPs, determined in the general South African Afrikaner population, were 0.62 : 0.38, for the nucleotide 566 C : G polymorphism, respectively, and 0.7 : 0.3 for the nucleotide 1253 T : C polymorphism, respectively.
Since both the PFHBI and ICCD families exhibit evidence of the phenomenon of genetic anticipation,5,6 pathological expansion of a triplet repeat in affected individuals could contribute to the disease phenotype. It has previously been shown that an expansion of an unstable triplet repeat in either the coding or non-coding sequences of a gene could be responsible for genetic disorders exhibiting anticipation.17 In silico searches encompassing the 5′ UTR, 3′ UTR and coding regions of KCNA7 in AC008687 sequences did not, however, detect any triplet repeats. The HRC gene is another plausible positional candidate for the PFHB1 locus by virtue of its expression in the cardiac luminal sarcoplasmic reticulum, where it may play a role in calcium ion homeostasis.4.6 Ongoing investigations have revealed that pathological expansion of two triplet repeats (GAT)n and (GAG)n within exon 1 of this gene have been excluded as a possible cause of PFHB1 (our unpublished results) and of ICCD.6
In conclusion, despite the evidence that KCNA7 maps within the PFHB1 locus, is expressed in heart and has IKur channel properties, no pathogenic mutations were identified in the coding region or splice sites that could be shown to be causative of this cardiac conduction disorder. Nevertheless, the dataset reported here on characterisation of human KCNA7, provision of the corrected mouse cDNA sequence and the mouse channel's electrophysiological properties provide an important resource for investigations involving this attractive candidate gene in other genetic cardiac conditions, such as ICCD.
While this report was under review, another report on the characterisation of KCNA7 was published (Kashuba V, Kvasha S, Protopopov A et al: Initial isolation and analysis of the human Kul.7 (KCNA7) gene, a member of the voltage-gated potassium channel gene family. Gene 2001; 268: 115–122.
References
Vatta M, Li H, Towbin JA . Molecular biology of arrhythmic syndromes Curr Opin Cardiol 2000 15: 12–22
Schott J-J, Alshinawi C, Kyndt F et al. Cardiac conduction defects associate with mutations in SCN5A Nature Genet 1999 23: 20–21
Tan HL, Bink-Boelkens MTE, Bezzina CR et al. A sodium-channel mutation causes isolated cardiac conduction disease Nature 2001 409: 1043–1047
Brink PA, Ferreira A, Moolman JC, Weymar HW, van der Merwe P-L, Corfield VA . Gene for progressive familial heart block type I maps to chromosome 19q13 Circulation 1995 91: 1633–1640
Brink AJ, Torrington M . Progressive familial heart block – two types S Afr Med J 1977 52: 53–59
de Meeus A, Stephan E, Debrus S et al. An isolated cardiac conduction disease maps to chromosome 19q Circ Res 1995 77: 735–740
Kalman K, Nguyen A, Tseng-Crank J et al. Genomic organization, chromosomal localization, tissue distribution, and biophysical characterization of a novel mammalian Shaker-related voltage-gated potassium channel, Kv1.7 J. Biol. Chem 1998 273: 5851–5857
Chandy KG, Gutman GA (eds). Voltage-gated K+ channel genes. Ligand and voltage-gated ion channels Handbook of Receptors and Channels 1994 Boca Raton: CRC Press pp 1–76
Chomczynski P, Sacchi N . Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction Anal Biochem 1987 162: 156–159
Orita M, Suzuki Y, Sekiya T, Hayashi K . Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction Genomics 1989 5: 874–879
Chandy K G, Williams C, Spencer RH et al. A family of three mouse K+ channel genes with intronless coding regions Science 1990 247: 973
Papazian DM, Schwarz TL, Tempel BL, Jan YN, Jan LY . Cloning of genomic and complementary DNA from Shaker, a putative potassium channel gene from Drosophila Science 1987 237: 749–753
Fedida D, Wible B, Wang Z et al. Identity of a novel delayed rectifier current from human heart with a cloned K+ channel current Circ. Res 1993 73: 210–216
Feng J, Xu D, Wang Z, Nattel S . Ultrarapid delayed rectifier current inactivation in human atrial myocytes: properties and consequences Am. J. Physiol 1998 275: H1717–H1725
Rampe D, Wible B, Fedida D, Dage RC, Brown AM . Verapamil blocks a rapidly activating delayed rectifier K+ channel cloned from the human heart Mol. Pharmacol 1993 44: 642–648
Feng J, Wible B, Li GR, Wang Z, Nattel S . Antisense oligodeoxynucleotides directed against Kv1.5 mRNA specifically inhibit ultrarapid delayed rectifier K+ currents in cultured adult human atrial myocytes Circ. Res 1997 80: 572–579
Mandel JL . Human genetics: Breaking the rule of three Nature 1997 384: 874–879
Acknowledgements
We acknowledge Dr Hanlie Moolman-Smook for heart-derived RNA samples and Dr Alan Christoffels for assistance with the bioinformatic analyses. This research was supported in part by the South African Medical Research Council, by a Wellcome Trust Research leave Fellowship (Z Arieff) and grant MH59222 to G Chandy. Sequence data from this article have been deposited with GenBank Data Libraries under Accession No. AF315818 and NCBI's dbSNP under Accession Nos. AF315818-566 and AF315818-1253.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bardien-Kruger, S., Wulff, H., Arieff, Z. et al. Characterisation of the human voltage-gated potassium channel gene, KCNA7, a candidate gene for inherited cardiac disorders, and its exclusion as cause of progressive familial heart block I (PFHBI). Eur J Hum Genet 10, 36–43 (2002). https://doi.org/10.1038/sj.ejhg.5200739
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5200739
Keywords
This article is cited by
-
Computational prognostic evaluation of Alzheimer’s drugs from FDA-approved database through structural conformational dynamics and drug repositioning approaches
Scientific Reports (2023)
-
Post-operative atrial fibrillation examined using whole-genome RNA sequencing in human left atrial tissue
BMC Medical Genomics (2017)
-
Investigation of Pathogenic Genes in Chinese sporadic Hypertrophic Cardiomyopathy Patients by Whole Exome Sequencing
Scientific Reports (2015)
-
A Quantitative Trait Locus for SBP Maps Near KCNB1 and PTGIS in a Population Isolate
American Journal of Hypertension (2009)
-
Novel truncated isoform of SK3 potassium channel is a potent dominant-negative regulator of SK currents: implications in schizophrenia
Molecular Psychiatry (2003)