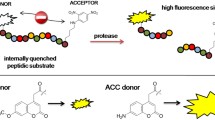
A step-by-step evaluation of dual-labeled FRET substrates for the protease calpain is reported. The study led to cell permeable selections, with optimized specificity and effectiveness for the target enzyme, and improved stability to non-specific degrading enzymes.
Similar content being viewed by others
Introduction
The application of solid-phase synthesis and novel dye and quencher derivatives for the development of a highly sensitive interference-free high throughput in vitro assay for the SARS-coronavirus 3CL protease has recently been described (Hamill et al., 2006). The development of protease assays in a cell-based format, either for pharmaceutical or academic research, poses significantly more substantial hurdles. These include:
-
(i)
Efficient transport of substrate into cells without affecting cellular metabolism. It should be noted that “transfection” reagents that improve cell permeability may weaken the integrity of the membrane and influence cellular activity. Covalent attachment of transport peptides (Vives et al., 1997) can lead to uneven distribution and concentration in the nucleus.
-
(ii)
Efficient and specific cleavage of the substrate within the cell.
-
(iii)
Persistence of fluorophore within the cell after cleavage.
-
(iv)
Sensitive detection by laser or light microscopy detection systems.
-
(v)
Fluorophore compatibility with simultaneous detection of other tracking dyes, e.g. by the green calcium indicator Fluo-4FF.
-
(vi)
Stability to ubiquitous non-specific proteolytic activities (e.g. the proteasome).
The enzyme calpain is a Ca2+-dependent protease that participates in the execution of apoptotic and necrotic forms of cell death (Lankiewicz et al., 2000; Goll et al., 2003; Tan et al., 2006). Calpain overactivation is a feature of numerous neurodegenerative conditions, including ischemia/reperfusion brain injury (Markgraf et al., 1998), Parkinson’s disease (Crocker et al., 2003), Huntington’s disease (Gafni and Ellerby, 2002), and Alzheimer’s disease (Saito et al., 1993). Modification of calpain activity via specific inhibitors provides a potential method of therapeutic intervention in such cases. In addition, the mechanisms, time course, and spatial distribution of calpain activation is poorly understood and is normally inferred from immunodetection of a single calpain breakdown product, such as cleaved α-spectrin. A prime complication to developing effective assays for this enzyme is that it cleaves a variety of substrate proteins of little obvious sequence similarity. It is believed that calpain substrates are targeted primarily by recognition of loop sequences on protein surfaces. For these reasons the development of a useful probe for kinetic detection of calpain activity in cells is both an important and challenging problem. A variety of fluorescent resonance energy transfer (FRET) substrates have been advocated as suitable probes (Mittoo et al., 2003; Tompa et al., 2004; Cuerrier et al., 2005), but these fall far short of the specified criteria.
The combination of CAL Fluor™ dyes with Black Hole Quenchers™ (BHQ’s) is a well-established technology for nucleotide assays (Johanssen and Cook, 2003). A particularly advantageous feature is the readiness of the BHQ quencher, paired with a variety of fluorophores, to adopt ground state complexes, involving a close interaction of the chromophores. This results in highly efficient quenching (Johanssen et al., 2002). The CAL Fluor Orange 560 fluorophore was selected because (1) the Ex/Em values (540/561 nm) are well suited to the optical system of the laser scanning confocal microscope employed, and (2) it allows for simultaneous monitoring of other cellular parameters (e.g. intracellular Ca2+) using fluorescent reporters that are excited at shorter wavelengths. A small fluorogenic substrate library was prepared with the CAL Fluor Orange 560/BHQ-2 fluorophore/quencher pair; it comprised calpain cleavage sequences derived from cellular substrates as well as previously described artificial peptide substrates. These probes were then analyzed in vitro for calpain susceptibility and specificity. Further modification has lead to specific cell permeable probes that allow for investigation of calpain activation in cells.
Materials and Methods
Peptide Synthesis
Fmoc-LinkerAm-Champion-1 resin, and all dye and quencher reagents were from Biosearch Technologies, Inc. Fmoc-amino acids were from NovaBiochem. Standard automated peptide synthesis methods were employed (as described in Biancalana et al., 2001). The addition of dye labeled intermediates and fluorophores was performed with 20–50 mg of resin in disposable 2 mL polypropylene syringes equipped with a coarse PE end filter (Porex, GA). Final cleavage of products was with TFA/water/triisopropylsilane (95:2.5:2.5; v/v) for 2 h, and the products were isolated by evaporation. It should be noted that the BHQ function is sensitive to reduction, especially by thiol reagents, and careful testing is required for any construct to confirm adequate stability during cleavage and assay.
General Procedure for Addition of Dye Carboxylates
The peptide resin, 20–50 mg (8–20 μmol), was placed in the syringe reactor, and the resin swollen in DMF. The dye (10 mg, typically 20 μmol) and HOAt (10 mg, 60 μmol, 1,2,3-triazolo-[4,5-b]pyridin-3-ol, 1-hydroxyl-7-azabenzotriazole, TCI America T1673) were placed in a screw capped 1.5 mL vial, DMF (100 μL) and DIPCDI (diisopropylcarbodiimide 10 μL, 65 μmol) were added, the vial sealed, and vortexed vigorously for 2 min. The dye solution was then diluted with DMF (200 μL), mixed and admitted to the reactor. This was covered with foil and shaken overnight. The resin was washed with DMF (at least 10× until eluant became colorless), then treated with 0.5 M acetic anhydride + 0.75 M HOBt in DMF (30 min) to cap residual amino groups (step was omitted for final dye modification before TFA cleavage).
General Procedure for Addition of Dye p-Nitrophenyl carbonates
Performed as above but with HOBt (10 mg) and diisopropylethylamine (10 μL) replacing HOAt and DIPCDI, respectively, as activators.
Fmoc-Lys(BHQ-2)-BioLinker Resin, for C-terminal PEG Variants
3,6,9-Trioxaundecane,1-11-dioic acid (Fluka, 4.8 g, 20 mmol) in dichloromethane (50 mL) was stirred and cooled in an ice-bath. N, N’-dicyclohexylcarbodiimide (4.2 g, 20.4 mmol) was added, the flask re-sealed, and stirring continued overnight. The precipitated urea was removed by filtration, and the filtrate evaporated and dried in vacuo to yield the corresponding anhydride (4.6 g). Fmoc-LinkerAm-OH (2.5 g, 5 mmol) and HOBt (1 g, 7.4 mmol) were dissolved in DMF (20 mL) and BOP (2.2 g, 5 mmol) and N-methylmorpholine (0.6 mL, 5 mmol) were added with mixing. After 2 min this was added to a suspension of 4-methylbenzhydrylamine resin (5 g, 0.6 mmol/g) swollen in DMF (30 mL) and the sealed flask was shaken overnight. The beads were isolated by filtration, washed repeatedly, first with DMF, then dichloromethane and finally methanol, then dried to constant weight. The LinkerAm resin (2 g, ∼1.1 mmol) was swollen in DMF, treated with 30% piperidine in DMF (20 mL; for 5 min, then repeated for 15 min), and the beads were repeatedly washed with DMF to remove piperidine. The above prepared anhydride (1.5 g, 6.8 mmol) and HOBt (1 g, 7.4 mmol) were dissolved in DMF (15 mL) and added to the washed resin, the suspension was shaken overnight, and washed with DMF to give a ninhydrin negative product. Carbonyl diimidazole (1.62 g, 10 mmol) was added to the resin suspended in DMF (10 mL) and shaken for 2 h in a tightly sealed container. The resin was washed very quickly 5× with DMF, then 4,7,10-trioxa-1-13-tridecanediamine (10 mL) and HOBt (3 g) in DMF (10 mL) were rapidly added. The reaction was shaken in a tightly sealed container overnight. The resin was washed repeatedly, first with DMF, then dichloromethane and finally methanol to yield the highly ninhydrin positive amino-BioLinker resin. This resin (140 mg, ∼56 μmol) was swollen in DMF in a syringe reactor, and coupled for 2 h with Fmoc-Lys(BHQ-2)-OH (35 mg, 0.04 mmol), HOBt (10 mg), BOP (30 mg, 66 μmol), and di-isopropylethylamine (20 μmol, 115 μmol) in DMF (for 2 h). After washing with DMF, the resin was capped by reaction with 0.5 M acetic anhydride + 0.75 M HOBt in DMF for 30 min. The resin was then washed and dried (note that cleavage with 95% TFA/5% water for 2 h yielded a single component on HPLC with the desired absorption spectrum and expected m/e).
Enzyme Stability/Digestion Studies
-
(i)
Microtiter plate format. Performed as described elsewhere (Hammill et al., 2006). Note that the signal/noise ratio (S/N) values are more precisely defined as S-N/N, calculated by subtraction from the digested sample of the background signal without enzyme prior to division. For Table I, stock solutions of peptides were prepared (in 1:1 acetonitrile/water), diluted in buffer (0.05 M Tris–HCl buffer pH 7.9 containing 10 mM CaCl2) and arrayed in triplicate, then treated with test enzymes, including Pronase E (protease from Streptomyces griseus, Sigma) as a positive control. Plates were read on a fluorescent plate reader (Tecan, Model Saffire, Austria) at 30 min, cleavage extent indicated as ++ in Table I defined as >75% cleavage, + 10–50% cleavage, − <10% cleavage.
-
(ii)
Digests monitored by real time spectrofluorometry (all calpain and Table II results). Peptide cleavage was monitored kinetically with excitation at 540 nm and emission at 561 nm (10 nm slit widths) using a Perkin Elmer LS50 B fluorimeter at 27 °C with active stirring. FRET substrates were added to 30 mM Tris–HCl buffer pH 7.5 containing 100 μM CaCl2 and 1.5 mM DTT from peptide stocks prepared in DMSO. Human erythrocyte calpain I (Calbiochem) was added from once-thawed aliquots stored at –80 °C and trypsin, α-chymotrypsin, and Pronase E (all from Sigma) were prepared fresh for each experiment.
Cell Based Assays
Primary hippocampal neurons were prepared from 1–2 pairs of E18 rat hippocampi (BrainBits™, LLC, Springfield, IL) by papain dissociation followed by gentle trituration and used at 9–12 days in vitro (DIV). Briefly, hippocampi were washed in 2 mL of Hibernate E™ medium (BrainBits™) and then digested with 2 mg/mL papain (Worthington) in Hibernate E™ for 30 min at 37 °C. Tissue was dispersed manually by 5–10 strokes with a 1 mL pipette and cells were plated onto poly-d-lysine-coated Lab-Tek 8-well chambered cover glasses at a density of 1 × 105 cells/well. Cells were initially plated in Neurobasal medium (Invitrogen) containing B27 supplement, 0.5 mM l-glutamine, 25 μM glutamate, 1% fetal bovine serum, and 1% penicillin/streptomycin. Neurons were maintained at 37 °C in an incubator with a humidified atmosphere of 5% CO2/95% air. On days 4 and 7 after plating, half of the medium was replaced with fresh medium lacking glutamate and serum. Cortical neurons were prepared from 1–2 pairs of E18 rat cortices (BrainBits™) by the same procedure and used at 5 DIV.
Live cell imaging of neurons was performed in a Pascal confocal system (Carl Zeiss AG, Oberkochen, Germany) using an Axiovert 100 M inverted microscope equipped with a 20× air objective and argon (488 nm) and helium/neon (543 nm) lasers. Neurons were loaded with cA or cXe as indicated in figure legends and imaged in a temperature-controlled enclosure at 37 °C. These probes were excited at 543 nm and emitted fluorescence was collected between 560 and 615 nm. Images were acquired at 1–2 min intervals for the duration of each experiment.
Results and Discussion
The Merrifield method of solid phase peptide synthesis (Merrifield, 1963) is ideally suited to the task of synthesizing dye-labeled peptides. First, complex constructs may be assembled in a stepwise manner with efficient incorporation of valuable intermediates. Second, as an intrinsic property of the method, purification is simplified because excess reagents used in fluorophore and quencher addition steps, that would normally be difficult to separate by solution-phase methods, are simply washed away.
Our basic synthesis method (Fig. 1) for the incorporation of novel Black Hole Quenchers™, (Cook et al., 2006) via the corresponding derivative Fmoc-Lys(BHQ-2)-OH (Carter et al., 2004), allows both incorporation at the C-terminus of any desired FRET substrate, as well as integration with preassembled “transporter” sequences or other C-terminal modifications. CAL Fluor dyes provide chemically and enzymatically stable, isomer free variants of standard xanthene dyes. CAL Fluor Orange 560 (Fig. 1, hereafter referred to by the abbreviation CFO) is available with a variety of biocompatible aliphatic spacer arm selections, as shown, and described elsewhere (Reddington and Lyttle, 2005). An isosteric version of the “short” spacer CFO-1 dye (designated as CFO-u1) is incorporated by a urethane bond (formed with a p-nitrophenyl carbonate precursor). Additionally, this variation has been incorporated via the corresponding Fmoc-Lys(CFO-u1)-OH derivative, which allows extension or modification to the amino terminus of the construct. The dye carboxylate forms (CFO-1, CFO-3) are preferentially attached via DIPCDI/HOAt-mediated coupling, although HATU and other reagents are also effective (Carpino et al., 1994).
(a) Schematic representation of the synthesis of FRET calpain substrates incorporating the CAL Fluor Orange 560 dye, C-extension represents either defined PEG d-Ala-Arg addition, and LinkerAM is an amide-producing linker frequently referred to as Rink linker. (b) Different spacer arm forms of the CAL Fluor Orange 560 dye.
An important previous publication (Tompa et al., 2004) derived, from several calpain cleavage sites within natural proteins, a “hybrid” form that was believed to be an optimal, stable and specific substrate (designated cB when provided in our format). The Bradley group (Mittoo et al., 2003) developed a substrate based on the α-spectrin calpain cleavage site, whereas a peptide library screening approach arrived at yet another substrate that was deemed kinetically superior to previous designs (Cuerrier, 2005). A selection of these and other known cleavage-susceptible sequences of proteins were synthesized in CFO-1/BHQ-2 FRET format; Table I shows a partial listing of the most interesting of these targets. These peptides were assessed as calpain substrates, analyzed for their stability to chymotrypsin and trypsin, two of the major proteolytic activities of the cellular proteasome, and tested for the signal/noise ratio (S/N, the ratio of any fluorescence signal (S), to the background Signal, N, measured in wells lacking digesting enzyme; a measure of expected assay sensitivity) obtained on complete digestion by the poorly specific Pronase E protease mixture.
Since many highly hydrophobic sequences (such as cG, cJ, cM) were very poorly soluble, we used extensions at the C-terminus (Fig. 1) to increase aqueous compatibility. The first modification was by addition of a 27-atom C-terminal PEG extension (Cook et al., 1994), which could conveniently be incorporated in this format by a simple two-step route (Rose and Vizzavona, 1999). The second variation was by addition of d-Ala-Arg (aR). A similar solubility-improving ruse (by addition of RRK tails) has been used in substrate libraries (Thomas et al., 2006). The PEG modification appeared highly compatible with in vitro assays, whereas aR forms proved to give inferior results. The variant of cG which incorporated the aR C-terminus and Ac-K(CFO-u1) at the N-terminus was a very poor probe, indeed (Table I). Both of these C-terminal modifications proved to inhibit membrane transport. Formal interchange of dye and quencher was equally undesirable (data not shown).
On the basis of the high stability of cA and cJ to potentially non-specific enzymatic activities, these substrates were initially chosen for evaluation in cells. cA, but not cJ, proved to enter cells without the addition of a transporter sequence, and fluorescence was observed upon activation of calpain pathways by the addition of the calcium ionophore ionomycin (Fig. 2a, full characterization of cA is to be published elsewhere). Of even greater interest, there was no obvious decline in signal after 2 h (data not shown), suggesting that on cleavage the fluorescent fragment generated in the cell is rendered incapable of crossing the membrane. In support of this idea, the synthetic peptide CFO-GSG-OH (corresponding to the predicted fluorescent cleavage product) was found to be cell impermeable (Fig. 2b). A further non-quenched analog of cA was prepared with K(BHQ-2) replaced by nor-leucine, a formal elimination of the epsilon N and BHQ moieties. Remarkably, this peptide also could not enter cells. These findings strongly suggest that the BHQ-2 modification is actively involved in transporting the probe across the membrane. On cleavage, the CFO fragment then lacks this transport facilitating feature, and is retained. It is also noteworthy that introduction of the known HIV tat protein Arg-rich transporter sequence, GRKKRRQRRRPQ (Vives et al., 1997), as a C-terminal extension to cA, produced a variant which afforded an approx. 5× greater sensitivity of detection in cell assays (data not shown).
Intracellular fluorescence of cA following treatment of primary hippocampal neurons with ionomycin. In (a), hippocampal neurons (transmission image, left) were preloaded with cA (1 μM) for 30 min in a Ca2+-free experimental buffer and treatment with ionomycin (5 μM) followed by the addition of CaCl2 (0.2 mM) lead to the appearance of intracellular cA fluorescence (right). In (b), hippocampal neurons (transmission image, left) were exposed to CFO-GSG-free acid (10 μM), the predicted fluorescent calpain cleavage product of cA. Fluorescence remained extracellular and was excluded from cell bodies (right). Dark areas in B correspond to cell bodies while bright spots correspond to probe binding to cellular debris.
Nevertheless, although cA and specified variants provided good stability and transport properties, detailed kinetic investigation showed that calpain digestions proceeded only partially, and addition of fresh enzyme, or pronase E, restarted digestion (Fig. 3, cA). One plausible explanation supposes that either the substrate or the digestion products are inhibiting the enzyme, yet we were unable to demonstrate this phenomenon with expected cleavage products.
Since the PLFAER substrate (Table I, cW) was far more sensitive to calpain digestion compared to cA, we proceeded to prepare and evaluate the set of variants shown in Table II. The relative merits of these substrates as probes for calpain and their relative stabilities to trypsin and chymotrypsin were assessed spectrofluorometrically in vitro. PLFAER (cW) was confirmed to be a far superior calpain substrate compared to cA (Fig. 3). As expected, the poor stability of cW to trypsin was dramatically improved in cX by addition of a Pro residue between the Arg residue and the Lys(BHQ-2) (Fig. 4). Remarkably, this addition also improved the peptide as a calpain substrate. Taking cX as a parent, the effect of substitution of the F residue was examined (Table II). Improved stability to chymotrypsin was expected for G, L, V (cXa to cXc), or cyclohexyl-alanine (cXi) variants (Fig. 5a–c, g). cXa was rather similar as a calpain substrate to its parent, but its chymotrypsin lability was affected remarkably little. Substitution with chA (cXi) also had little effect on proteolysis by either enzyme (Fig. 5g), whereas Val in this position dramatically reduced the effectiveness of calpain processing (cXc, Fig. 5c). cXb, the Phe transformed to Leu analog, was at least as an effective calpain substrate as its parent, with considerably improved chymotryptic stability (Fig. 5b). Optimization of this series was completed by examining the effect of modifying the CAL Fluor dye composition of cXb by variations of the CFO linker component. This phase was embarked on with no great expectations, since all versions were known to be stable to chymotrypsin, either in the native dye, or when attached to substrates. Indeed, cXf afforded little improvement (Fig. 5g), but cXg (Fig. 5f) was significantly improved. cXe, bearing the CFO-3 variation (Fig. 1), attached by a far longer spacer, proved not only very much more stable to chymotrypsin but was also an improved calpain substrate (Fig. 5d depicts the kinetic profiles shown by the three enzymes). The reason for the improved chymotrypsin stability of cXe is unclear. The observation that all peptides shown in Table II give very high S/N values, whatever the sequence or form of the CFO dyes, suggests that all adopt a ground state complex in which the dye and quencher are in close proximity. This form results in efficient static quenching that augments the standard dynamic Forster effect (which on its own decreases dramatically with increasing separation). It is tempting to speculate that the longer flexible CFO-3 spacer incorporated in cXe promotes formation of a calpain-preferred/chymotrypsin unfavorable conformation of the substrate peptide.
Spectrofluorometric monitoring of calpain I-, chymotrypsin- and trypsin-mediated cleavage of cX variants described in Table 2 (conditions as per legend to Fig. 3). (a) shows data from digests of cXa, (b) shows cXb, (c) shows cXc, (d) shows cXe, (e) shows cXf, (f) shows cXg, (g) shows cXi.
cXe, like cA, proved to enter cells without a transporter sequence (Fig. 6). However, individual cells accumulated cXe to different degrees and cXe fluorescence in some cells could only be visualized by increasing the laser intensity. While it is possible that cell-to-cell variation in cXe fluorescence reflects a difference in the level of basal protease activity, we observed similar signal variability of permanently labeled fluorescent peptides (data not shown). This suggests that there are intrinsic differences in the ability of individual cells to accumulate exogenous peptides. Continuing studies are examining the effects of C-terminal modifications to enhance cell permeability of cA and cXe. Detailed studies are also proceeding on the effectiveness of these probes in identifying calpain activity in cells in response to various stimuli. Results are still preliminary and will be reported elsewhere.
Intracellular fluorescence of cXe following treatment of primary cortical neurons with ionomycin. Cortical neurons (transmission image, left) were preloaded with cXe (2 μM) for 40 min, washed to remove non-internalized cXe, then treated with ionomycin (5 μM). This resulted in increased intracellular fluorescence that was variable and accumulated in nuclei as cells underwent apoptotic rounding.
Conclusions
This study shows the usefulness of Merrifield’s method of SPPS in preparing and optimizing complex FRET calpain substrate sequences bearing novel fluorophores and quenchers. Particular noteworthy is the fact that the bis-diazo Black Hole Quencher (BHQ-2) not only provides excellent quenching of the CAL Fluor Orange 560 dye (however configured); but also has a very positive influence on these peptides by promoting cross-membrane transport.
Developing suitable FRET peptide substrates as probes of select enzymes in cells, or in vivo, is a complex problem, and requires careful optimization. Two excellent candidate probes have been discovered which appear to satisfy the specified criteria of transportability, selectivity, signal persistence, and stability.
REFERENCES
Biancalana S., Hudson D., Songster M. F., Thompson S. A. (2001) Lett. Peptide Sci. 7:291–297
Carpino, L. A., El-Faham, A., Truran, G. A., Minor, C. A., Kates, S. A., Griffin, G. W., Shroff, H., Triolo, S. A., and Albericio, F.: 1994, in: R. Epton (ed.), Innovations and Perspectives in Solid-Phase Synthesis, Proceedings of the Third International Symposium, Mayflower Pubs, Oxford, pp. 95–104
Carter, T. G., Cook, R. M., Hudson, D., Johansson, M. K., Lyttle, M. H., Reddington, M. and Walton, T.: 2004, in R. Epton (ed.), Innovations and Perspectives in Solid-Phase Synthesis, Proceedings of the Eighth International Symposium, Mayflower Pubs., London, pp. 103–106
Cook R. M., Adams J. H., Hudson D. (1994) Tetrahedron Lett. 35:6777–6780
Cook, R. M., Lyttle, M. and Dick, D.: 2006, US Patent US 7,019,129 B1
Crocker S. J., Smith P. D., Jackson-Lewis V., Lamba W. R., Hayley S. P., Grimm E., Callaghan S. M., Slack R. S., Melloni E., Przedborski S., Robertson G. S., Anisman H., Merali Z., Park D. S. (2003) J. Neurosci. 23:4081–4091
Cuerrier D., Moldoveanu T., Davies P. L. (2005) J. Biol. Chem. 280:40632–40641
Fischer S., Vandekerckhove J., Ampe C., Traub P., Weber K. (1986) Biol. Chem. Hoppe Seyler 367:1147–1152
Gafni J., Ellerby L. M. (2002) J. Neurosci. 22:4842–4849
Goll D. E., Thompson V. F., Li H., Wei W., Cong J. (2003) Physiol. Rev. 83: 731–801
Hamill P., Hudson D., Kao R. Y., Chow P., Raj M., Xu H., Richler M. J., Jean F. (2006) Biol. Chem. 387:1063–1074
Johanssen M. K., Cook R. M. (2003) Chem. Eur. J. 9:3466
Johanssen M. K., Fidder H., Dick D., Cook R. M. (2002) J. Am. Chem. Soc. 124:6950
Kakkar R., Raju R. V., Sharma R. K. (1998) Arch. Biochem. Biophys. 358:320–328
Lankiewicz S., Marc L. C., Truc B. N., Krohn A. J., Poppe M., Cole G. M., Saido T. C., Prehn J. H. (2000) J. Biol. Chem. 275:17064–17071
Markgraf C. G., Velayo N. L., Johnson M. P., McCarty D. R., Medhi S., Koehl J. R., Chmielewski P. A., Linnik M. D. (1998) Stroke 29:152–158
Merrifield R. B. (1963) J. Am. Chem. Soc. 85:2149
Mittoo S., Sundstrom L. E., Bradley M. (2003) Anal. Biochem. 319:234–238
Reddington, M. and Lyttle, M.: 2005, US Patent Application US 2005/0170363 A1
Rose K., Vizzavona J. (1999) J. Am. Chem. Soc. 121:7034–7038
Saito K., Elce J. S., Hamos J. E., Nixon R. A. (1993) Proc. Natl. Acad. Sci. USA 90:2628–2632
Tan Y., Wu C., De V. T., Greer P. A. (2006) J. Biol. Chem. 281:17689–17698
Thomas D. A., Francis P., Smith C., Ratcliffe S., Ede N. J., Kay C., Wayne G., Martin S. L., Moore K., Amour A., Hooper N. M. (2006) Proteomics 6:2112–2120
Tompa P., Buzder-Lantos P., Tantos A., Farkas A., Szilagyi A., Banoczi Z., Hudecz F., Friedrich P. (2004) J. Biol. Chem. 279:20775–20785
Vives E., Granier C., Prevot P., Lebleu B. (1997) Lett. Peptide Sci. 4:429–436
ACKNOWLEDGMENTS
The authors gratefully acknowledge Ronald M. Cook for his support, and Mark Reddington for helpful discussions and development of many of the CAL Fluor dye spacer variations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Polster, B.M., Arze, R., Lyttle, M.H. et al. Solid Phase Synthesis of Dual Labeled Peptides: Development of Cell Permeable Calpain Specific Substrates. Int J Pept Res Ther 13, 83–91 (2007). https://doi.org/10.1007/s10989-006-9049-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-006-9049-9