Abstract
Coronaviruses pose a serious threat to public health. Tremendous efforts are dedicated to advance reliable and effective detection of coronaviruses. Currently, the coronavirus disease 2019 (COVID-19) diagnosis mainly relies on the detection of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genetic materials by using reverse transcription-polymerase chain reaction (RT-PCR) assay. However, simpler and more rapid and reliable alternatives are needed to meet high demand during the pandemic. Biosensor-based diagnosis approaches become alternatives for selectively and rapidly detecting virus particles because of their biorecognition elements consisting of biomaterials that are specific to virus biomarkers. Here, we summarize biorecognition materials, including antibodies and antibody-like molecules, that are designed to recognize SARS-CoV-2 biomarkers and the advances of recently developed biosensors for COVID-19 diagnosis. The design of biorecognition materials or layers is crucial to maximize biosensing performances, such as high selectivity and sensitivity of virus detection. Additionally, the recent representative achievements in developing bioelectronics for sensing coronavirus are included. This review includes scholarly articles, mainly published in 2020 and early 2021. In addition to capturing the fast development in the fields of applied materials and biodiagnosis, the outlook of this rapidly evolving technology is summarized. Early diagnosis of COVID-19 could help prevent the spread of this contagious disease and provide significant information to medical teams to treat patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The coronavirus disease 2019 (COVID-19), which is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has been a major global threat since late 2019. COVID-19 has been declared as a critical pandemic in 2020. As of early 2021, the report from www.worldometers.info/coronavirus/ shows that over 95,000,000 cases of COVID-19 have been confirmed around the world. 2,032,634 deaths were reported, attributable to SARS-CoV-2 infection (as of January 17, 2021) [1]. Reportedly, 40% to 45% of people infected with SARS-CoV-2 have no symptoms of COVID-19 [2,3,4]. This could allow the silent spread of the virus that causes uncontrolled transmission of SARS-CoV-2 throughout the globe [5]. Apart from asymptomatic patients, COVID-19 patients have a large variety of symptoms, leading to the difficulty to determine the infection [6]. Undoubtedly, researchers need to explore fast, accurate, and cost-effective approaches, such as biosensors, to detect viruses [7,8,9].
Early diagnosis of COVID-19 can help prevent the spread of this contagious disease. Thus, identifying viral biological markers (i.e., viral genetic material, viral proteins, or host immune responses to infection) and discovering their biorecognition molecules, which specifically bind to those markers, are important to enabling the development of a variety of methods for SARS-CoV-2 detections either in the laboratory setting or the point-of-care (POC) testing [10, 11]. The COVID-19 outbreak pushes the development of biosensors and bioelectronics forward, even more, to the advanced diagnosis technology for detecting viruses [12]. Although the nucleic acid amplification tests (NAATs), such as the real-time reverse transcription-polymerase chain reaction (RT-PCR), are recommended by the World Health Organization (WHO) as a standard approach to detect unique SARS-CoV-2 genome [13], this PCR-based method has some limitations; for example, the cost for instrumentation and operation limits the affordability. Rapid and accurate diagnosis is ideal for public strategic requirements to control the COVID-19 crisis [14, 15]. In addition, RT-PCR relies on special analysts and centralized laboratory in the hospital. Inevitably, this challenge stimulates researchers to develop new alternative strategies.
Advanced materials enable the design of high-performance biosensors and bioelectronics. For example, nanomaterials, such as graphene and gold nanoparticles, provide a large active surface-to-volume ratio, enhancing the efficient immobilization and conductivity. Utilizing advanced materials allows the development of nano/biosensors with high sensitivity and other preferable analytical performances for the detection of biochemicals in clinical applications, offering new alternatives to benchtop-based complex instruments [16,17,18]. Therefore, a broad range of advanced functional materials is applied to support the fabrication of SARS-CoV-2 sensing systems. A main group of the analyte indicating the presence of viruses is the genome (target). The preconcentration of targets is almost always mandatory in order to achieve the limit of detection of the developed protocols. An example of favored strategies is using genome amplification [19, 20]. Apart from the genome target, protein-based biomarkers are also important targets used to indicate SARS-CoV-2 infection. Compared with targeting genetic materials from viruses, the main limitation of detecting non-genetic targets (such as proteins or antibodies) is an extremely low concentration. Some analytical challenges here are considered. Challenges are peculiar to trace bioanalysis when dealing with low concentrations of targets and small volumes of samples. If we can detect viral biomarkers at the early stage of SARS-CoV-2 infection, which usually presents in ultralow concentrations of viral targets, by using fast and reliable approaches, it is essential to strengthening the way to slow down the spreading of viruses or, importantly, save the life of the patient effectively.
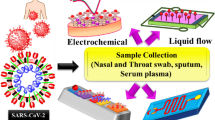
In this review, we aim to describe the current advances of biosensor platforms for COVID-19 diagnosis (Fig. 1). Only articles written in English are considered. In addition to several published review articles [21, 22], we expand our discussion focusing on biological recognition elements and bioelectronics. Starting with the basic biological structure and cell entry mechanism of SARS-CoV-2, we summarize the possible antigens and biomarkers, such as viral ribonucleic acid (RNA) and proteins, or human immune response to the infection, which can be used to identify the presence of SARS-CoV-2 in humans. Subsequently, we introduce the biorecognition elements, such as nucleic acid probes, antibodies, and antibody-like molecules, that have been applied to specifically bind to those viral antigens and biomarkers. Although antibodies are primarily used for the detection of viral proteins, there is the possibility of using antibody-like molecules. The advantages of antibody-like molecules over antibodies are described. Moreover, we demonstrate the current advancement of using biorecognition elements for diagnosis of COVID-19 from patient samples. Lastly, we show recent examples of the integration between viral biomarkers and the advent of biorecognition elements in biosensors based on electrochemical bioelectronics to rapidly detect SARS-CoV-2 in patient samples.
Illustrations of the SARS-CoV-2 and the concept of using biosensing technologies for effective COVID-19 diagnosis, including a sampling, b coronavirus structure, c biodiagnostic tools, and d modern clinical systems, which seeks to deliver important data and allows fast and efficient managements of the individual and public health
2 The structure of SARS-CoV-2 and entering mechanism
Understanding the pathogenesis of SARS-CoV-2 could lead to appropriate diagnosis and therapeutics. The novel coronavirus, SARS-CoV-2, is closely related to betacoronavirus [23, 24] that had caused two outbreaks at the beginning of the twenty-first century, e.g., severe acute respiratory syndrome (SARS) caused by SARS-CoV in 2002 [25, 26] and Middle East respiratory syndrome (MERS) caused by MERS-CoV in 2012 [27, 28]. Similar to other coronaviruses, SARS-CoV-2, which is a positive-sense single-stranded RNA (+ssRNA) virus, shares ~80% and 50% genome sequence identity with SARS-CoV and MERS-CoV, respectively, and its genome consists of ~30,000 base pairs (bp) in length encoding four main structural proteins, e.g., spike (S), nucleocapsid (N), membrane (M), envelop (E), nonstructural proteins, and accessory proteins (as shown in Fig. 2a) [29,30,31,32]. Among these proteins, the sequence of the SARS-CoV-2 S protein is more diverse with about 77% identity to SARS-CoV, while other proteins are highly evolutionary conserved [33]. Therefore, distinct domains within the S protein are used by different coronaviruses to recognize a variety of attachments and entry receptors [34].
The structure and cell entry mechanism of SARS-CoV-2. a Four structural proteins of SARS-CoV-2 include spike (S), nucleocapsid (N), membrane (M), and envelop (E) proteins. To enter host cells, the SARS-CoV-2 S protein binds to host angiotensin-converting enzyme 2 (ACE2) receptors (shown in red). Subsequently, the surface protease, e.g., transmembrane protease serine 2 (TMPRSS2, shown in purple), cleaves at the S1/S2 boundary which triggers a series of conformational changes that lead to fusion between the viral envelope and the target host cell membrane. b Schematic of the SARS-CoV-2 S protein. The total length is 1273 amino acids which consist of a signal peptide (residue: 1–13) located at the N-terminus, the S1 subunit (residue: 14–685, shown in yellow), and the S2 subunit (residue: 686–1273, shown in green). In the S1 subunit, there are an N-terminal domain (residue: 14–305) and a receptor-binding domain (RBD; residue: 319–541, highlighted in orange). In the S2 subunit, there are the fusion peptide (FP; residue: 788–806), heptapeptide repeat sequence 1 and 2 (residue: 912–984 for HR1, and 1163–1213 for HR2), transmembrane domain (TM; residue: 1213–1237), and cytoplasm domain (residue: 1237–1273). Using the RBD, the trimeric spike molecule binds to ACE2
The engagement between the SARS-CoV-2 S protein and a host cell receptor initiates virus entering the host cell. The SARS-CoV-2 S protein consists of 1273 amino acids (Fig. 2b), including a signal peptide (residues 1–13), an N-terminal subunit (S1; residues 14–685) that mediates receptor binding, and a C-terminal subunit (S2; residues 686–1273) that mediates fusion between virus and the membrane of the host cell [31, 35] [36]. For viral attachment to the surface of host cells, the receptor-binding domain (RBD; residues 319–541) of the S1 subunit binds to the host angiotensin-converting enzyme 2 (ACE2) receptor with high affinity (dissociation constant, KD = 14.7 nM [37]). For viral entry, with facilitating of the host protease, e.g., transmembrane serine protease 2 (TMPRSS2) on the surface of host cells for the S protein priming, it cleaves the boundary between S1 and S2 that triggers a conformational change in the S2 subunit and allows the fusion peptide (residues 788–806) of the S2 subunit to fuse to the host cells, resulting in the release of the viral genome into the host cells [38, 39]. The SARS-CoV-2 genetic materials then utilize the molecular machinery of the host cell to replicate themselves and release the new virus particles into the host organism. Understanding the structure and function of SARS-CoV-2 allows researchers to design the effective approaches for reliable and rapid diagnosis of COVID-19 and to develop the potential treatments to meet the high demand during the pandemic.
3 SARS-CoV-2 biomarkers and biorecognition elements
Several viral antigens and biomarkers are identified to be used as an indicator for SARS-CoV-2 infection [40]. Generally, the amount of SARS-CoV-2 presenting in patients depends on the window period and different types of patient samples, including nasopharyngeal swabs, sputum, urine, or stool. The mean incubation period for COVID-19 is typically 5–6 days, and the infectiousness could start about 11–12 days before symptom onset and peaked at onset [41,42,43]. To diagnose COVID-19, several samples from patients are collected to extract the viral genetic materials which are later determined by RT-PCR. Particularly, the viral loads in nasopharyngeal swabs and sputum samples reached the maximum level at around 5–6 days after symptom onset (~104 to 107 copies mL–1) [44]. Currently, the molecular diagnostics is the key methods that have been applied to detect viral biological markers, including viral genetic material (i.e., RNA) or viral proteins (i.e., S or N proteins) in patient samples [45]. The viral genetic material is typically targeted because it can be amplified to enhance the detection sensitivity. Conversely, viral proteins are difficult for direct detection because of their trace amount [46]. In addition to viral biological markers, the host immune responses to infection are another indicator for COVID-19 prognosis [47]. Thus, detecting viral RNA, proteins, or human immunoglobulins with biorecognition elements (e.g., antibodies, antibody-like molecules, and nucleic acid probes) that specifically bind to those biomarkers enables the possibility to develop a variety of methods for SARS-CoV-2 detection [16]. The viral biomarkers with their recognition molecules are discussed in the following section.
For the SARS-CoV-2 RNA, it is currently used as a major biomarker for COVID-19 diagnosis, and its standard recognition molecule is nucleic acid probes. Due to the advent of next-generation sequencing (NGS) and bioinformatics, the whole genome of SARS-CoV-2 has been identified that enables the design of the SARS-CoV-2-specific primers (short nucleic acid sequence) to amplify a unique sequence of SARS-CoV-2 genome using RT-PCR [31]. The common targets of the SARS-CoV-2 genome include the genes encoding the E, N, and S proteins, the open reading frame 1ab, and the RNA-dependent RNA polymerase (RdRP) gene, of which each of them differently affects the specificity of RT-PCR [46, 48]. For example, the S gene and RdRP gene are commonly used to differentiate SARS-CoV-2 from other coronaviruses, while the E and N genes are used to identify coronaviruses because these genes are more conserved among coronaviruses that may have cross-reaction [31, 44]. Consequently, the whole-genome sequences of SARS-CoV-2 allow researchers to design specific probes to target and amplify viral genetic material in the RT-PCR process.
For the SARS-CoV-2 proteins, the N and S proteins are typically used as the antigens or biomarkers for viral detection, and their biorecognition molecules can be either antibodies or antibody-like molecules. The N protein, which is a multifunction RNA-binding protein involved in viral RNA replication and assembly, expresses in high levels in infected cells at the early stage of SARS-CoV-2 infection [49]. Moreover, the S protein, which is a large type-I transmembrane glycoprotein covering the surface of SARS-CoV-2, binds to host ACE2 to facilitate the viral entry into target cells [35, 50]. Targeting either N or S protein by biorecognition molecules could be useful to developing diagnostic systems. For example, monoclonal antibodies (mAbs, Fig. 3a) have been mainly used to recognize antigens, and other antibody-like molecules (see Fig. 3b–f), such as antigen-binding fragment (Fab), single-chain variable fragment (scFv), single-domain antibody (nanobody), and monobody, can also bind to antigens. The binding specificity and affinity between antibodies or antibody-like molecules and target antigens can be improved through protein engineering techniques which consist of the following steps (Fig. 3g). Firstly, the library containing a diversity of variants is generated. In this step, the library of mAbs is isolated from B-cell samples of virus-infected patients, or the synthetic library of biorecognition molecules is diversified by mutagenesis. Secondly, these libraries are screened/selected against the target molecule through the display technology, especially the phage display that is widely used for in vitro selection [56]. Lastly, the function of selected variants is verified by a double-antibody sandwich enzyme-linked immunosorbent assay (ELISA) or affinity determination, before use. Thus, utilizing protein engineering techniques, the binding affinity of biorecognition molecules can be improved with high specificity against the viral markers. The examples of improving the binding of biorecognition molecules (antibody and antibody-like molecule) against viral proteins (e.g., N or S protein) are described in the following paragraph.
Antibodies and antibody-like molecules and schematic overview of their selection pipeline. The structure and size of selected biorecognition molecules. a Human immunoglobulin G (IgG, ~150 kDa) antibody consists of two heavy (blue) and two light (pink) chains connected by disulfide bonds (S–S). Both chains consist of several constant domains (CL and CH1–CH3) and the variable domains (VL and VH). The variable domains contain the complementarity-determining regions (CDRs), which determine antibody-binding specificity. b The antigen-binding fragment (Fab, ~55 kDa) is composed of each VH, CH1, VL, and CL domains. c The single-chain variable fragment (scFv, ~28 kDa) is composed of each VH and VL domains connected via a linker. d The camelid heavy-chain antibody (HcAbs, ~96 kDa) is made up of only two heavy chains; each of which consists of two constant (CH2 and CH3) domains and a single variable (VHH, ~12–15 kDa) domain. e Structure of the nanobody (e.g., vhhGFP4, PDB ID:3OGO; [51]). f Structure of the monobody (~10 kDa) (e.g., PDB ID: 1TTG; [52]). g Selection of biorecognition molecules using the phage display technology. (Top) For antibody-based scaffolds, an antibody library is generated using lymphocytes (B cells) isolated from either human or camelids (immunized by viral proteins). Subsequently, lymphocyte RNA encoding recognition protein binders is transcribed into cDNA by RT-PCR. A binder phage library is then created in the M13 phage that allows protein binders to be presented on the phage surface. The protein binder library undergoes a panning process [53]. Several cycles of panning are conducted for the selection of high-affinity binders. After that, all selected binders will be verified. (Bottom) For non-antibody-based scaffolds, synthetic peptide libraries are used, and the same procedure is repeated. In addition to the phage display, the yeast surface display can be applied to screen desired molecules [54]. a–d and g are adapted with permission from [55]. Copyright 2018, The Company of Biologists
Recently, several mAbs and antibody-like molecules have been produced to specifically target SARS-CoV-2 protein markers or antigens (Table 1). For instance, the anti-SARS-CoV-2 antibody library derived from peripheral blood samples of COVID-19 patients was generated by phage display technology and screened against the SARS-CoV-2 N protein. This led to JSo8 antibody with high binding affinity that can be used for a rapid diagnostic test kit for clinical use [57]. Although the N protein is highly conserved among coronaviruses and the sensitivity in detection of antibodies against the N protein is reported to be higher than the S protein for early detection of infection [64, 65], most of anti-SARS-CoV-2 antibodies have been developed to target the viral S protein as shown in Table 1. This is because the S protein is on the surface of virus and is involved in viral entry by binding to the host receptor; the S protein becomes the main target for biorecognition molecules. Moreover, anti-S-protein antibodies are used to defend a cell from virus infection by neutralizing its biological effect. This type of antibody is called neutralize antibody (nAb). For instance, nAb for SARS-CoV, such as CR3022, also has potential cross-neutralizing activity against SARS-CoV-2 infection due to the sequence identity of the S protein (77%) between SARS-CoV and SARS-CoV-2 [33, 61]. Despite of using an individual viral antigen, anti-SARS-CoV-2 antibodies can be selected using recombinant antigens, such as N, S, RBD, or ectodomain of the S protein [66, 67].
In addition to mAb, antibody-like molecules, such as nanobody (derived from camelid single-chain antibodies (VHH)) and monobody (derived from human fibronectin domain III), are an alternative binding protein against viral proteins because of their smaller size, being easier to be produced (in yeast or bacteria), and better binding affinity (KD in the range of pM–nM) than those of mAbs [68]. For example, a yeast surface-display library of synthetic nanobody sequences was constructed and screened against a mutant form of the SARS-CoV-2 S protein [39]. After several rounds of selection and optimization, a trivalent form of Nb6 binds to the S protein with femtomolar affinity. Another example of nanobody library is that it can be constructed from a llama immunized with the recombinant RBD, and this library was selected using a proteomic strategy to achieve anti-RBD nanobodies with picomolar to femtomolar affinity [62]. Apart from the nanobody, the monobody library can be synthetically constructed and screened against the SARS-CoV-2 S1 subunit, resulting in anti-S1 monobodies with sub-nanomolar to nanomolar affinity [63]. These monobodies were then applied to capture SARS-CoV-2 particles from the nasal swab samples of patients. Representative of biorecognition molecules designed to target SARS-CoV-2 antigens are summarized in Table 1.
For the host immune responses to SARS-CoV-2 infection, immunoglobulin M (IgM) and IgG provide a defense mechanism, and they can be used as another indicator of SARS-CoV-2 infection for the serology test [47]. During viral infections, IgM is the first antibody to be produced by the immune system that can be detected in the first week after infection and reached its peak after 2 weeks before reduced to near-background level. IgG, on the other hand, is produced after 1 week of infection, reached its peak in 3 weeks, and remained at a high level for long-term immunity and immunological memory [69,70,71]. In other words, the detection of IgM antibodies could indicate recent exposure to SARS-CoV-2, while the detection of IgG antibodies could indicate virus exposure some time ago. Hence, the SARS-CoV-2 antigens are generally used to target IgM or IgG. Chemiluminescence immunoassay (CLIA) is an example that utilizes the magnetic beads coated with SARS-CoV-2 antigens (e.g., N and S proteins) to detect IgM or IgG in serum, and the amount of detected IgM or IgG is quantified by the CLIA analyzer [72]. However, the sequential production of IgM and IgG in COVID-19 patients is still inconclusive because their production can be time-dependent in different patients which raises diagnostic problems [73]. Recently, immunoglobulin A (IgA) antibodies have been found to be a promising biomarker for the detection of early SARS-CoV-2-specific humoral responses as it can be frequently detected before the presence of IgG [73, 74].
4 Recent biosensing approaches for detecting SARS-CoV-2
In the present, the COVID-19 diagnosis mainly relies on the detection of SARS-CoV-2 RNA by real-time RT-PCR assay. Several test kits based on RT-PCR have been developed and commercially available to target various SARS-CoV-2 genes with relatively high sensitivity [46, 75, 76]. However, these test kits still suffer from limitations, including long turnaround times, complicated operation, expensive equipment, and high false-negative rate [47, 48, 77]. Therefore, the simple and cost-effective diagnostic methods are needed to accurately and rapidly screen patients during the pandemic. In this section, we summarize the recently developed molecular diagnostic methods that use different virus biological markers as a target, such as viral RNA, proteins, or human IgM/IgG.
For detecting viral RNA, isothermal amplification techniques, such as loop-mediated isothermal amplification (LAMP), have been applied to detect viral genome with similar sensitivity as standard PCR. In LAMP, the target DNA sequence is specifically amplified using three pairs of primers at a constant temperature ranging from 60 to 65 °C [78]. The positive detection can be visually quantified based on colorimetry, fluorimetry, and turbidity. Besides DNA amplification, reverse transcription LAMP (RT-LAMP) is used for amplifying specific RNA sequences that allow the direct detection of viral RNA [79]. Furthermore, combining LAMP or RT-LAMP with CRISPR, the DNA-endonuclease-targeted-CRISPR-trans-reporter (DETECTR) can further increase the specificity during amplification (Fig. 4a) [81]. The SARS-CoV-2 DETECTR assay provides a visualization of the result and fast (<40 min) detection of SARS-CoV-2 from a patient sample with 95% sensitivity and 100% specificity [80].
Applications of biorecognition elements in detecting SARS-CoV-2. a Schematic of SARS-CoV-2 DETECTR workflow. Viral RNA is extracted from COVID-19 patient samples and used as an input to DETECTR (LAMP preamplification and Cas12-based detection). The readout of this DETECTR can be visualized by a lateral flow strip. A positive result requires detection of at least one of the two SARS-CoV-2 viral gene targets (N gene or E gene). RNase P gene is used as a control, and QC represents quality control. Adapted with permission from [80], Copyright 2020, Springer Nature, b A schematic representation of the sandwich ELISA using the monobody. The monobody can replace antibody for the sandwich ELISA. The S1 subunit of SARS-CoV-2 S protein is added to the monobody-immobilized microplate. After the S1 subunit is bound with the monobody and unbound molecules are washed away, the binding between the S1 subunit and the monobody is identified by detecting monobody-HRP. All possible combinations (nine S1-binding monobodies) are tested as shown in the inset table. Nus represents Nus-Tag fused at the C terminus of the monobody. HRP is horseradish peroxidase enzyme that is used to amplify signal in photometric assays by catalyzing the conversion chemiluminescent substrates. Adapted from [63], Copyright, The Authors, some rights reserved; exclusive licensee AAAS. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC)
For detecting viral proteins and human immunoglobulins, the detection of these proteins provides an alternative diagnosis for COVID-19. One of the assays that have been applied to detect viral proteins is ELISA. In this assay, the specific binding between antibody and viral proteins (immobilized on a surface of a plate) can be detected with another additional tracer antibody that produces a colorimetric or fluorescent-based readout [82]. Thus, antibodies are important for targeting virus in this assay. Similarly, antibody-like molecules which recognize viral proteins can be used as an alternative to antibodies. For example, the recently engineered monobodies, which specifically bind to the S1 subunit of SARS-CoV-2, were used in ELISA assay, and the result was comparable as using antibody (Fig. 4b) [63]. These monobodies can be also coated on the magnetic beads to pull down and enrich SARS-CoV-2 virus particles from patient nasal swab samples for RT-PCR. In addition, human immunoglobulins, e.g., IgG or IgM can be quantitatively detected by a multiplex label-free antigen microarray on the Arrayed Imaging Reflectometry (AIR) platform [83].
Another approach to detect viral proteins is biosensors of which recognition domains are generally made of antibodies recognizing viral antigens, but transduction systems can be different. For instance, a nanoplasmonic resonance sensor utilizes the immobilized SARS-CoV-2 mAbs to detect SARS-CoV-2 virus particles in one step (< 15 min) with the virus range of 0 to 107 vp mL–1 [84]. In addition to detecting virus particles, an opto-microfluidic sensing platform, of which sensing unit is based on localized surface plasmon resonance, was developed to rapidly detect antibodies specific to the SARS-CoV-2 S protein in human plasma (< 30 min) with the limit of detection of ~ 0.08 ng mL–1 [85]. Additionally, engineering the surface of quartz crystals can form a sensitive transducer to detect biotargets [86, 87]. This concept can be integrated with a quartz crystal microbalance for sensing SARS-CoV-2 [88]. For instance, the S1 protein bears hydrophobic and positive charged surfaces. Therefore, it can interact with hydrophobic ends and carboxylated moieties which can be attached on the quartz crystal microbalance. This binding event causes the change of mass of the microbalance, thus providing sensitive analytical signals. However, nonspecific adsorption is challenging. Anti-S protein (as a bioreceptor) is a candidate to be functionalized on the quartz crystal surface to ensure high specificity to SARS-CoV-2. Moreover, electrochemical biosensors whose biorecognition element was made of membrane-engineered mammalian cells bearing the human chimeric spike S1 antibody can detect the SARS-CoV-2 S1 subunit of S protein (~3 min) with a detection limit of 1 fg mL–1 [89]. Similarly, a field-effect transistor (FET)-based biosensor utilizes immobilized SARS-CoV-2 mAbs on a graphene sheet to detect SARS-CoV-2 S proteins at concentrations of 1 fg mL–1 in phosphate-buffered saline and 100 fg mL–1 in clinical transport medium [90]. More detail of the bio-FET and its application will be elaborated in Sect. 5. Since similarity between coronaviruses, simultaneously detecting multiple SARS-CoV-2 biomarkers, could provide rapid and low-cost COVID-19 diagnosis. The SARS-CoV-2 RapidPlex is a graphene-based multiplexed telemedicine platform to detect SARS-CoV-2 N protein, inflammatory biomarker C-reactive protein (CRP), and specific immunoglobulins against SARS-CoV-2 S1 protein by coating specific anti-N protein mAbs, anti-CRP mAbs, and purified SARS-CoV-2 S1 proteins, respectively (detection time can be as low as 1 min, see Fig. 5a). Different expression levels of these antigens can be used to track the infection progression the COVID-19 (Table 2). The details of the bioelectrode designs and electrochemical detection systems will be discussed further in the following section.
Examples of electrochemical sensors for COVID-19 diagnosis. a A wireless graphene-based telemedicine platform (i) an image of a disposable and flexible graphene array, (ii) an illustration of the graphene sensor array layout, (iii) schematic illustrations of the detection of SARS-CoV-2 viral proteins, antibodies (IgG and IgM), and inflammatory biomarker C-reactive protein (CRP) in blood and saliva (also see Table 2), (iv) an image of the electrochemical sensing system connected to a printed circuit board for signal processing and wireless communication, and (v) data transmission via wireless to a user interface. WE, CE, and RE: stand for working electrode, counter electrode, and reference electrode. Adopted with permission from [91], Copyright 2020, Elsevier, b A schematic illustration of SARS-CoV-2 detection using the screen-printed electrode; the electrochemical detection process using a smartphone along with differential pulse voltammograms for different concentrations of artificial target for the SARS-CoV-2 biosensor. Adopted with permission from [92], Copyright 2020, Elsevier, c A schematic diagram of the COVID-19 field-effect transistor (FET) based biosensor. Adapted with permission from [90, 93], Copyright 2020, American Chemical Society. This permission is granted for the duration of the World Health Organization (WHO) declaration of COVID-19 as a global pandemic, d The detection principle of the COVID-19 electrochemical paper-based analytical device in human serum sample along with square wave voltammetric responses tested with different concentrations of the SARS-CoV-2 spike protein. Adopted with permission from [94], Copyright 2020, Elsevier
5 Emerging electrochemical bioelectronics toward COVID-19 diagnosis
POC biosensors offers rapid and on-site detection of viruses and infection risks [95, 96]. Two key features need to be considered for POC biosensors, including portability and sensitivity. Portable sensing devices open unique opportunities to speedily on-site detect viruses or track relevant symptoms by analyzing body fluids (such as saliva and blood) or viruses spread in our surroundings [95, 97]. Besides, bioelectronics are crucial to strengthening modern digital health initiatives, enabling the immediate management because they are ready to provide measurable electrical signals that are convenient to connect to digital systems [98,99,100]. Electrochemical detection approaches, for example, are extensively applied to POC applications. In the case of virus detection, electrochemical sensors can be designed to analyze viral protein-based biomarkers, apart from detection of viral genetic material. However, unlike genome materials which can be amplified, it is needed to engineer the sensor to provide a very sensitive and low limit of detection for detecting protein-based biomarkers.
Expanding from the previous discussion about viral biomarkers and biorecognition elements in Sects. 3 and 4, the basis of many electrochemical immunosensors relies on the specific virus-antibody recognition. The binding interaction between the immobilized capturing molecule (i.e., antibody) as a bioreceptor on the solid supports (i.e., electrode) and the target virus (or the relevant biocomponent from the virus) can form a stable complex. This recognition event can be designed to cause electrochemical changes. This event is detectable by using an electrical transducer. In terms of electrical transducing parameters, the interaction occurring on the sensing electrode can be converted into four main characteristic responses, depending on the detection methods, including potentiometric (i.e., the potential change of an indicator electrode) [101], voltammetric/amperometric (i.e., the associated current as a function of potential) [102, 103], conductometric (i.e., the conductivity or resistance) [104], and impedimetric (i.e., the impedance of a system) signals with respect to the target analyte concentrations [105]. Moreover, this can be extended to a microelectronic-based transducer, such as a FET-based biosensor. The binding of viruses or their antigens can induce the change in the surface potential of the device. This event causes the change in conductance of the FET channel, allowing the measurable signal to indicate the presence of viruses [90, 106]. Examples of electrochemical bioelectronics toward COVID-19 diagnosis are discussed in this section.
A recent example of electrochemical sensors for rapid COVID-19 diagnosis is shown in Fig. 5a [91]. The sensor design relies on sandwich- and indirect-based immunosensing mechanisms. The SARS-CoV-2 N protein, specific immunoglobulins against the SARS-CoV-2 S protein (S1) (S1-IgM and S1-IgG), and CRP are biomarkers used to indicate the virus. Four working electrodes are patterned on a flexible polyimide substrate. The detection of SARS-CoV-2 N protein and CRP employs double-sandwich and sandwich assays by immobilizing capture antibody on the graphene electrode. These targets can interact with the bottom biorecognition layer immobilized on the graphene surface. Afterwards, the detector antibody (DAb) or secondary detector antibody (DAb2) attached with horseradish peroxidase (HRP) will bind specifically on the top of the targets. Apart from the detection of the N protein and CRP, the indirect assay system can be used to monitor the concentration of S1-IgM and S1-IgG. Antibodies specific to IgG and IgM are attached with HRP enzymes to provide electrochemical signals. The unreacted sites are also blocked with bovine serum albumin (BSA) in order to obviate the nonspecific binding of other molecules presented in the real analysis system. By applying the amperometric technique at a low potential of –0.2 V (versus Ag/AgCl) toward the sensing electrode in the presence of the hydroquinone (HQ) and hydrogen peroxide (H2O2), the HRP unit catalyzes the electrochemical reactions, resulting in a measurable electrochemical signal. Due to the selective biological recognition of the viral antigen-antibody binding events in the system, the DAb2-labeled HRP component is related to the number of targets. Therefore, the viral concentration can be determined by observing the electrochemical signal output generated by the HRP probe. COVID-19-positive and COVID-19-negative serum and saliva samples were tested in this work, suggesting the possibility to use electrochemical sensors as promising diagnostic tools. A customized four-channel electronic device can record amperometric currents from four working electrodes. These data can send out wirelessly to a user device. This is a representative promise for supporting remote healthcare management. Note that a variety of biomarkers, such as the N protein, anti-S protein immunoglobulins, and CRP, can indicate the presence of coronavirus in different status. For instance, COVID-19 patients have a high C-reactive protein CRP level (Table 2) [91, 107].
Significant factors to be considered in developing the base of the electrode are the rate of heterogeneous electron transfer and double layer capacitance as they control the response time, sensitivity, limit of detection, and signal-to-noise ratio [14, 108, 109]. Several materials, such as common carbon-based nanomaterials [110, 111], gold [112], and platinum [113], can be employed. Unique properties of nanomaterials, e.g., low resistivity, durable affinity towards bioreceptor immobilization, and large surface area, ensure high performances of nucleic acid-based or protein-based biosensors [114, 115]. Examples of nanomaterials are carbon nanomaterials [116], gold nanoparticles [117], magnetic nanoparticles [118], silica nanoparticles [119], quantum dots [120], and hybrid nanostructures [121]. The choice of linker is also crucial because it is required to ensure the stable immobilization of specific bioreceptors such as antibodies or proteins, to nanomaterials and minimize non-specific adsorptions. A molecular linker consisting of π-electron system and a carboxylic group is used to attach the bioreceptor on the electrode. The approach used for the bioreceptor immobilization is one of the significant aspects in offering a successful affinity-based bioelectrode.
Another example of electrochemical sensor for SARS-CoV-2 relies on supersandwich-type biorecognition, as shown in Fig. 5b [92]. This biosensor was designed to detect RNA present in SARS-CoV-2. In this example, graphene oxide is functionalized with macrocyclic oligomer calixarene, i.e., p-sulfocalix [8]arene (SCX8). The resulting calixarene/carbon composite is also attached with gold and toluidine blue materials to provide a high electrochemical signal. With the presence of the target RNA from the virus, the RNA target can bind specifically with the capture probe which is attached on the gold/Fe3O4 nanoparticles via gold and thiol on the capture probe. Note that Fe3O4 nanoparticles are designed to allow magnetic separation [118]. Sequentially, the resulting target-capture probe combination binds with SCX8 attached on gold/toluidine blue/graphene composite (Au@SCX8-RGO-toluidine blue nanocomposites). Host-guest recognition between p-sulfocalix [8]arene and toluidine blue is chosen to ensure reliable and clear electrical signal [122]. Labeled signal probe and auxiliary probe are also used to bind with Au@SCX8-RGO-toluidine blue nanocomposites. The 5′- and 3′-ends of target sequence are matching to the capture probe and the label probe, respectively. The auxiliary probe is designed to give a lengthy shape by binding with the label probe areas. Therefore, the sandwich configuration consists of (1) the capture probe, (2) RNA target in the middle of sandwich, and (3) the labeled signal probe. Hence, with the presence of the target (even 10−12 M), the distinct differential pulse voltammetric peak related to the electrochemical toluidine blue redox probe can be observed. The amplifying signal strategy by combining the label probe with toluidine blue and other materials suggests a way to address the common challenge pertaining to low analytical signal. The clear peak current can be measured as an analytical signal. This approach is highly sensitive that no extra steps (e.g., nucleic acid amplification or reverse transcription) are required. In addition, the toluidine blue signal can be read by a smartphone.
A transistor that employs an electric field to regulate the flow of current can be modified to fabricate a sensitive biosensor [123]. An example of a FET-based biosensor for monitoring SARS-CoV-2 was demonstrated as shown in Fig. 5c [90, 93]. Graphene-based material is used to immobilize the SARS-CoV-2 spike antibody by using 1-pyrenebutyric acid N-hydroxysuccinimide ester as a linker. This immobilized coronavirus spike antibody is a recognition on the drain–source surface. Upon the addition of the target, real-time responses with a wide dynamic range can be obtained. When testing the developed biosensor with MERS-CoV S proteins, the signal is absent. This test of immobilized antibody against the other viruses on the graphene-based transistor proves the high specificity to the target (i.e., SARS-CoV-2 antigen).
Another representative is based on a label-free paper-based electrochemical device, as shown in Fig. 5d [94]. The mechanism for the detection of the SARS-CoV-2 antibody relies on the binding event of the antibody target with the SARS-CoV-2 S protein receptor-binding domain (RBD), which is attached on the detection zone of the graphene-based electrode sensor. The immobilization of the RBD is obtained by using 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide/N-hydroxysuccinimide (EDC/NHS) coupling on the carboxylated graphene surface. Upon the addition of antibodies existing in the serum sample, the target will bind with the biorecognition layer. This event causes the blockage of electrochemically active surface. Therefore, when scanning square-wave voltammetry in the redox probe solution (i.e., [Fe(CN)6]3−/4−), the signal of the redox probe is turned off, indicating the presence of SARS-CoV-2 IgG or and SARS-CoV-2 IgM. This hindering process is sensitive as the system can monitor the target with the detection limit of ~0.1 ng mL–1 immunoglobulins (IgG and IgM). The same concept is also extended for the detection of viral antigen. The paper-based sensor was designed for sensing SARS-CoV-2 S protein. In this case, SARS-CoV-2 IgM was used to be immobilized on the detection area. The result shows that the electrochemical sensor for monitoring the SARS-CoV-2 RBD provides a dynamic range of 1–1000 ng mL–1 with a detection limit of 0.11 ng mL–1. Even though the detection limit of this sensor is low, the target in the sample obtained from nasopharyngeal swab is as low as picograms per milliliter. Therefore, further improvement is still required.
6 Conclusions and prospects
The COVID-19 pandemic is an ongoing critical global challenge since December 2019. The battle against COVID-19 requires efforts to deal with public health management. Although “social distancing” is important to flatten the curve of COVID-19 spreading, society, and economy cannot be completely stopped. Therefore, developing materials and systems to support effective COVID-19 detection is vital. Such requirements challenge multidisciplinary researchers to develop simple, high-throughput, and accurate diagnostic tools.
Tremendous progress has been made to develop diagnosis assay/kits that can accurately and rapidly detect viral biomarkers especially in asymptomatic and/or early-stage patients for reducing the spread of this disease. RT-PCR typically serves as a routine method for the detection of SARS-CoV-2 genetic material; however, it is costly, time consuming, labor intensive, and requires specialized laboratory equipment. Biosensors, on the other hand, provide low-cost, rapid, and sensitive detection of SARS-CoV-2 virus particles [124, 125]. General platforms of biosensors for SARS-CoV-2 detection involve three important components, including (1) the target biomarkers of virus (e.g., viral RNA, viral proteins, or human immunoglobulins), (2) identification methods (based on biorecognition materials, e.g., antibodies, antibody-like molecules, or nucleic acid probes), and (3) the transduction systems for signal amplification (based on electrical, electrochemical, optical, surface plasmon resonance, and fluorescent signals) [45]. Using RNA as a target can be inconvenient because it requires the additional RNA extraction step from patient sample. Using viral protein as a target, on the other hand, allows direct detection in patient sample. Importantly, the binding affinity between biorecognition molecules and their target viral proteins needs to be improved to increase specificity of biosensor to target SARS-CoV-2. The mAbs are usually used as the biorecognition molecules in many detection assays and biosensors because they are highly specific to the target antigen. However, mAbs are complex molecules with 150 kDa that leads to high manufacturing cost and difficult production process. Alternatively, antibody-like molecules are small and easy to produce (in bacteria or yeast). With the advent of protein engineering techniques (e.g., rational design and directed evolution), these molecules can be engineered to be highly specific to target biomarkers. Therefore, antibody-like molecules could be a promising alternative biorecognition unit to mAbs for the detection of viral proteins.
Many diagnosis assays/kits for the detection of SARS-CoV-2 have been approved under Emergency Use Authorization (EUA) by The United States Food and Drug Administration (FDA) to be commercialized since 2020. These in vitro diagnostic kits are used to detect different SARS-CoV-2 biomarkers, such as the Xpert Xpress SARS-CoV-2 (Cepheid) and cobas SARS-CoV-2 kits (Roche Molecular Systems, Inc) to detect viral nucleic acids by RT-PCR, BD Veritor System for Rapid Detection of SARS-CoV-2 (Becton, Dickinson and Company) and BinaxNOW COVID-19 Ag Card (Abbott Diagnostics Scarborough, Inc.) to detect the N protein in nasal swabs, Assure COVID-19 IgG/IgM Rapid Test Device (Assure Tech. (Hangzhou) Co., Ltd.) and RightSign COVID-19 IgG/IgM Rapid Test Cassette (Hangzhou Biotest Biotech Co., Ltd.) to detect IgM and IgG antibodies specific to SARS-CoV-2 in human venous whole blood sample and fingerstick whole blood. More examples of in vitro diagnostic kits for COVID-19 can be found on the FDA website, https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/vitro-diagnostics-euas [76].
This review also includes examples of coronavirus detection using electrochemical approaches. Electrochemical sensing strategies represent attractive POC testing alternatives. Apart from conventional gold-standard methods which have limitations for speedy on-site diagnosis, it is crucial to develop the sensing device that enables the rapid assessment and promotes the telemedicine testing for field applications. In this regard, the development of a wireless telemedicine sensing platform offers fast communications of the identified real-time patient’s infection information/status delivered to the medical care system via mobile health platforms, regardless of the location. The state of the art in such bioelectronics and wireless technologies thus may fill the need for widespread COVID-19 testing and surpasses the testing backlogs. Although the non-invasive detection of the sensing platform in human saliva sample is safe and user-friendly for at-home use, the validation of the relationship between saliva and serum concentrations over the course of the SARS-CoV-2 infection is still required to achieve an accurate assay. There still remain much to be done as accurate measurements at a trace level of targets are required and crucial.
The requirement to obtain simple and rapid analysis while using small sample volumes draws attention to electrochemical sensors. We describe some representatives of recent technology to detect coronavirus. Nanomaterials are powerful to support the fabrication of electrodes and allow reliable immobilization of biorecognition layers on the electrode surface. Functional materials are also essential to enhance analytical performances, such as high signal-to-noise ratio, sensitivity, and stability. Integrated electrode arrays provide multiplex detection of multicomponents (targets) which indicates the COVID-19 disease in patients. Moreover, the integration of other technologies, such as microfluidic-integrated systems and microelectronics, can enable the high-throughput and fully automated sampling-handling detection and telemedicine fashion. With the achievements in developing bioelectronics, it is expected that the obtained diagnostic tools through smart micro total analysis systems will not only support the acquisition of signals indicating the presence of viral targets, but also data processing, storage, and the extraction of specific features. The important data will then be sent out to the central government or public healthcare organization to regulate the pandemic.
References
COVID-19 Coronavirus Pandemic. 2021 [cited Access January 18, 2021]; Available from: https://www.worldometers.info/coronavirus/
D.P. Oran, E.J. Topol, Prevalence of Asymptomatic SARS-CoV-2 infection. Ann. Intern. Med. 173, 362–367 (2020). https://doi.org/10.7326/M20-3012
C.-C. Lai, T.-P. Shih, W.-C. Ko, H.-J. Tang, P.-R. Hsueh, Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents 55, 105924 (2020). https://doi.org/10.1016/j.ijantimicag.2020.105924
K.-H. Liang, T.-J. Chang, M.-L. Wang, P.-H. Tsai, T.-H. Lin, C.-T. Wang, D.-M. Yang, Novel biosensor platforms for the detection of coronavirus infection and severe acute respiratory syndrome coronavirus 2. J. Chin. Med. Assoc. 83, 701–703 (2020)
M. Gandhi, D.S. Yokoe, D.V. Havlir, Asymptomatic transmission, the Achilles’ heel of current strategies to control Covid-19. N. Engl. J. Med. 382, 2158–2160 (2020). https://doi.org/10.1056/NEJMe2009758
A. Aydin, G. Cebi, Z.E. Demirtas, H. Erkus, A. Kucukay, M. Ok, L. Sakalli, S. Alpdagtas, O. Gunduz, C.B. Ustundag, Combating COVID-19 with tissue engineering: a review. Emerg. Mater. (2020). https://doi.org/10.1007/s42247-020-00138-6
B.V. Ribeiro, T.A.R. Cordeiro, G.R.O. e Freitas, L.F. Ferreira, D.L. Franco, Biosensors for the detection of respiratory viruses: a review. Talanta Open 2, 100007 (2020). https://doi.org/10.1016/j.talo.2020.100007
S.A. Hashemi, N.G. Golab Behbahan, S. Bahrani, S.M. Mousavi, A. Gholami, S. Ramakrishna, M. Firoozsani, M. Moghadami, K.B. Lankarani, N. Omidifar, Ultra-sensitive viral glycoprotein detection NanoSystem toward accurate tracing SARS-CoV-2 in biological/non-biological media. Biosens. Bioelectron. 171, 112731 (2021). https://doi.org/10.1016/j.bios.2020.112731
T. Ozer, B.J. Geiss, C.S. Henry, Review—chemical and biological sensors for viral detection. J. Electrochem. Soc. 167, 037523 (2020). https://doi.org/10.1149/2.0232003jes
D. van Bockel, C.M. Munier, S. Turville, S.G. Badman, G. Walker, A.O. Stella, A. Aggarwal, M. Yeang, A. Condylios, A.D. Kelleher, T.L. Applegate, A. Vallely, D. Whiley, W. Rawlinson, P. Cunningham, J. Kaldor, R. Guy, Evaluation of commercially available viral transport medium (VTM) for SARS-CoV-2 inactivation and use in point-of-care (POC) testing. Viruses 12, 1208 (2020). https://doi.org/10.3390/v12111208
S. Ahmad, N. Ali, M. Kausar, H. Misbah, A. Wahid, Road toward rapid-molecular point of care test to detect novel SARS-coronavirus 2019 (COVID-19): review from updated literature. Allergol. Immunopathol. 48, 518–520 (2020). https://doi.org/10.1016/j.aller.2020.06.001
M.A. AlMaadeed, Emergent materials and industry 4.0 contribution toward pandemic diseases such as COVID-19. Emerg. Mater. 3, 107–108 (2020). https://doi.org/10.1007/s42247-020-00102-4
Diagnostic testing for SARS-CoV-2: interim guidance, 11 September 2020. 2020 Last Update [cited Access January 18, 2021]; Available from: https://apps.who.int/iris/handle/10665/334254 License: CC BY-NC-SA 3.0 IGO
I. Jeerapan, T. Sonsa-ard, D. Nacapricha, Applying nanomaterials to modern biomedical electrochemical detection of metabolites, electrolytes, and pathogens. Chemosensors 8, 71 (2020). https://doi.org/10.3390/chemosensors8030071
A. Demeke Teklemariam, M. Samaddar, M.G. Alharbi, R.R. Al-Hindi, A.K. Bhunia, Biosensor and molecular-based methods for the detection of human coronaviruses: a review. Mol. Cell. Probes 54, 101662 (2020). https://doi.org/10.1016/j.mcp.2020.101662
B. Purohit, P.R. Vernekar, N.P. Shetti, P. Chandra, Biosensor nanoengineering: Design, operation, and implementation for biomolecular analysis. Sensors Int. 1, 100040 (2020). https://doi.org/10.1016/j.sintl.2020.100040
M. Srivastava, N. Srivastava, P.K. Mishra, B.D. Malhotra, Prospects of nanomaterials-enabled biosensors for COVID-19 detection. Sci. Total Environ. 754, 142363 (2021). https://doi.org/10.1016/j.scitotenv.2020.142363
Prakash, R. and P. Chandra, Nanobiomaterial engineering: concepts and their applications in biomedicine and diagnostics. 2020: Springer.
K. Chrzastek, D.-h. Lee, D. Smith, P. Sharma, D.L. Suarez, M. Pantin-Jackwood, D.R. Kapczynski, Use of sequence-independent, single-primer-amplification (SISPA) for rapid detection, identification, and characterization of avian RNA viruses. Virology 509, 159–166 (2017). https://doi.org/10.1016/j.virol.2017.06.019
Y.H. Baek, J. Um, K.J.C. Antigua, J.-H. Park, Y. Kim, S. Oh, Y.-i. Kim, W.-S. Choi, S.G. Kim, J.H. Jeong, B.S. Chin, H.D.G. Nicolas, J.-Y. Ahn, K.S. Shin, Y.K. Choi, J.-S. Park, M.-S. Song, Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerg. Microbes Infect. 9, 998–1007 (2020). https://doi.org/10.1080/22221751.2020.1756698
T. Ji, Z. Liu, G. Wang, X. Guo, S.A. Khan, C. Lai, H. Chen, S. Huang, S. Xia, B. Chen, H. Jia, Y. Chen, Q. Zhou, Detection of COVID-19: a review of the current literature and future perspectives. Biosens. Bioelectron. 166, 112455 (2020). https://doi.org/10.1016/j.bios.2020.112455
N. Ravi, D.L. Cortade, E. Ng, S.X. Wang, Diagnostics for SARS-CoV-2 detection: a comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens. Bioelectron. 165, 112454 (2020). https://doi.org/10.1016/j.bios.2020.112454
W.S. Lee, A.K. Wheatley, S.J. Kent, B.J. DeKosky, Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 5, 1185–1191 (2020). https://doi.org/10.1038/s41564-020-00789-5
T.M. Wassenaar, Y. Zou, 2019_nCoV/SARS-CoV-2: rapid classification of betacoronaviruses and identification of Traditional Chinese Medicine as potential origin of zoonotic coronaviruses. 70, 342–348 (2020). https://doi.org/10.1111/lam.13285
C. Drosten, S. Günther, W. Preiser, S. van der Werf, H.-R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R.A.M. Fouchier, A. Berger, A.-M. Burguière, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J.-C. Manuguerra, S. Müller, V. Rickerts, M. Stürmer, S. Vieth, H.-D. Klenk, A.D.M.E. Osterhaus, H. Schmitz, H.W. Doerr, Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1967–1976 (2003). https://doi.org/10.1056/NEJMoa030747
N.S. Zhong, B.J. Zheng, Y.M. Li, L.L.M. Poon, Z.H. Xie, K.H. Chan, P.H. Li, S.Y. Tan, Q. Chang, J.P. Xie, X.Q. Liu, J. Xu, D.X. Li, K.Y. Yuen, J.S.M. Peiris, Y. Guan, Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet 362, 1353–1358 (2003). https://doi.org/10.1016/S0140-6736(03)14630-2
A.M. Zaki, S. van Boheemen, T.M. Bestebroer, A.D.M.E. Osterhaus, R.A.M. Fouchier, Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820 (2012). https://doi.org/10.1056/NEJMoa1211721
A. Banerjee, K. Baid, K. Mossman, Molecular pathogenesis of Middle East Respiratory Syndrome (MERS) coronavirus. Curr. Clin. Microbiol. Rep. 6, 139–147 (2019). https://doi.org/10.1007/s40588-019-00122-7
L. Chen, W. Liu, Q. Zhang, K. Xu, G. Ye, W. Wu, Z. Sun, F. Liu, K. Wu, B. Zhong, Y. Mei, W. Zhang, Y. Chen, Y. Li, M. Shi, K. Lan, Y. Liu, RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg. Microbes Infect. 9, 313–319 (2020). https://doi.org/10.1080/22221751.2020.1725399
S. Jiang, C. Hillyer, L. Du, Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 41, 355–359 (2020). https://doi.org/10.1016/j.it.2020.03.007
P. Zhou, X.-L. Yang, X.-G. Wang, B. Hu, L. Zhang, W. Zhang, H.-R. Si, Y. Zhu, B. Li, C.-L. Huang, H.-D. Chen, J. Chen, Y. Luo, H. Guo, R.-D. Jiang, M.-Q. Liu, Y. Chen, X.-R. Shen, X. Wang, X.-S. Zheng, K. Zhao, Q.-J. Chen, F. Deng, L.-L. Liu, B. Yan, F.-X. Zhan, Y.-Y. Wang, G.-F. Xiao, Z.-L. Shi, A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). https://doi.org/10.1038/s41586-020-2012-7
R. Lu, X. Zhao, J. Li, P. Niu, B. Yang, H. Wu, W. Wang, H. Song, B. Huang, N. Zhu, Y. Bi, X. Ma, F. Zhan, L. Wang, T. Hu, H. Zhou, Z. Hu, W. Zhou, L. Zhao, J. Chen, Y. Meng, J. Wang, Y. Lin, J. Yuan, Z. Xie, J. Ma, W.J. Liu, D. Wang, W. Xu, E.C. Holmes, G.F. Gao, G. Wu, W. Chen, W. Shi, W. Tan, Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395, 565–574 (2020). https://doi.org/10.1016/S0140-6736(20)30251-8
Y. Zhou, Y. Hou, J. Shen, Y. Huang, W. Martin, F. Cheng, Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discover. 6, 14 (2020). https://doi.org/10.1038/s41421-020-0153-3
A.C. Walls, Y.-J. Park, M.A. Tortorici, A. Wall, A.T. McGuire, D. Veesler, Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292.e6 (2020). https://doi.org/10.1016/j.cell.2020.02.058
Y. Huang, C. Yang, X.-f. Xu, W. Xu, S.-w. Liu, Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 41, 1141–1149 (2020). https://doi.org/10.1038/s41401-020-0485-4
J. Huo, A. Le Bas, R.R. Ruza, H.M.E. Duyvesteyn, H. Mikolajek, T. Malinauskas, T.K. Tan, P. Rijal, M. Dumoux, P.N. Ward, J. Ren, D. Zhou, P.J. Harrison, M. Weckener, D.K. Clare, V.K. Vogirala, J. Radecke, L. Moynié, Y. Zhao, J. Gilbert-Jaramillo, M.L. Knight, J.A. Tree, K.R. Buttigieg, N. Coombes, M.J. Elmore, M.W. Carroll, L. Carrique, P.N.M. Shah, W. James, A.R. Townsend, D.I. Stuart, R.J. Owens, J.H. Naismith, Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 27, 846–854 (2020). https://doi.org/10.1038/s41594-020-0469-6
D. Wrapp, N. Wang, K.S. Corbett, J.A. Goldsmith, C.-L. Hsieh, O. Abiona, B.S. Graham, J.S. McLellan, Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020). https://doi.org/10.1126/science.abb2507
M. Hoffmann, H. Kleine-Weber, S. Schroeder, N. Krüger, T. Herrler, S. Erichsen, T.S. Schiergens, G. Herrler, N.-H. Wu, A. Nitsche, M.A. Müller, C. Drosten, S. Pöhlmann, SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8 (2020). https://doi.org/10.1016/j.cell.2020.02.052
M. Schoof, B. Faust, R.A. Saunders, S. Sangwan, V. Rezelj, N. Hoppe, M. Boone, C.B. Billesbølle, C. Puchades, C.M. Azumaya, H.T. Kratochvil, M. Zimanyi, I. Deshpande, J. Liang, S. Dickinson, H.C. Nguyen, C.M. Chio, G.E. Merz, M.C. Thompson, D. Diwanji, K. Schaefer, A.A. Anand, N. Dobzinski, B.S. Zha, C.R. Simoneau, K. Leon, K.M. White, U.S. Chio, M. Gupta, M. Jin, F. Li, Y. Liu, K. Zhang, D. Bulkley, M. Sun, A.M. Smith, A.N. Rizo, F. Moss, A.F. Brilot, S. Pourmal, R. Trenker, T. Pospiech, S. Gupta, B. Barsi-Rhyne, V. Belyy, A.W. Barile-Hill, S. Nock, Y. Liu, N.J. Krogan, C.Y. Ralston, D.L. Swaney, A. García-Sastre, M. Ott, M. Vignuzzi, P. Walter, A. Manglik, An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science 370, 1473 (2020). https://doi.org/10.1126/science.abe3255
S. Mahapatra, P. Chandra, Clinically practiced and commercially viable nanobio engineered analytical methods for COVID-19 diagnosis. Biosens. Bioelectron. 165, 112361 (2020). https://doi.org/10.1016/j.bios.2020.112361
Q. Li, X. Guan, P. Wu, X. Wang, L. Zhou, Y. Tong, R. Ren, K.S.M. Leung, E.H.Y. Lau, J.Y. Wong, X. Xing, N. Xiang, Y. Wu, C. Li, Q. Chen, D. Li, T. Liu, J. Zhao, M. Liu, W. Tu, C. Chen, L. Jin, R. Yang, Q. Wang, S. Zhou, R. Wang, H. Liu, Y. Luo, Y. Liu, G. Shao, H. Li, Z. Tao, Y. Yang, Z. Deng, B. Liu, Z. Ma, Y. Zhang, G. Shi, T.T.Y. Lam, J.T. Wu, G.F. Gao, B.J. Cowling, B. Yang, G.M. Leung, Z. Feng, Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 382, 1199–1207 (2020). https://doi.org/10.1056/NEJMoa2001316
X. He, E.H.Y. Lau, P. Wu, X. Deng, J. Wang, X. Hao, Y.C. Lau, J.Y. Wong, Y. Guan, X. Tan, X. Mo, Y. Chen, B. Liao, W. Chen, F. Hu, Q. Zhang, M. Zhong, Y. Wu, L. Zhao, F. Zhang, B.J. Cowling, F. Li, G.M. Leung, Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 26, 672–675 (2020). https://doi.org/10.1038/s41591-020-0869-5
S.A. Lauer, K.H. Grantz, Q. Bi, F.K. Jones, Q. Zheng, H.R. Meredith, A.S. Azman, N.G. Reich, J. Lessler, The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Intern. Med. 172, 577–582 (2020). https://doi.org/10.7326/M20-0504
D.K.W. Chu, Y. Pan, S.M.S. Cheng, K.P.Y. Hui, P. Krishnan, Y. Liu, D.Y.M. Ng, C.K.C. Wan, P. Yang, Q. Wang, M. Peiris, L.L.M. Poon, Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 66, 549–555 (2020). https://doi.org/10.1093/clinchem/hvaa029
S. Iravani, Nano- and biosensors for the detection of SARS-CoV-2: challenges and opportunities. Mater Adv. 1, 3092–3103 (2020). https://doi.org/10.1039/D0MA00702A
W. Feng, A.M. Newbigging, C. Le, B. Pang, H. Peng, Y. Cao, J. Wu, G. Abbas, J. Song, D.-B. Wang, M. Cui, J. Tao, D.L. Tyrrell, X.-E. Zhang, H. Zhang, X.C. Le, Molecular diagnosis of COVID-19: challenges and research needs. Anal. Chem. 92, 10196–10209 (2020). https://doi.org/10.1021/acs.analchem.0c02060
Z. Li, Y. Yi, X. Luo, N. Xiong, Y. Liu, S. Li, R. Sun, Y. Wang, B. Hu, W. Chen, Y. Zhang, J. Wang, B. Huang, Y. Lin, J. Yang, W. Cai, X. Wang, J. Cheng, Z. Chen, K. Sun, W. Pan, Z. Zhan, L. Chen, F. Ye, Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 92, 1518–1524 (2020). https://doi.org/10.1002/jmv.25727
J.F.-W. Chan, C.C.-Y. Yip, K.K.-W. To, T.H.-C. Tang, S.C.-Y. Wong, K.-H. Leung, A.Y.-F. Fung, A.C.-K. Ng, Z. Zou, H.-W. Tsoi, G.K.-Y. Choi, A.R. Tam, V.C.-C. Cheng, K.-H. Chan, O.T.-Y. Tsang, K.-Y. Yuen, Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse rranscription-PCR assay validated <em>In Vitro</em> and with clinical specimens. J. Clin. Microbiol. 58, e00310–e00320 (2020). https://doi.org/10.1128/JCM.00310-20
W. Liu, L. Liu, G. Kou, Y. Zheng, Y. Ding, W. Ni, Q. Wang, L. Tan, W. Wu, S. Tang, Z. Xiong, S. Zheng, Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 58, e00461–e00420 (2020). https://doi.org/10.1128/JCM.00461-20
J. Sui, W. Li, A. Murakami, A. Tamin, L.J. Matthews, S.K. Wong, M.J. Moore, A.S.C. Tallarico, M. Olurinde, H. Choe, L.J. Anderson, W.J. Bellini, M. Farzan, W.A. Marasco, Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. U. S. A. 101, 2536–2541 (2004). https://doi.org/10.1073/pnas.0307140101
M.H. Kubala, O. Kovtun, K. Alexandrov, B.M. Collins, Structural and thermodynamic analysis of the GFP:GFP-nanobody complex. Protein Sci. 19, 2389–2401 (2010). https://doi.org/10.1002/pro.519
A.L. Main, T.S. Harvey, M. Baron, J. Boyd, I.D. Campbell, The three-dimensional structure of the tenth type III module of fibronectin: an insight into RGD-mediated interactions. Cell 71, 671–678 (1992). https://doi.org/10.1016/0092-8674(92)90600-H
G.P. Smith, V.A. Petrenko, Phage display. Chem. Rev. 97, 391–410 (1997). https://doi.org/10.1021/cr960065d
P. Limsakul, Q. Peng, Y. Wu, M.E. Allen, J. Liang, A.G. Remacle, T. Lopez, X. Ge, B.K. Kay, H. Zhao, A.Y. Strongin, X.-L. Yang, S. Lu, Y. Wang, Directed evolution to engineer monobody for FRET biosensor assembly and imaging at live-cell surface. Cell Chemical Biology 25, 370–379.e4 (2018). https://doi.org/10.1016/j.chembiol.2018.01.002
S. Harmansa, M. Affolter, Protein binders and their applications in developmental biology. Development 145, dev148874 (2018). https://doi.org/10.1242/dev.148874
G.P. Smith, Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228, 1315–1317 (1985). https://doi.org/10.1126/science.4001944
L. Zhang, B. Zheng, X. Gao, L. Zhang, H. Pan, Y. Qiao, G. Suo, F. Zhu, Development of patient-derived human monoclonal antibodies against nucleocapsid protein of severe acute respiratory syndrome coronavirus 2 for coronavirus disease 2019 diagnosis. Front. Immunol. 11, 595970 (2020). https://doi.org/10.3389/fimmu.2020.595970
B. Ju, Q. Zhang, J. Ge, R. Wang, J. Sun, X. Ge, J. Yu, S. Shan, B. Zhou, S. Song, X. Tang, J. Yu, J. Lan, J. Yuan, H. Wang, J. Zhao, S. Zhang, Y. Wang, X. Shi, L. Liu, J. Zhao, X. Wang, Z. Zhang, L. Zhang, Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584, 115–119 (2020). https://doi.org/10.1038/s41586-020-2380-z
T. Noy-Porat, E. Makdasi, R. Alcalay, A. Mechaly, Y. Levy, A. Bercovich-Kinori, A. Zauberman, H. Tamir, Y. Yahalom-Ronen, M.A. Israeli, E. Epstein, H. Achdout, S. Melamed, T. Chitlaru, S. Weiss, E. Peretz, O. Rosen, N. Paran, S. Yitzhaki, S.C. Shapira, T. Israely, O. Mazor, R. Rosenfeld, A panel of human neutralizing mAbs targeting SARS-CoV-2 spike at multiple epitopes. Nat. Commun. 11, 4303 (2020). https://doi.org/10.1038/s41467-020-18159-4
C. Wang, W. Li, D. Drabek, N.M.A. Okba, R. van Haperen, A.D.M.E. Osterhaus, F.J.M. van Kuppeveld, B.L. Haagmans, F. Grosveld, B.-J. Bosch, A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 11, 2251 (2020). https://doi.org/10.1038/s41467-020-16256-y
X. Tian, C. Li, A. Huang, S. Xia, S. Lu, Z. Shi, L. Lu, S. Jiang, Z. Yang, Y. Wu, T. Ying, Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 9, 382–385 (2020). https://doi.org/10.1080/22221751.2020.1729069
Y. Xiang, S. Nambulli, Z. Xiao, H. Liu, Z. Sang, W.P. Duprex, D. Schneidman-Duhovny, C. Zhang, Y. Shi, Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science 370, 1479 (2020). https://doi.org/10.1126/science.abe4747
T. Kondo, Y. Iwatani, K. Matsuoka, T. Fujino, S. Umemoto, Y. Yokomaku, K. Ishizaki, S. Kito, T. Sezaki, G. Hayashi, H. Murakami, Antibody-like proteins that capture and neutralize SARS-CoV-2. Sci. Adv. 6, eabd3916 (2020). https://doi.org/10.1126/sciadv.abd3916
S.F. Ahmed, A.A. Quadeer, M.R. McKay, Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses 12, 254 (2020). https://doi.org/10.3390/v12030254
P.D. Burbelo, F.X. Riedo, C. Morishima, S. Rawlings, D. Smith, S. Das, J.R. Strich, D.S. Chertow, R.T. Davey Jr., J.I. Cohen, Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus 2 in patients with coronavirus disease 2019. J. Infect. Dis. 222, 206–213 (2020). https://doi.org/10.1093/infdis/jiaa273
Y. Chen, X. Tong, Y. Li, B. Gu, J. Yan, Y. Liu, H. Shen, R. Huang, C. Wu, A comprehensive, longitudinal analysis of humoral responses specific to four recombinant antigens of SARS-CoV-2 in severe and non-severe COVID-19 patients. PLoS Pathog. 16, e1008796 (2020). https://doi.org/10.1371/journal.ppat.1008796
L. Premkumar, B. Segovia-Chumbez, R. Jadi, D.R. Martinez, R. Raut, A.J. Markmann, C. Cornaby, L. Bartelt, S. Weiss, Y. Park, C.E. Edwards, E. Weimer, E.M. Scherer, N. Rouphael, S. Edupuganti, D. Weiskopf, L.V. Tse, Y.J. Hou, D. Margolis, A. Sette, M.H. Collins, J. Schmitz, R.S. Baric, A.M. de Silva, The receptor-binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci Immunol. 5, eabc8413 (2020). https://doi.org/10.1126/sciimmunol.abc8413
S. Muyldermans, Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 82, 775–797 (2013). https://doi.org/10.1146/annurev-biochem-063011-092449
R. Racine, G.M. Winslow, IgM in microbial infections: taken for granted? Immunol. Lett. 125, 79–85 (2009). https://doi.org/10.1016/j.imlet.2009.06.003
H.-K. Lee, B.-H. Lee, S.-H. Seok, M.-W. Baek, H.-Y. Lee, D.-J. Kim, Y.-R. Na, K.-J. Noh, S.-H. Park, D.N. Kumar, H. Kariwa, M. Nakauchi, S.-J. Heo, J.-H. Park, Production of specific antibodies against SARS-coronavirus nucleocapsid protein without cross reactivity with human coronaviruses 229E and OC43. J. Vet. Sci. 11, 165–167 (2010)
Z. Wan, X. Zhang, X. Yan, IFA in testing specific antibody of SARS coronavirus. South China J. Prevent. Med. 29, 36–37 (2003)
Hou, H., T. Wang, B. Zhang, Y. Luo, L. Mao, F. Wang, S. Wu, and Z. Sun, Detection of IgM and IgG antibodies in patients with coronavirus disease. Clin Transl Immunol.. 2019. 9, e1136 (2020) https://doi.org/10.1002/cti2.1136
H. Ma, W. Zeng, H. He, D. Zhao, D. Jiang, P. Zhou, L. Cheng, Y. Li, X. Ma, T. Jin, Serum IgA, IgM, and IgG responses in COVID-19. Cell. Mol. Immunol. 17, 773–775 (2020). https://doi.org/10.1038/s41423-020-0474-z
D. Sterlin, A. Mathian, M. Miyara, A. Mohr, F. Anna, L. Claër, P. Quentric, J. Fadlallah, H. Devilliers, P. Ghillani, C. Gunn, R. Hockett, S. Mudumba, A. Guihot, C.-E. Luyt, J. Mayaux, A. Beurton, S. Fourati, T. Bruel, O. Schwartz, J.-M. Lacorte, H. Yssel, C. Parizot, K. Dorgham, P. Charneau, Z. Amoura, G. Gorochov, IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. 13, eabd2223 (2020). https://doi.org/10.1126/scitranslmed.abd2223
V.M. Corman, O. Landt, M. Kaiser, R. Molenkamp, A. Meijer, D.K. Chu, T. Bleicker, S. Brünink, J. Schneider, M.L. Schmidt, D.G. Mulders, B.L. Haagmans, B. van der Veer, S. van den Brink, L. Wijsman, G. Goderski, J.-L. Romette, J. Ellis, M. Zambon, M. Peiris, H. Goossens, C. Reusken, M.P. Koopmans, C. Drosten, Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25, 2000045 (2020). https://doi.org/10.2807/1560-7917.ES.2020.25.3.2000045
In Vitro Diagnostics EUAs. Food and Drug Administration [cited Access January 18, 2021]; Available from: https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/vitro-diagnostics-euas
T. Ai, Z. Yang, H. Hou, C. Zhan, C. Chen, W. Lv, Q. Tao, Z. Sun, L. Xia, Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014. Cases 296, E32–E40 (2020). https://doi.org/10.1148/radiol.2020200642
T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, T. Hase, Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28, e63–e63 (2000). https://doi.org/10.1093/nar/28.12.e63
L. Yu, S. Wu, X. Hao, X. Dong, L. Mao, V. Pelechano, W.-H. Chen, X. Yin, Rapid Detection of COVID-19 Coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin. Chem. 66, 975–977 (2020). https://doi.org/10.1093/clinchem/hvaa102
J.P. Broughton, X. Deng, G. Yu, C.L. Fasching, V. Servellita, J. Singh, X. Miao, J.A. Streithorst, A. Granados, A. Sotomayor-Gonzalez, K. Zorn, A. Gopez, E. Hsu, W. Gu, S. Miller, C.Y. Pan, H. Guevara, D.A. Wadford, J.S. Chen, C.Y. Chiu, CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 38, 870–874 (2020). https://doi.org/10.1038/s41587-020-0513-4
J.S. Chen, E. Ma, L.B. Harrington, M. Da Costa, X. Tian, J.M. Palefsky, J.A. Doudna, CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439 (2018). https://doi.org/10.1126/science.aar6245
L.J. Carter, L.V. Garner, J.W. Smoot, Y. Li, Q. Zhou, C.J. Saveson, J.M. Sasso, A.C. Gregg, D.J. Soares, T.R. Beskid, S.R. Jervey, C. Liu, Assay techniques and test development for COVID-19 diagnosis. ACS Central Sci 6, 591–605 (2020). https://doi.org/10.1021/acscentsci.0c00501
D.J. Steiner, J.S. Cognetti, E.P. Luta, A.M. Klose, J. Bucukovski, M.R. Bryan, J.J. Schmuke, P. Nguyen-Contant, M.Y. Sangster, D.J. Topham, B.L. Miller, Array-based analysis of SARS-CoV-2, other coronaviruses, and influenza antibodies in convalescent COVID-19 patients. Biosens. Bioelectron. 169, 112643 (2020). https://doi.org/10.1016/j.bios.2020.112643
L. Huang, L. Ding, J. Zhou, S. Chen, F. Chen, C. Zhao, J. Xu, W. Hu, J. Ji, H. Xu, G.L. Liu, One-step rapid quantification of SARS-CoV-2 virus particles via low-cost nanoplasmonic sensors in generic microplate reader and point-of-care device. Biosens. Bioelectron. 171, 112685 (2021). https://doi.org/10.1016/j.bios.2020.112685
R. Funari, K.-Y. Chu, A.Q. Shen, Detection of antibodies against SARS-CoV-2 spike protein by gold nanospikes in an opto-microfluidic chip. Biosens. Bioelectron. 169, 112578 (2020). https://doi.org/10.1016/j.bios.2020.112578
A. Alassi, M. Benammar, D. Brett, Quartz crystal microbalance electronic interfacing systems: a review. Sensors 17, 2799 (2017). https://doi.org/10.3390/s17122799
Zainuddin, A.A., A.N. Nordin, M.A.M. Asri, R. Ab Rahim, C. Guines, M. Chatras, A. Pothier, and W.C. Mak. Development of integrated electrochemical-quartz crystal microbalance biosensor arrays: towards ultrasensitive, multiplexed and rapid point-of-care dengue detection. in BioDevices. 2019.
L.M. Pandey, Design of engineered surfaces for prospective detection of SARS-CoV-2 using quartz crystal microbalance-based techniques. Expert Rev. Proteomics 17, 425–432 (2020). https://doi.org/10.1080/14789450.2020.1794831
S. Mavrikou, G. Moschopoulou, V. Tsekouras, S. Kintzios, Development of a portable, ultra-rapid and ultra-sensitive cell-based biosensor for the direct detection of the SARS-CoV-2 S1 spike protein antigen. Sensors 20 (2020). https://doi.org/10.3390/s20113121
G. Seo, G. Lee, M.J. Kim, S.-H. Baek, M. Choi, K.B. Ku, C.-S. Lee, S. Jun, D. Park, H.G. Kim, S.-J. Kim, J.-O. Lee, B.T. Kim, E.C. Park, S.I. Kim, Correction to rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano 14, 12257–12258 (2020). https://doi.org/10.1021/acsnano.0c06726
R.M. Torrente-Rodríguez, H. Lukas, J. Tu, J. Min, Y. Yang, C. Xu, H.B. Rossiter, W. Gao, SARS-CoV-2 RapidPlex: a graphene-based multiplexed telemedicine platform for rapid and low-cost COVID-19 diagnosis and monitoring. Matter 3, 1981–1998 (2020). https://doi.org/10.1016/j.matt.2020.09.027
H. Zhao, F. Liu, W. Xie, T.-C. Zhou, J. OuYang, L. Jin, H. Li, C.-Y. Zhao, L. Zhang, J. Wei, Y.-P. Zhang, C.-P. Li, Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sensors Actuators B Chem. 327, 128899 (2021). https://doi.org/10.1016/j.snb.2020.128899
G. Seo, G. Lee, M.J. Kim, S.-H. Baek, M. Choi, K.B. Ku, C.-S. Lee, S. Jun, D. Park, H.G. Kim, S.-J. Kim, J.-O. Lee, B.T. Kim, E.C. Park, S.I. Kim, Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano 14, 5135–5142 (2020). https://doi.org/10.1021/acsnano.0c02823
A. Yakoh, U. Pimpitak, S. Rengpipat, N. Hirankarn, O. Chailapakul, S. Chaiyo, Paper-based electrochemical biosensor for diagnosing COVID-19: detection of SARS-CoV-2 antibodies and antigen. Biosens. Bioelectron. 176, 112912 (2020). https://doi.org/10.1016/j.bios.2020.112912
A. Parihar, P. Ranjan, S.K. Sanghi, A.K. Srivastava, R. Khan, Point-of-care biosensor-based diagnosis of COVID-19 holds promise to combat current and future pandemics. ACS Appl Bio Mater 3, 7326–7343 (2020). https://doi.org/10.1021/acsabm.0c01083
Mahapatra, S., A. Baranwal, B. Purohit, S. Roy, S.K. Mahto, and P. Chandra, Advanced biosensing methodologies for ultrasensitive detection of human coronaviruses, in Diagnostic strategies for COVID-19 and other coronaviruses. 2020, Springer. p. 19-36.
H. Zhang, B.L. Miller, Immunosensor-based label-free and multiplex detection of influenza viruses: state of the art. Biosens. Bioelectron. 141, 111476 (2019). https://doi.org/10.1016/j.bios.2019.111476
P. Chandra, Miniaturized label-free smartphone assisted electrochemical sensing approach for personalized COVID-19 diagnosis. Sensors Int. 1, 100019 (2020). https://doi.org/10.1016/j.sintl.2020.100019
I. Jeerapan, S. Poorahong, Review—flexible and stretchable electrochemical sensing systems: materials, energy sources, and integrations. J. Electrochem. Soc. 167, 037573 (2020). https://doi.org/10.1149/1945-7111/ab7117
S. Menon, M.R. Mathew, S. Sam, K. Keerthi, K.G. Kumar, Recent advances and challenges in electrochemical biosensors for emerging and re-emerging infectious diseases. J. Electroanal. Chem. 878, 114596 (2020). https://doi.org/10.1016/j.jelechem.2020.114596
A.-L. Sun, A potentiometric immunosensor for enterovirus 71 based on bis-MPA-COOH dendrimer-doped AgCl nanospheres with a silver ion-selective electrode. Analyst 143, 487–492 (2018). https://doi.org/10.1039/C7AN01305A
R. Singh, S. Hong, J. Jang, Label-free detection of influenza viruses using a reduced graphene oxide-based electrochemical immunosensor integrated with a microfluidic platform. Sci. Rep. 7, 42771 (2017). https://doi.org/10.1038/srep42771
G. Martins, J.L. Gogola, L.H. Budni, B.C. Janegitz, L.H. Marcolino-Junior, M.F. Bergamini, 3D-printed electrode as a new platform for electrochemical immunosensors for virus detection. Anal. Chim. Acta 1147, 30–37 (2020). https://doi.org/10.1016/j.aca.2020.12.014
H. Liu, Y. Yang, P. Chen, Z. Zhong, Enhanced conductometric immunoassay for hepatitis B surface antigen using double-codified nanogold particles as labels. Biochem. Eng. J. 45, 107–112 (2009). https://doi.org/10.1016/j.bej.2009.03.002
Q.Y. Siew, E.L. Pang, H.-S. Loh, M.T.T. Tan, Highly sensitive and specific graphene/TiO2 impedimetric immunosensor based on plant-derived tetravalent envelope glycoprotein domain III (EDIII) probe antigen for dengue diagnosis. Biosens. Bioelectron. 176, 112895 (2020). https://doi.org/10.1016/j.bios.2020.112895
S. Hideshima, H. Hayashi, H. Hinou, S. Nambuya, S. Kuroiwa, T. Nakanishi, T. Momma, S.-I. Nishimura, Y. Sakoda, T. Osaka, Glycan-immobilized dual-channel field effect transistor biosensor for the rapid identification of pandemic influenza viral particles. Sci. Rep. 9, 11616 (2019). https://doi.org/10.1038/s41598-019-48076-6
C. Wu, X. Chen, Y. Cai, J.A. Xia, X. Zhou, S. Xu, H. Huang, L. Zhang, X. Zhou, C. Du, Y. Zhang, J. Song, S. Wang, Y. Chao, Z. Yang, J. Xu, X. Zhou, D. Chen, W. Xiong, L. Xu, F. Zhou, J. Jiang, C. Bai, J. Zheng, Y. Song, Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 180, 934–943 (2020). https://doi.org/10.1001/jamainternmed.2020.0994
S. Chen, Y. Liu, J. Chen, Heterogeneous electron transfer at nanoscopic electrodes: importance of electronic structures and electric double layers. Chem. Soc. Rev. 43, 5372–5386 (2014). https://doi.org/10.1039/C4CS00087K
H. Beitollahi, F. Movahedifar, S. Tajik, S. Jahani, A review on the effects of introducing CNTs in the modification process of electrochemical. Sensors 31, 1195–1203 (2019). https://doi.org/10.1002/elan.201800370
A.A. Lahcen, S. Rauf, T. Beduk, C. Durmus, A. Aljedaibi, S. Timur, H.N. Alshareef, A. Amine, O.S. Wolfbeis, K.N. Salama, Electrochemical sensors and biosensors using laser-derived graphene: a comprehensive review. Biosens. Bioelectron. 168, 112565 (2020). https://doi.org/10.1016/j.bios.2020.112565
V. Schroeder, S. Savagatrup, M. He, S. Lin, T.M. Swager, Carbon nanotube chemical sensors. Chem. Rev. 119, 599–663 (2019). https://doi.org/10.1021/acs.chemrev.8b00340
M. Alagiri, P. Rameshkumar, A. Pandikumar, Gold nanorod-based electrochemical sensing of small biomolecules: a review. Microchim. Acta 184, 3069–3092 (2017). https://doi.org/10.1007/s00604-017-2418-6
L. Jiang, J. Han, F. Li, J. Gao, Y. Li, Y. Dong, Q. Wei, A sandwich-type electrochemical immunosensor based on multiple signal amplification for α-fetoprotein labeled by platinum hybrid multiwalled carbon nanotubes adhered copper oxide. Electrochim. Acta 160, 7–14 (2015). https://doi.org/10.1016/j.electacta.2015.02.050
M. Santhanam, I. Algov, L. Alfonta, DNA/RNA electrochemical biosensing devices a future replacement of PCR methods for a fast epidemic containment. Sensors 20 (2020). https://doi.org/10.3390/s20164648
S.A. Lim, M.U. Ahmed, Electrochemical immunosensors and their recent nanomaterial-based signal amplification strategies: a review. RSC Adv. 6, 24995–25014 (2016). https://doi.org/10.1039/C6RA00333H
C. Yang, M.E. Denno, P. Pyakurel, B.J. Venton, Recent trends in carbon nanomaterial-based electrochemical sensors for biomolecules: a review. Anal. Chim. Acta 887, 17–37 (2015). https://doi.org/10.1016/j.aca.2015.05.049
P.A. Rasheed, N. Sandhyarani, Electrochemical DNA sensors based on the use of gold nanoparticles: a review on recent developments. Microchim. Acta 184, 981–1000 (2017). https://doi.org/10.1007/s00604-017-2143-1
M. Pastucha, Z. Farka, K. Lacina, Z. Mikušová, P. Skládal, Magnetic nanoparticles for smart electrochemical immunoassays: a review on recent developments. Microchim. Acta 186, 312 (2019). https://doi.org/10.1007/s00604-019-3410-0
A. Walcarius, Silica-based electrochemical sensors and biosensors: recent trends. Cur. Opin. Electrochem. 10, 88–97 (2018). https://doi.org/10.1016/j.coelec.2018.03.017
M.A. Farzin, H. Abdoos, A critical review on quantum dots: from synthesis toward applications in electrochemical biosensors for determination of disease-related biomolecules. Talanta 224, 121828 (2021). https://doi.org/10.1016/j.talanta.2020.121828
S. Zhang, R. Geryak, J. Geldmeier, S. Kim, V.V. Tsukruk, Synthesis, assembly, and applications of hybrid nanostructures for biosensing. Chem. Rev. 117, 12942–13038 (2017). https://doi.org/10.1021/acs.chemrev.7b00088
H.J. Kim, M.H. Lee, L. Mutihac, J. Vicens, J.S. Kim, Host–guest sensing by calixarenes on the surfaces. Chem. Soc. Rev. 41, 1173–1190 (2012). https://doi.org/10.1039/C1CS15169J
Poghossian, A., M. Jablonski, D. Molinnus, C. Wege, and M.J. Schöning, Field-effect sensors for virus detection: from ebola to SARS-CoV-2 and plant viral enhancers. Front. Plant Sci. 11, 598103 (2020). https://doi.org/10.3389/fpls.2020.598103
M. Asif, M. Ajmal, G. Ashraf, N. Muhammad, A. Aziz, T. Iftikhar, J. Wang, H. Liu, The role of biosensors in coronavirus disease-2019 outbreak. Cur. Opin. Electrochem. 23, 174–184 (2020). https://doi.org/10.1016/j.coelec.2020.08.011
G. Qiu, Z. Gai, Y. Tao, J. Schmitt, G.A. Kullak-Ublick, J. Wang, Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano 14, 5268–5277 (2020). https://doi.org/10.1021/acsnano.0c02439
Funding
I.J. was supported by the Faculty of Science Research Fund 2021 (contract number: 264003), Prince of Songkla University, Hat Yai, Thailand.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Limsakul, P., Charupanit, K., Moonla, C. et al. Advances in emergent biological recognition elements and bioelectronics for diagnosing COVID-19. emergent mater. 4, 231–247 (2021). https://doi.org/10.1007/s42247-021-00175-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42247-021-00175-9