Abstract
Photodeoxygenation of dibenzothiophene S-oxide and its derivatives have been used to generate atomic oxygen [O(3P)] to examine its effect on proteins, nucleic acids, and lipids. The unique reactivity and selectivity of O(3P) have shown distinct oxidation products and outcomes in biomolecules and cell-based studies. To understand the scope of its global impact on the cell, we treated MDA-MB-231 cells with 2,8-diacetoxymethyldibenzothiophene S-oxide and UV-A light to produce O(3P) without targeting a specific cell organelle. Cellular responses to O(3P)-release were analyzed using cell viability and cell cycle phase determination assays. Cell death was observed when cells were treated with higher concentrations of sulfoxides and UV-A light. However, significant differences in cell cycle phases due to UV-A irradiation of the sulfoxide were not observed. We further performed RNA-Seq analysis to study the underlying biological processes at play, and while UV-irradiation itself influenced gene expression, there were 9 upregulated and 8 downregulated genes that could be attributed to photodeoxygenation.
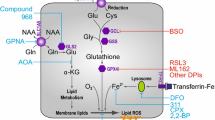
Graphical abstract








Similar content being viewed by others
Notes
Data for VC_UV and VC_No UV have been presented in ref [25]
References
Bucher, G., & Scaiano, J. C. (1994). Laser flash photolysis of pyridine N-Oxide: Kinetic studies of atomic oxygen [O(3P)] in solution. Journal of Physical Chemistry, 98(1), 12411–12413.
Lucien, E., & Greer, A. (2001). Electrophilic oxidant produced in the photodeoxygenation of 1,2-benzodiphenylene sulfoxide. Journal of Organic Chemistry, 66(13), 4576–4579. https://doi.org/10.1021/jo010009z
Gregory, D. D., Wan, Z., & Jenks, W. S. (1997). Photodeoxygenation of dibenzothiophene sulfoxide: Evidence for a unimolecular S-O cleavage mechanism. Journal of the American Chemical Society, 119(1), 94–102. https://doi.org/10.1021/ja962975i
Thomas, K. B., & Greer, A. (2003). Gauging the significance of atomic oxygen [O(3P] in sulfoxide photochemistry. A method for hydrocarbon oxidation. Journal of Organic Chemistry, 68(5), 1886–1891. https://doi.org/10.1021/jo0266487
Omlid, S. M., Zhang, M., Isor, A., & McCulla, R. D. (2017). Thiol reactivity toward atomic oxygen generated during the photodeoxygenation of dibenzothiophene S-oxide. Journal of Organic Chemistry, 82(24), 13333–13341. https://doi.org/10.1021/acs.joc.7b02428
Rastogi, R. P., Richa, K., Kumar, A., Tyagi, M. B., & Sinha, R. P. (2010). Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. Journal of Nucleic Acids, 2010, 592980. https://doi.org/10.4061/2010/592980
Pattison, D. I., Rahmanto, A. S., & Davies, M. J. (2012). Photo-oxidation of proteins. Photochemical and Photobiological Sciences, 11, 38–53. https://doi.org/10.1039/c1pp05164d
Roy, S. (2017). Impact of UV radiation on genome stability and human health. Advances in Experimental Medicine and Biology, 996, 207–219. https://doi.org/10.1007/978-3-319-56017-5_17
Zheng, X., Baumann, S. M., Chintala, S. M., Galloway, K. D., Slaughter, J. B., & McCulla, R. D. (2016). Photodeoxygenation of dinaphthothiophene, benzophenanthrothiophene, and benzonaphthothiophene: S-oxides. Photochemical and Photobiological Sciences, 15(6), 791–800. https://doi.org/10.1039/c5pp00466g
Chintala, S. M., Petroff, J. T., Barnes, A., & McCulla, R. D. (2019). Photodeoxygenation of phenanthro[4,5-bcd]thiophene S-oxide, triphenyleno[1,12-bcd]thiophene S-oxide and perylo[1,12-bcd]thiophene S-oxide. Journal of Sulfur Chemistry, 40(5), 503–515. https://doi.org/10.1080/17415993.2019.1615065
Korang, J., Grither, W. R., & McCulla, R. D. (2010). Photodeoxygenation of dibenzothiophene S-oxide derivatives in aqueous media. Journal of the American Chemical Society, 132(12), 4466–4476. https://doi.org/10.1021/ja100147b
Omlid, S., Isor, A., Sulkowski, K., Chintala, S., Petroff, J., & McCulla, R. (2018). Synthesis of aromatic disulfonic acids for water-soluble dibenzothiophene derivatives. Synthesis, 50(12), 2359–2366. https://doi.org/10.1055/s-0036-1591969
Isor, A., Chartier, B. V., Abo, M., Currens, E. R., Weerapana, E., & McCulla, R. D. (2021). Identifying cysteine residues susceptible to oxidation by photoactivatable atomic oxygen precursors using a proteome-wide analysis. RSC Chemical Biology, 2(2), 577–591. https://doi.org/10.1039/d0cb00200c
Zhang, M., Ravilious, G. E., Hicks, L. M., Jez, J. M., & McCulla, R. D. (2012). Redox switching of adenosine-5′-phosphosulfate kinase with photoactivatable atomic oxygen precursors. Journal of the American Chemical Society, 134(41), 16979–16982. https://doi.org/10.1021/ja3078522
Korang, J., Emahi, I., Grither, W. R., Baumann, S. M., Baum, D. A., & McCulla, R. D. (2013). Photoinduced DNA cleavage by atomic oxygen precursors in aqueous solutions. RSC Advances, 3(30), 12390–12397. https://doi.org/10.1039/c3ra41597j
Bourdillon, M. T., Ford, B. A., Knulty, A. T., Gray, C. N., Zhang, M., Ford, D. A., & McCulla, R. D. (2014). Oxidation of plasmalogen, low-density lipoprotein and raw 264.7 cells by photoactivatable atomic oxygen precursors. Photochemistry and Photobiology, 90(2), 386–393. https://doi.org/10.1111/php.12201
Petroff, J. T., Isor, A., Chintala, S. M., Albert, C. J., Franke, J. D., Weinstein, D., Omlid, S. M., Arnatt, C. K., Ford, D. A., & McCulla, R. D. (2020). In vitro oxidations of low-density lipoprotein and RAW 264.7 cells with lipophilic O(3P)-precursors. RSC Advances, 10(44), 26553–26565. https://doi.org/10.1039/D0RA01517B
Patrick, G. L. (2013). Prodrugs to improve membrane permeability. An introduction to medicinal chemistry (5th ed., p. 259). Oxford University Press.
Kalyanaraman, B., Cheng, G., Hardy, M., Ouari, O., Bennett, B., & Zielonka, J. (2018). Teaching the basics of reactive oxygen species and their relevance to cancer biology: Mitochondrial reactive oxygen species detection, redox signaling, and targeted therapies. Redox Biology, 15, 347–362. https://doi.org/10.1016/j.redox.2017.12.012
Fleury, C., Mignotte, B., & Vayssière, J. L. (2002). Mitochondrial reactive oxygen species in cell death signaling. Biochimie, 84(2–3), 131–141. https://doi.org/10.1016/S0300-9084(02)01369-X
Redza-Dutordoir, M., & Averill-Bates, D. A. (2016). Activation of apoptosis signalling pathways by reactive oxygen species. Biochimica et Biophysica Acta, 1863(12), 2977–2992. https://doi.org/10.1016/j.bbamcr.2016.09.012
Circu, M. L., & Aw, T. Y. (2010). Reactive oxygen species, cellular redox systems, and apoptosis. Free Radical Biology and Medicine, 48(6), 749–762. https://doi.org/10.1016/j.freeradbiomed.2009.12.022
Petroff, J. T., Skubic, K. N., Arnatt, C. K., & McCulla, R. D. (2018). Asymmetric dibenzothiophene sulfones as fluorescent nuclear stains. Journal of Organic Chemistry, 83(22), 14063–14068. https://doi.org/10.1021/acs.joc.8b01931
Petroff, J. T., Grady, S., Arnatt, C. K., & McCulla, R. D. (2020). Dibenzothiophene sulfone derivatives as plasma membrane dyes. Photochemistry and Photobiology, 96(1), 67–73. https://doi.org/10.1111/php.13175
Isor, A., O’Dea, A. T., Petroff, J. T., Skubic, K. N., Grady, S. F., Arnatt, C. K., & McCulla, R. D. (2020). Synthesis of triphenylphosphonium dibenzothiophene S-oxide derivatives and their effect on cell cycle as photodeoxygenation-based cytotoxic agents. Bioorganic Chemistry, 105(2020), 104442–104454. https://doi.org/10.1016/j.bioorg.2020.104442
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B., Tinevez, J. Y., White, D. J., Hartenstein, V., Eliceiri, K., Tomancak, P., & Cardona, A. (2012). Fiji: An open-source platform for biological-image analysis. Nature Methods, 9(7), 676–682. https://doi.org/10.1038/nmeth.2019
Callegari, A. J., & Kelly, T. J. (2007). Shedding light on the DNA damage checkpoint. Cell Cycle, 6(6), 660–666. https://doi.org/10.4161/cc.6.6.3984
R Core Team. (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria URL http://www.R-projectorg/
Kolde, R. (2015). Pheatmap : Pretty Heatmaps. R package version 1.0.12. http://cran.r-project.org/web/packages/pheatmap/index.html.
Online Mendelian Inheritance in Man, OMIM. (2021). McKusick–Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, MD) and National Center for Biotechnology Information, National Library of Medicine (Bethesda, MD). https://omim.org/.
Png, K. J., Halberg, N., Yoshida, M., & Tavazoie, S. F. (2012). A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature, 481(7380), 190–194. https://doi.org/10.1038/nature10661
Polesskaya, A., Cuvellier, S., Naguibneva, I., Duquet, A., Moss, E. G., & Harel-Bellan, A. (2007). Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes and Development, 21(9), 1125–1138. https://doi.org/10.1101/gad.415007
Yu, J., Vodyanik, M. A., Smuga-Otto, K., Antosiewicz-Bourget, J., Frane, J. L., Tian, S., Nie, J., Jonsdottir, G. A., Ruotti, V., Stewart, R., Slukvin, I. I., & Thomson, J. A. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science, 318(5858), 1917–1920. https://doi.org/10.1126/science.1151526
Takefuji, M., Asano, H., Mori, K., Amano, M., Kato, K., Watanabe, T., Morita, Y., Katsumi, A., Itoh, T., Takenawa, T., Hirashiki, A., Izawa, H., Nagata, K., Hirayama, H., Takatsu, F., Naoe, T., Yokota, M., & Kaibuchi, K. (2010). Mutation of ARHGAP9 in patients with coronary spastic angina. Journal of Human Genetics, 55(1), 42–49. https://doi.org/10.1038/jhg.2009.120
Furukawa, Y., Kawasoe, T., Daigo, Y., Nishiwaki, T., Ishiguro, H., Takahashi, M., Kitayama, J., & Nakamura, Y. (2001). Isolation of a novel human gene, ARHGAP9, encoding a Rho-GTPase activating protein. Biochemical and Biophysical Research Communications, 284(3), 643–649. https://doi.org/10.1006/bbrc.2001.5022
Blighe, K., Rana, S., & Lewis, M. (2020). Enhanced volcano: Publication-ready volcano plots with enhanced colouring and labeling. version 1.12.0. https://github.com/kevinblighe/EnhancedVolcano.
Gentleman, R. C., Carey, V. J., Bates, D. M., Bolstad, B., Dettling, M., Dudoit, S., Ellis, B., Gautier, L., Ge, Y., Gentry, J., Hornik, K., Hothorn, T., Huber, W., Iacus, S., Irizarry, R., Leisch, F., Li, C., Maechler, M., Rossini, A. J., … Zhang, J. (2004). Bioconductor: Open software development for computational biology and bioinformatics. Genome Biology, 5(10), R80. https://doi.org/10.1186/gb-2004-5-10-r80
Chen, J., Bardes, E. E., Aronow, B. J., & Jegga, A. G. (2009). ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Research, 37, W305–W311. https://doi.org/10.1093/nar/gkp427
Chen, J., Aronow, B. J., & Jegga, A. G. (2009). Disease candidate gene identification and prioritization using protein interaction networks. BMC Bioinformatics, 10(1), 73. https://doi.org/10.1186/1471-2105-10-73
Chen, J., Xu, H., Aronow, B. J., & Jegga, A. G. (2007). Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinformatics, 8(1), 392. https://doi.org/10.1186/1471-2105-8-392
Lyall, R., Nikoloski, Z., & Gechev, T. (2020). Comparative analysis of ROS network genes in extremophile eukaryotes. International Journal of Molecular Sciences. https://doi.org/10.3390/ijms21239131
Carbon, S., Ireland, A., Mungall, C. J., Shu, S., Marshall, B., & Lewis, S. (2009). AmiGO: Online access to ontology and annotation data. Bioinformatics, 25(2), 288–289. https://doi.org/10.1093/bioinformatics/btn615
Riss, T. L., Moravec, R. A., Niles, A. L., Duellman, S., Benink, H. A., Worzella, T. J., & Minor, L., et al. (2004). Cell viability assays. In S. Markossian, A. Grossman, & K. Brimacombe (Eds.), Assay guidance manual. Eli Lilly and Company and the National Center for Advancing Translational Sciences.
Edgar, R. (2002). Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research, 30(1), 207–210. https://doi.org/10.1093/nar/30.1.207
Barrett, T., Wilhite, S. E., Ledoux, P., Evangelista, C., Kim, I. F., Tomashevsky, M., Marshall, K. A., Phillippy, K. H., Sherman, P. M., Holko, M., Yefanov, A., Lee, H., Zhang, N., Robertson, C. L., Serova, N., Davis, S., & Soboleva, A. (2013). NCBI GEO: Archive for functional genomics data sets-Update. Nucleic Acids Research, 41, D991–D995. https://doi.org/10.1093/nar/gks1193
Huber, W., Carey, V. J., Gentleman, R., Anders, S., Carlson, M., Carvalho, B. S., Bravo, H. C., Davis, S., Gatto, L., Girke, T., Gottardo, R., Hahne, F., Hansen, K. D., Irizarry, R. A., Lawrence, M., Love, M. I., MacDonald, J., Obenchain, V., Oles, A. K., … Morgan, M. (2015). Orchestrating high-throughput genomic analysis with Bioconductor. Nature Methods, 12(2), 115–121. https://doi.org/10.1038/nmeth.3252
Kanehisa, M., & Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research, 28(1), 27–30. https://doi.org/10.1093/nar/28.1.27
Kanehisa, M. (2019). Toward understanding the origin and evolution of cellular organisms. Protein Science, 28(11), 1947–1951. https://doi.org/10.1002/pro.3715
Kanehisa, M., Furumichi, M., Sato, Y., Ishiguro-Watanabe, M., & Tanabe, M. (2021). KEGG: Integrating viruses and cellular organisms. Nucleic Acids Research, 49(D1), D545–D551. https://doi.org/10.1093/nar/gkaa970
Nishimura, D. (2001). BioCarta. Biotech Software and Internet Report, 2(3), 117–120. https://doi.org/10.1089/152791601750294344
Karp, P. D., Billington, R., Caspi, R., Fulcher, C. A., Latendresse, M., Kothari, A., Keseler, I. M., Krummenacker, M., Midford, P. E., Ong, Q., Ong, W. K., Paley, S. M., & Subhraveti, P. (2019). The BioCyc collection of microbial genomes and metabolic pathways. Briefings in Bioinformatics, 20(4), 1085–1093. https://doi.org/10.1093/bib/bbx085
Fabregat, A., Korninger, F., Viteri, G., Sidiropoulos, K., Marin-Garcia, P., Ping, P., Wu, G., Stein, L., D’Eustachio, P., & Hermjakob, H. (2018). Reactome graph database: Efficient access to complex pathway data. PLoS Computational Biology, 14(1), e1005968. https://doi.org/10.1371/journal.pcbi.1005968
Fabregat, A., Sidiropoulos, K., Viteri, G., Forner, O., Marin-Garcia, P., Arnau, V., D’Eustachio, P., Stein, L., & Hermjakob, H. (2017). Reactome pathway analysis: A high-performance in-memory approach. BMC Bioinformatics, 18(1), 142. https://doi.org/10.1186/s12859-017-1559-2
Fabregat, A., Sidiropoulos, K., Viteri, G., Marin-Garcia, P., Ping, P., Stein, L., D’Eustachio, P., & Hermjakob, H. (2018). Reactome diagram viewer: Data structures and strategies to boost performance. Bioinformatics, 34(7), 1208–1214. https://doi.org/10.1093/bioinformatics/btx752
Jassal, B., Matthews, L., Viteri, G., Gong, C., Lorente, P., Fabregat, A., Sidiropoulos, K., Cook, J., Gillespie, M., Haw, R., Loney, F., May, B., Milacic, M., Rothfels, K., Sevilla, C., Shamovsky, V., Shorser, S., Varusai, T., Weiser, J., … D’Eustachio, P. (2020). The reactome pathway knowledgebase. Nucleic Acids Research, 48(D1), D498–D503. https://doi.org/10.1093/nar/gkz1031
Sidiropoulos, K., Viteri, G., Sevilla, C., Jupe, S., Webber, M., Orlic-Milacic, M., Jassal, B., May, B., Shamovsky, V., Duenas, C., Rothfels, K., Matthews, L., Song, H., Stein, L., Haw, R., D’Eustachio, P., Ping, P., Hermjakob, H., & Fabregat, A. (2017). Reactome enhanced pathway visualization. Bioinformatics, 33(21), 3461–3467. https://doi.org/10.1093/bioinformatics/btx441
Wu, G., & Haw, R. (2017). Functional interaction network construction and analysis for disease discovery. Methods in Molecular Biology, 1558, 235–253. https://doi.org/10.1007/978-1-4939-6783-4_11
Dahlquist, K. D., Salomonis, N., Vranizan, K., Lawlor, S. C., & Conklin, B. R. (2002). GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nature Genetics, 31(1), 19–20. https://doi.org/10.1038/ng0502-19
Liberzon, A., Birger, C., Thorvaldsdottir, H., Ghandi, M., Mesirov, J. P., & Tamayo, P. (2015). The molecular signatures database (MSigDB) hallmark gene set collection. Cell Systems, 1(6), 417–425. https://doi.org/10.1016/j.cels.2015.12.004
Liberzon, A., Subramanian, A., Pinchback, R., Thorvaldsdottir, H., Tamayo, P., & Mesirov, J. P. (2011). Molecular signatures database (MSigDB) 3.0. Bioinformatics, 27(12), 1739–1740. https://doi.org/10.1093/bioinformatics/btr260
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., Paulovich, A., Pomeroy, S. L., Golub, T. R., Lander, E. S., & Mesirov, J. P. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences, 102(43), 15545–15550. https://doi.org/10.1073/pnas.0506580102
Petri, V., Shimoyama, M., Hayman, G. T., Smith, J. R., Tutaj, M., de Pons, J., et al. (2011). The rat genome database pathway portal. Database. https://doi.org/10.1093/database/bar010Suns.
Acknowledgements
This work was supported by the National Science Foundation CHE-1900417.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There are no conflicts to declare.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Isor, A., O’Dea, A.T., Grady, S.F. et al. Effects of photodeoxygenation on cell biology using dibenzothiophene S-oxide derivatives as O(3P)-precursors. Photochem Photobiol Sci 20, 1621–1633 (2021). https://doi.org/10.1007/s43630-021-00136-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43630-021-00136-5