Abstract
In bottom–up self-assembly, DNA nanotechnology plays a vital role in the development of novel materials and promises to revolutionize nanoscale manufacturing technologies. DNA shapes exhibit many versatile characteristics, such as their addressability and programmability, which can be used for determining the organization of nanoparticles. Furthermore, the precise design of DNA tiles and origami provides a promising technique to synthesize various complex desired architectures. These nanoparticle-based structures with targeted organizations open the possibility to specific applications in sensing, optics, catalysis, among others. Here we review progress in the development and design of DNA shapes for the self-assembly of nanoparticles and discuss the broad range of applications for these architectures.




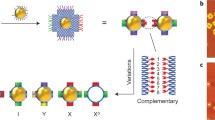
Reproduced from He et al. [36], with permission, copyright 2008, Nature Publishing Group





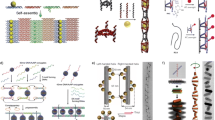
a, b Reproduced from Rothemund [8], with permission, copyright 2006, Nature Publishing Group





Reproduced from Tian et al. [19] with permission, copyright 2016, Nature Publishing Group






Similar content being viewed by others
References
Seeman NC (1982) Nucleic acid junctions and lattices. J Theor Biol 99(2):237–247. https://doi.org/10.1016/0022-5193(82)90002-9
Holliday R (1964) A mechanism for gene conversion in fungi. Genet Res. https://doi.org/10.1017/s0016672300001233
Kallenbach NR, Ma RI, Seeman NC (1983) An immobile nucleic-acid junction constructed from oligonucleotides. Nature 305(5937):829–831. https://doi.org/10.1038/305829a0
Fu TJ, Seeman NC (1993) DNA double-crossover molecules. Biochemistry 32(13):3211–3220. https://doi.org/10.1021/bi00064a003
LaBean TH, Yan H, Kopatsch J, Liu FR, Winfree E, Reif JH, Seeman NC (2000) Construction, analysis, ligation, and self-assembly of DNA triple crossover complexes. J Am Chem Soc 122(9):1848–1860. https://doi.org/10.1021/ja993393e
Chen JH, Seeman NC (1991) Synthesis from DNA of a molecule with the connectivity of a cube. Nature 350(6319):631–633. https://doi.org/10.1038/350631a0
Zhang YW, Seeman NC (1994) Construction of a DNA-truncated octahedron. J Am Chem Soc 116(5):1661–1669. https://doi.org/10.1021/ja00084a006
Rothemund PWK (2006) Folding DNA to create nanoscale shapes and patterns. Nature 440(7082):297–302. https://doi.org/10.1038/nature04586
Alivisatos AP, Johnsson KP, Peng X, Wilson TE, Loweth CJ, Bruchez MP, Schultz PG (1996) Organization of ‘nanocrystal molecules’ using DNA. Nature 382(6592):609–611. https://doi.org/10.1038/382609a0
Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ (1996) A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 382(6592):607–609. https://doi.org/10.1038/382607a0
Nykypanchuk D, Maye MM, van der Lelie D, Gang O (2008) DNA-guided crystallization of colloidal nanoparticles. Nature 451(7178):549–552. https://doi.org/10.1038/nature06560
Park SY, Lytton-Jean AKR, Lee B, Weigand S, Schatz GC, Mirkin CA (2008) DNA-programmable nanoparticle crystallization. Nature 451(7178):553–556. https://doi.org/10.1038/nature06508
Xiong HM, van der Lelie D, Gang O (2008) DNA linker-mediated crystallization of nanocolloids. J Am Chem Soc 130(8):2442–2443. https://doi.org/10.1021/ja710710j
Xiong H, van der Lelie D, Gang O (2009) Phase behavior of nanoparticles assembled by DNA linkers. Phys Rev Lett 102(1):015504. https://doi.org/10.1103/PhysRevLett.102.015504
Vial S, Nykypanchuk D, Yager KG, Tkachenko AV, Gang O (2013) Linear mesostructures in DNA–nanorod self-assembly. ACS Nano 7(6):5437–5445. https://doi.org/10.1021/nn401413b
Jones MR, Macfarlane RJ, Lee B, Zhang J, Young KL, Senesi AJ, Mirkin CA (2010) DNA–nanoparticle superlattices formed from anisotropic building blocks. Nat Mater 9(11):913–917. https://doi.org/10.1038/nmat2870
Macfarlane RJ, Lee B, Jones MR, Harris N, Schatz GC, Mirkin CA (2011) Nanoparticle superlattice engineering with DNA. Science 334(6053):204–208. https://doi.org/10.1126/science.1210493
Liu W, Tagawa M, Xin HL, Wang T, Emamy H, Li H, Yager KG, Starr FW, Tkachenko AV, Gang O (2016) Diamond family of nanoparticle superlattices. Science 351(6273):582–586. https://doi.org/10.1126/science.aad2080
Tian Y, Zhang Y, Wang T, Xin HL, Li H, Gang O (2016) Lattice engineering through nanoparticle-DNA frameworks. Nat Mater 15(6):654–661. https://doi.org/10.1038/nmat4571
Zhang T, Hartl C, Frank K, Heuer-Jungemann A, Fischer S, Nickels PC, Nickel B, Liedl T (2018) 3D DNA origami crystals. Adv Mater 30(28):e1800273. https://doi.org/10.1002/adma.201800273
Pal S, Deng Z, Ding B, Yan H, Liu Y (2010) DNA-origami-directed self-assembly of discrete silver-nanoparticle architectures. Angew Chem Int Ed Engl 49(15):2700–2704. https://doi.org/10.1002/anie.201000330
Wang YL, Mueller JE, Kemper B, Seeman NC (1991) Assembly and characterization of 5-arm and 6-arm DNA branched junctions. Biochemistry 30(23):5667–5674. https://doi.org/10.1021/bi00237a005
Winfree E, Liu FR, Wenzler LA, Seeman NC (1998) Design and self-assembly of two-dimensional DNA crystals. Nature 394(6693):539–544. https://doi.org/10.1038/28998
He Y, Tian Y, Ribbe AE, Mao C (2006) Highly connected two-dimensional crystals of DNA six-point-stars. J Am Chem Soc 128(50):15978–15979. https://doi.org/10.1021/ja0665141
Zheng J, Birktoft JJ, Chen Y, Wang T, Sha R, Constantinou PE, Ginell SL, Mao C, Seeman NC (2009) From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature 461(7260):74–77. https://doi.org/10.1038/nature08274
Ma RI, Kallenbach NR, Sheardy RD, Petrillo ML, Seeman NC (1986) 3-Arm nucleic-acid junctions are flexible. Nucleic Acids Res 14(24):9745–9753. https://doi.org/10.1093/nar/14.24.9745
Wang X, Seeman NC (2007) Assembly and characterization of 8-arm and 12-arm DNA branched junctions. J Am Chem Soc 129(26):8169–8176. https://doi.org/10.1021/ja0693441
Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH (2003) DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science 301(5641):1882–1884. https://doi.org/10.1126/science.1089389
He Y, Chen Y, Liu HP, Ribbe AE, Mao CD (2005) Self-assembly of hexagonal DNA two-dimensional (2D) arrays. J Am Chem Soc 127(35):12202–12203. https://doi.org/10.1021/ja0541938
He Y, Mao C (2006) Balancing flexibility and stress in DNA nanostructures. Chem Commun (Camb) 9:968–969. https://doi.org/10.1039/b513962g
Ding BQ, Sha RJ, Seeman NC (2004) Pseudohexagonal 2D DNA crystals from double crossover cohesion. J Am Chem Soc 126(33):10230–10231. https://doi.org/10.1021/ja047486u
Mao CD, Sun WQ, Seeman NC (1999) Designed two-dimensional DNA Holliday junction arrays visualized by atomic force microscopy. J Am Chem Soc 121(23):5437–5443. https://doi.org/10.1021/ja9900398
Hong F, Jiang S, Wang T, Liu Y, Yan H (2016) 3D framework DNA origami with layered crossovers. Angew Chem Int Ed 55(41):12832–12835. https://doi.org/10.1002/anie.201607050
Kuzuya A, Wang R, Sha R, Seeman NC (2007) Six-helix and eight-helix DNA nanotubes assembled from half-tubes. Nano Lett 7(6):1757–1763. https://doi.org/10.1021/nl070828k
Shih WM, Quispe JD, Joyce GF (2004) A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature 427(6975):618–621. https://doi.org/10.1038/nature02307
He Y, Ye T, Su M, Zhang C, Ribbe AE, Jiang W, Mao C (2008) Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature 452(7184):198-U141. https://doi.org/10.1038/nature06597
Zhang C, Su M, He Y, Zhao X, Fang P-A, Ribbe AE, Jiang W, Mao C (2008) Conformational flexibility facilitates self-assembly of complex DNA nanostructures. Proc Natl Acad Sci USA 105(31):10665–10669. https://doi.org/10.1073/pnas.0803841105
Ong LL, Hanikel N, Yaghi OK, Grun C, Strauss MT, Bron P, Lai-Kee-Him J, Schueder F, Wang B, Wang P, Kishi JY, Myhrvold C, Zhu A, Jungmann R, Bellot G, Ke Y, Yin P (2017) Programmable self-assembly of three-dimensional nanostructures from 10,000 unique components. Nature 552(7683):72–77. https://doi.org/10.1038/nature24648
Ke Y, Ong LL, Sun W, Song J, Dong M, Shih WM, Yin P (2014) DNA brick crystals with prescribed depths. Nat Chem 6(11):994–1002. https://doi.org/10.1038/nchem.2083
Ke Y, Ong LL, Shih WM, Yin P (2012) Three-dimensional structures self-assembled from DNA bricks. Science 338(6111):1177–1183. https://doi.org/10.1126/science.1227268
Zhang C, Li X, Tian C, Yu G, Li Y, Jiang W, Mao C (2014) DNA nanocages swallow gold nanoparticles (AuNPs) to form AuNP@DNA cage core–shell structures. ACS Nano 8(2):1130–1135. https://doi.org/10.1021/nn406039p
Li Y, Liu Z, Yu G, Jiang W, Mao C (2015) Self-assembly of molecule-like nanoparticle clusters directed by DNA nanocages. J Am Chem Soc 137(13):4320–4323. https://doi.org/10.1021/jacs.5b01196
Li HY, Park SH, Reif JH, LaBean TH, Yan H (2004) DNA-templated self-assembly of protein and nanoparticle linear arrays. J Am Chem Soc 126(2):418–419. https://doi.org/10.1021/ja0383367
Xiao SJ, Liu FR, Rosen AE, Hainfeld JF, Seeman NC, Musier-Forsyth K, Kiehl RA (2002) Self assembly of metallic nanoparticle arrays by DNA scaffolding. J Nanopart Res 4(4):313–317. https://doi.org/10.1023/a:1021145208328
Le JD, Pinto Y, Seeman NC, Musier-Forsyth K, Taton TA, Kiehl RA (2004) DNA-templated self-assembly of metallic nanocomponent arrays on a surface. Nano Lett 4(12):2343–2347. https://doi.org/10.1021/nl048635+
Pinto YY, Le JD, Seeman NC, Musier-Forsyth K, Taton TA, Kiehl RA (2005) Sequence-encoded self-assembly of multiple-nanocomponent arrays by 2D DNA scaffolding. Nano Lett 5(12):2399–2402. https://doi.org/10.1021/nl0515495
Zhang JP, Liu Y, Ke YG, Yan H (2006) Periodic square-like gold nanoparticle arrays templated by self-assembled 2D DNA nanogrids on a surface. Nano Lett 6(2):248–251. https://doi.org/10.1021/nl052210l
Sharma J, Chhabra R, Liu Y, Ke YG, Yan H (2006) DNA-templated self-assembly of two-dimensional and periodical gold nanoparticle arrays. Angew Chem Int Ed 45(5):730–735. https://doi.org/10.1002/anie.200503208
Zheng J, Constantinou PE, Micheel C, Alivisatos AP, Kiehl RA, Seeman NC (2006) Two-dimensional nanoparticle arrays show the organizational power of robust DNA motifs. Nano Lett 6(7):1502–1504. https://doi.org/10.1021/nl060994c
Sharma J, Chhabra R, Cheng A, Brownell J, Liu Y, Yan H (2009) Control of self-assembly of DNA tubules through integration of gold nanoparticles. Science 323(5910):112–116. https://doi.org/10.1126/science.1165831
Maye MM, Nykypanchuk D, van der Lelie D, Gang O (2007) DNA-regulated micro- and nanoparticle assembly. Small 3(10):1678–1682. https://doi.org/10.1002/smll.200700357
Maye MM, Nykypanchuk D, Cuisinier M, van der Lelie D, Gang O (2009) Stepwise surface encoding for high-throughput assembly of nanoclusters. Nat Mater 8(5):388–391. https://doi.org/10.1038/nmat2421
Song M, Ding Y, Zerze H, Snyder MA, Mittal J (2018) Binary superlattice design by controlling DNA-mediated interactions. Langmuir 34(3):991–998. https://doi.org/10.1021/acs.langmuir.7b02835
Zhang YG, Lu F, Yager KG, van der Lelie D, Gang O (2013) A general strategy for the DNA-mediated self-assembly of functional nanoparticles into heterogeneous systems. Nat Nanotechnol 8(11):865–872. https://doi.org/10.1038/nnano.2013.209
Zhang YG, Pal S, Srinivasan B, Vo T, Kumar S, Gang O (2015) Selective transformations between nanoparticle superlattices via the reprogramming of DNA-mediated interactions. Nat Mater 14(8):840–847. https://doi.org/10.1038/Nmat4296
Srinivasan B, Vo T, Zhang Y, Gang O, Kumar S, Venkatasubramanian V (2013) Designing DNA-grafted particles that self-assemble into desired crystalline structures using the genetic algorithm. Proc Natl Acad Sci USA 110(46):18431. https://doi.org/10.1073/pnas.1316533110
Vo T, Venkatasubramanian V, Kumar S, Srinivasan B, Pal S, Zhang Y, Gang O (2015) Stoichiometric control of DNA-grafted colloid self-assembly. Proc Natl Acad Sci USA 112(16):4982–4987. https://doi.org/10.1073/pnas.1420907112
Damasceno PF, Engel M, Glotzer SC (2012) Predictive self-assembly of polyhedra into complex structures. Science 337(6093):453–457. https://doi.org/10.1126/science.1220869
Lu F, Vo T, Zhang Y, Frenkel A, Yager KG, Kumar S, Gang O (2019) Unusual packing of soft-shelled nanocubes. Sci Adv 5(5):eaaw2399. https://doi.org/10.1126/sciadv.aaw2399
Lu F, Yager KG, Zhang Y, Xin H, Gang O (2015) Superlattices assembled through shape-induced directional binding. Nat Commun 6(1):6912. https://doi.org/10.1038/ncomms7912
O’Brien MN, Jones MR, Lee B, Mirkin CA (2015) Anisotropic nanoparticle complementarity in DNA-mediated co-crystallization. Nat Mater 14:833. https://doi.org/10.1038/nmat4293
Andersen ES, Dong M, Nielsen MM, Jahn K, Lind-Thomsen A, Mamdouh W, Gothelf KV, Besenbacher F, Kjems J (2008) DNA origami design of dolphin-shaped structures with flexible tails. ACS Nano 2(6):1213–1218. https://doi.org/10.1021/nn800215j
Qian L, Wang Y, Zhang Z, Zhao J, Pan D, Zhang Y, Liu Q, Fan C, Hu J, He L (2006) Analogic China map constructed by DNA. Chin Sci Bull 51(24):2973–2976. https://doi.org/10.1007/s11434-006-2223-9
Douglas SM, Chou JJ, Shih WM (2007) DNA-nanotube-induced alignment of membrane proteins for NMR structure determination. Proc Natl Acad Sci USA 104(16):6644–6648. https://doi.org/10.1073/pnas.0700930104
Mathieu F, Liao SP, Kopatscht J, Wang T, Mao CD, Seeman NC (2005) Six-helix bundles designed from DNA. Nano Lett 5(4):661–665. https://doi.org/10.1021/nl050084f
Kuzyk A, Schreiber R, Fan Z, Pardatscher G, Roller EM, Hogele A, Simmel FC, Govorov AO, Liedl T (2012) DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature 483(7389):311–314. https://doi.org/10.1038/nature10889
Shen X, Song C, Wang J, Shi D, Wang Z, Liu N, Ding B (2012) Rolling up gold nanoparticle-dressed DNA origami into three-dimensional plasmonic chiral nanostructures. J Am Chem Soc 134(1):146–149. https://doi.org/10.1021/ja209861x
Ke Y, Sharma J, Liu M, Jahn K, Liu Y, Yan H (2009) Scaffolded DNA origami of a DNA tetrahedron molecular container. Nano Lett 9(6):2445–2447. https://doi.org/10.1021/nl901165f
Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, Mamdouh W, Golas MM, Sander B, Stark H, Oliveira CLP, Pedersen JS, Birkedal V, Besenbacher F, Gothelf KV, Kjems J (2009) Self-assembly of a nanoscale DNA box with a controllable lid. Nature 459(7243):73–U75. https://doi.org/10.1038/nature07971
Dietz H, Douglas SM, Shih WM (2009) Folding DNA into twisted and curved nanoscale shapes. Science 325:725–730. https://doi.org/10.1126/science.1174251
Douglas SM, Dietz H, Liedl T, Hoegberg B, Graf F, Shih WM (2009) Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459(7245):414–418. https://doi.org/10.1038/nature08016
Douglas SM, Marblestone AH, Teerapittayanon S, Vazquez A, Church GM, Shih WM (2009) Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res 37(15):5001–5006. https://doi.org/10.1093/nar/gkp436
Ke Y, Douglas SM, Liu M, Sharma J, Cheng A, Leung A, Liu Y, Shih WM, Yan H (2009) Multilayer DNA origami packed on a square lattice. J Am Chem Soc 131(43):15903–15908. https://doi.org/10.1021/ja906381y
Ke Y, Voigt NV, Gothelf KV, Shih WM (2012) Multilayer DNA origami packed on hexagonal and hybrid lattices. J Am Chem Soc 134(3):1770–1774. https://doi.org/10.1021/ja209719k
Han D, Pal S, Nangreave J, Deng Z, Liu Y, Yan H (2011) DNA origami with complex curvatures in three-dimensional space. Science 332(6027):342–346. https://doi.org/10.1126/science.1202998
Yang Y, Han D, Nangreave J, Liu Y, Yan H (2012) DNA origami with double-stranded DNA as a unified scaffold. ACS Nano 6(9):8209–8215. https://doi.org/10.1021/nn302896c
Sobczak J-PJ, Martin TG, Gerling T, Dietz H (2012) Rapid folding of DNA into nanoscale shapes at constant temperature. Science 338(6113):1458–1461. https://doi.org/10.1126/science.1229919
Iinuma R, Ke Y, Jungmann R, Schlichthaerle T, Woehrstein JB, Yin P (2014) Polyhedra self-assembled from DNA tripods and characterized with 3D DNA-PAINT. Science 344(6179):65–69. https://doi.org/10.1126/science.1250944
Tian Y, Wang T, Liu W, Xin HL, Li H, Ke Y, Shih WM, Gang O (2015) Prescribed nanoparticle cluster architectures and low-dimensional arrays built using octahedral DNA origami frames. Nat Nanotechnol 10(7):637–644. https://doi.org/10.1038/nnano.2015.105
Veneziano R, Ratanalert S, Zhang K, Zhang F, Yan H, Chiu W, Bathe M (2016) Designer nanoscale DNA assemblies programmed from the top down. Science 352(6293):1534. https://doi.org/10.1126/science.aaf4388
Liu X, Zhang F, Jing X, Pan M, Liu P, Li W, Zhu B, Li J, Chen H, Wang L, Lin J, Liu Y, Zhao D, Yan H, Fan C (2018) Complex silica composite nanomaterials templated with DNA origami. Nature 559(7715):593–598. https://doi.org/10.1038/s41586-018-0332-7
Linh N, Doeblinger M, Liedl T, Heuer-Jungemann A (2019) DNA-origami-templated silica growth by sol–gel chemistry. Angew Chem Int Ed 58(3):912–916. https://doi.org/10.1002/anie.201811323
Schreiber R, Santiago I, Ardavan A, Turberfield AJ (2016) Ordering gold nanoparticles with DNA origami nanoflowers. ACS Nano 10(8):7303–7306. https://doi.org/10.1021/acsnano.6b03076
Zhao Z, Jacovetty EL, Liu Y, Yan H (2011) Encapsulation of gold nanoparticles in a DNA origami cage. Angew Chem Int Ed Engl 50(9):2041–2044. https://doi.org/10.1002/anie.201006818
Shen C, Lan X, Lu X, Meyer TA, Ni W, Ke Y, Wang Q (2016) Site-specific surface functionalization of gold nanorods using DNA origami clamps. J Am Chem Soc 138(6):1764–1767. https://doi.org/10.1021/jacs.5b11566
Stearns LA, Chhabra R, Sharma J, Liu Y, Petuskey WT, Yan H, Chaput JC (2009) Template-directed nucleation and growth of inorganic nanoparticles on DNA scaffolds. Angew Chem Int Ed Engl 48(45):8494–8496. https://doi.org/10.1002/anie.200903319
Bui H, Onodera C, Kidwell C, Tan Y, Graugnard E, Kuang W, Lee J, Knowlton WB, Yurke B, Hughes WL (2010) Programmable periodicity of quantum dot arrays with DNA origami nanotubes. Nano Lett 10(9):3367–3372. https://doi.org/10.1021/nl101079u
Chhabra R, Sharma J, Ke Y, Liu Y, Rinker S, Lindsay S, Yan H (2007) Spatially addressable multiprotein nanoarrays templated by aptamer-tagged DNA nanoarchitectures. J Am Chem Soc 129(34):10304–10305. https://doi.org/10.1021/ja072410u
Sharma J, Chhabra R, Andersen CS, Gothelf KV, Yan H, Liu Y (2008) Toward reliable gold nanoparticle patterning on self-assembled DNA nanoscaffold. J Am Chem Soc 130(25):7820–7821. https://doi.org/10.1021/ja802853r
Schreiber R, Do J, Roller EM, Zhang T, Schuller VJ, Nickels PC, Feldmann J, Liedl T (2014) Hierarchical assembly of metal nanoparticles, quantum dots and organic dyes using DNA origami scaffolds. Nat Nanotechnol 9(1):74–78. https://doi.org/10.1038/nnano.2013.253
Lin ZW, Xiong Y, Xiang ST, Gang O (2019) Controllable covalent-bound nanoarchitectures from DNA frames. J Am Chem Soc 141(17):6797–6801. https://doi.org/10.1021/jacs.9b01510
Liu W, Halverson J, Tian Y, Tkachenko AV, Gang O (2016) Self-organized architectures from assorted DNA-framed nanoparticles. Nat Chem 8(9):867–873. https://doi.org/10.1038/nchem.2540
Liu WY, Mahynski NA, Gang O, Panagiotopoulos AZ, Kumar SK (2017) Directionally interacting spheres and rods form ordered phases. ACS Nano 11(5):4950–4959. https://doi.org/10.1021/acsnano.7b01592
Tian Y, Lhermitte JR, Bai L, Vo T, Xin HL, Li H, Li R, Fukuto M, Yager KG, Kahn JS, Xiong Y, Minevich B, Kumar SK, Gang O (2020) Ordered three-dimensional nanomaterials using DNA-prescribed and valence-controlled material voxels. Nat Mater. https://doi.org/10.1038/s41563-019-0550-x
Emamy H, Gang O, Starr FW (2019) The stability of a nanoparticle diamond lattice linked by DNA. Nanomaterials (Basel) 9(5):661. https://doi.org/10.3390/nano9050661
Mastroianni AJ, Claridge SA, Alivisatos AP (2009) Pyramidal and chiral groupings of gold nanocrystals assembled using DNA scaffolds. J Am Chem Soc 131(24):8455–8459. https://doi.org/10.1021/ja808570g
Yan W, Xu L, Xu C, Ma W, Kuang H, Wang L, Kotov NA (2012) Self-assembly of chiral nanoparticle pyramids with strong R/S optical activity. J Am Chem Soc 134(36):15114–15121. https://doi.org/10.1021/ja3066336
Pal S, Deng Z, Wang H, Zou S, Liu Y, Yan H (2011) DNA directed self-assembly of anisotropic plasmonic nanostructures. J Am Chem Soc 133(44):17606–17609. https://doi.org/10.1021/ja207898r
Lan X, Chen Z, Dai G, Lu X, Ni W, Wang Q (2013) Bifacial DNA origami-directed discrete, three-dimensional, anisotropic plasmonic nanoarchitectures with tailored optical chirality. J Am Chem Soc 135(31):11441–11444. https://doi.org/10.1021/ja404354c
Shen X, Zhan P, Kuzyk A, Liu Q, Asenjo-Garcia A, Zhang H, de Abajo FJ, Govorov A, Ding B, Liu N (2014) 3D plasmonic chiral colloids. Nanoscale 6(4):2077–2081. https://doi.org/10.1039/c3nr06006c
Lan X, Lu X, Shen C, Ke Y, Ni W, Wang Q (2015) Au nanorod helical superstructures with designed chirality. J Am Chem Soc 137(1):457–462. https://doi.org/10.1021/ja511333q
Tian C, Cordeiro MAL, Lhermitte J, Xin HLL, Shani L, Liu MZ, Ma CL, Yeshurun Y, DiMarzio D, Gang O (2017) Supra-nanoparticle functional assemblies through programmable stacking. ACS Nano 11(7):7036–7048. https://doi.org/10.1021/acsnano.7b02671
Zhang H, Li M, Wang K, Tian Y, Chen J-S, Fountaine KT, DiMarzio D, Liu M, Cotlet M, Gang O (2020) Polarized single-particle quantum dot emitters through programmable cluster assembly. ACS Nano 14(2):1369–1378. https://doi.org/10.1021/acsnano.9b06919
Zhou C, Duan X, Liu N (2015) A plasmonic nanorod that walks on DNA origami. Nat Commun 6:8102. https://doi.org/10.1038/ncomms9102
Urban MJ, Zhou C, Duan X, Liu N (2015) Optically resolving the dynamic walking of a plasmonic walker couple. Nano Lett 15(12):8392–8396. https://doi.org/10.1021/acs.nanolett.5b04270
Kuzyk A, Schreiber R, Zhang H, Govorov AO, Liedl T, Liu N (2014) Reconfigurable 3D plasmonic metamolecules. Nat Mater 13(9):862–866. https://doi.org/10.1038/nmat4031
Kuzyk A, Yang Y, Duan X, Stoll S, Govorov AO, Sugiyama H, Endo M, Liu N (2016) A light-driven three-dimensional plasmonic nanosystem that translates molecular motion into reversible chiroptical function. Nat Commun 7:10591. https://doi.org/10.1038/ncomms10591
Kuzyk A, Urban MJ, Idili A, Ricci F, Liu N (2017) Selective control of reconfigurable chiral plasmonic metamolecules. Sci Adv. https://doi.org/10.1126/sciadv.1602803
Funck T, Nicoli F, Kuzyk A, Liedl T (2018) Sensing picomolar concentrations of RNA using switchable plasmonic chirality. Angew Chem Int Ed 57(41):13495–13498. https://doi.org/10.1002/anie.201807029
Shen C, Lan X, Zhu C, Zhang W, Wang L, Wang Q (2017) Spiral patterning of Au nanoparticles on Au nanorod surface to form chiral AuNR@AuNP helical superstructures templated by DNA origami. Adv Mater. https://doi.org/10.1002/adma.201606533
Urban MJ, Dutta PK, Wang P, Duan X, Shen X, Ding B, Ke Y, Liu N (2016) Plasmonic toroidal metamolecules assembled by DNA origami. J Am Chem Soc 138(17):5495–5498. https://doi.org/10.1021/jacs.6b00958
Shen X, Asenjo-Garcia A, Liu Q, Jiang Q, García de Abajo FJ, Liu N, Ding B (2013) Three-dimensional plasmonic chiral tetramers assembled by DNA origami. Nano Lett 13(5):2128–2133. https://doi.org/10.1021/nl400538y
Schreiber R, Luong N, Fan Z, Kuzyk A, Nickels PC, Zhang T, Smith DM, Yurke B, Kuang W, Govorov AO, Liedl T (2013) Chiral plasmonic DNA nanostructures with switchable circular dichroism. Nat Commun 4(1):2948. https://doi.org/10.1038/ncomms3948
Acuna GP, Möller FM, Holzmeister P, Beater S, Lalkens B, Tinnefeld P (2012) Fluorescence enhancement at docking sites of DNA-directed self-assembled nanoantennas. Science 338(6106):506. https://doi.org/10.1126/science.1228638
Thacker VV, Herrmann LO, Sigle DO, Zhang T, Liedl T, Baumberg JJ, Keyser UF (2014) DNA origami based assembly of gold nanoparticle dimers for surface-enhanced Raman scattering. Nat Commun 5:3448. https://doi.org/10.1038/ncomms4448
Kuhler P, Roller EM, Schreiber R, Liedl T, Lohmuller T, Feldmann J (2014) Plasmonic DNA-origami nanoantennas for surface-enhanced Raman spectroscopy. Nano Lett 14(5):2914–2919. https://doi.org/10.1021/nl5009635
Simoncelli S, Roller EM, Urban P, Schreiber R, Turberfield AJ, Liedl T, Lohmuller T (2016) Quantitative single-molecule surface-enhanced raman scattering by optothermal tuning of DNA origami-assembled plasmonic nanoantennas. ACS Nano 10(11):9809–9815. https://doi.org/10.1021/acsnano.6b05276
Ko SH, Du K, Liddle JA (2013) Quantum-dot fluorescence lifetime engineering with DNA origami constructs. Angew Chem Int Ed 52(4):1193–1197. https://doi.org/10.1002/anie.201206253
Samanta A, Zhou Y, Zou S, Yan H, Liu Y (2014) Fluorescence quenching of quantum dots by gold nanoparticles: a potential long range spectroscopic ruler. Nano Lett 14(9):5052–5057. https://doi.org/10.1021/nl501709s
Liu J, Geng Y, Pound E, Gyawali S, Ashton JR, Hickey J, Woolley AT, Harb JN (2011) Metallization of branched DNA origami for nanoelectronic circuit fabrication. ACS Nano 5(3):2240–2247. https://doi.org/10.1021/nn1035075
Schreiber R, Kempter S, Holler S, Schuller V, Schiffels D, Simmel SS, Nickels PC, Liedl T (2011) DNA origami-templated growth of arbitrarily shaped metal nanoparticles. Small 7(13):1795–1799. https://doi.org/10.1002/smll.201100465
Pilo-Pais M, Goldberg S, Samano E, Labean TH, Finkelstein G (2011) Connecting the nanodots: programmable nanofabrication of fused metal shapes on DNA templates. Nano Lett 11(8):3489–3492. https://doi.org/10.1021/nl202066c
Helmi S, Ziegler C, Kauert DJ, Seidel R (2014) Shape-controlled synthesis of gold nanostructures using DNA origami molds. Nano Lett 14(11):6693–6698. https://doi.org/10.1021/nl503441v
Sun W, Boulais E, Hakobyan Y, Wang WL, Guan A, Bathe M, Yin P (2014) Casting inorganic structures with DNA molds. Science 346(6210):1258361. https://doi.org/10.1126/science.1258361
Douglas SM, Bachelet I, Church GM (2012) A logic-gated nanorobot for targeted transport of molecular payloads. Science 335(6070):831–834. https://doi.org/10.1126/science.1214081
Liu M, Fu J, Hejesen C, Yang Y, Woodbury NW, Gothelf K, Liu Y, Yan H (2013) A DNA tweezer-actuated enzyme nanoreactor. Nat Commun 4:2127. https://doi.org/10.1038/ncomms3127
Linko V, Eerikainen M, Kostiainen MA (2015) A modular DNA origami-based enzyme cascade nanoreactor. Chem Commun (Camb) 51(25):5351–5354. https://doi.org/10.1039/c4cc08472a
Zhao Z, Fu J, Dhakal S, Johnson-Buck A, Liu M, Zhang T, Woodbury NW, Liu Y, Walter NG, Yan H (2016) Nanocaged enzymes with enhanced catalytic activity and increased stability against protease digestion. Nat Commun 7:10619. https://doi.org/10.1038/ncomms10619
Wang P, Rahman MA, Zhao Z, Weiss K, Zhang C, Chen Z, Hurwitz SJ, Chen ZG, Shin DM, Ke Y (2018) Visualization of the cellular uptake and trafficking of DNA origami nanostructures in cancer cells. J Am Chem Soc 140(7):2478–2484. https://doi.org/10.1021/jacs.7b09024
Kershner RJ, Bozano LD, Micheel CM, Hung AM, Fornof AR, Cha JN, Rettner CT, Bersani M, Frommer J, Rothemund PW, Wallraff GM (2009) Placement and orientation of individual DNA shapes on lithographically patterned surfaces. Nat Nanotechnol 4(9):557–561. https://doi.org/10.1038/nnano.2009.220
Hung AM, Micheel CM, Bozano LD, Osterbur LW, Wallraff GM, Cha JN (2010) Large-area spatially ordered arrays of gold nanoparticles directed by lithographically confined DNA origami. Nat Nanotechnol 5(2):121–126. https://doi.org/10.1038/nnano.2009.450
Chao J, Wang JB, Wang F, Ouyang XY, Kopperger E, Liu HJ, Li Q, Shi JY, Wang LH, Hu J, Wang LH, Huang W, Simmel FC, Fan CH (2019) Solving mazes with single-molecule DNA navigators. Nat Mater 18(3):273–279. https://doi.org/10.1038/s41563-018-0205-3
Kwon PS, Ren S, Kwon S-J, Kizer ME, Kuo L, Zhou F, Zhang F, Kim D, Fraser K, Kramer LD, Seeman NC, Dordick JS, Linhardt RJ, Chao J, Wang X (2020) Designer DNA architecture offers precise and multivalent spatial pattern-recognition for viral sensing and inhibition. Nat Chem 12(1):26–35. https://doi.org/10.1101/608380
Seeman NC, Gang O (2017) Three-dimensional molecular and nanoparticle crystallization by DNA nanotechnology. MRS Bull 42(12):904–912. https://doi.org/10.1557/mrs.2017.280
Zhang YN, Chao J, Liu HJ, Wang F, Su S, Liu B, Zhang L, Shi JY, Wang LH, Huang W, Wang LH, Fan CH (2016) Transfer of two-dimensional oligonucleotide patterns onto stereocontrolled plasmonic nanostructures through DNA-origami-based nanoimprinting lithography. Angew Chem Int Ed 55(28):8036–8040. https://doi.org/10.1002/anie.201512022
Acknowledgements
The work was supported by National Natural Science Foundation of China (Grant nos. 21971109 and 21834004), Jiangsu Youth Fund (Grant no. BK20180337), the Fundamental Research Funds for the Central Universities (Grant no. 14380151), and the US Department of Energy, Office of Basic Energy Sciences (Grant DE-SC0008772). The research carried out at the Center for Functional Nanomaterials, Brookhaven National Laboratory was supported by US Department of Energy, Office of Science Facility, under Contract No. DE-SC0012704. This work was also supported by the Program for Innovative Talents and Entrepreneur in Jiangsu (No. 133181), Shenzhen International Cooperation Research Project (No. GJHZ20180930090602235) and Nanjing Science and Technology Innovation Project for Oversea Scholars’ Merit Funding (No. 133170).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection “DNA Nanotechnology: From Structure to Functionality”; edited by Chunhai Fan, Yonggang Ke.
Rights and permissions
About this article
Cite this article
Ma, N., Minevich, B., Liu, J. et al. Directional Assembly of Nanoparticles by DNA Shapes: Towards Designed Architectures and Functionality. Top Curr Chem (Z) 378, 36 (2020). https://doi.org/10.1007/s41061-020-0301-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s41061-020-0301-0