Abstract
Loss of heterozygosity (LOH) involving chromosome 16q23.4 occurs frequently in breast tumors, which suggests that this region may contain a tumor suppressor gene. Since ZFP276 is located in this region, we have therefore cloned and performed mutation analysis of its coding region in 70 breast tumors. One silent polymorphism and two alterations predicted to result in amino acid changes were detected. Absence of the wild-type allele in tumors carrying the E530D variant suggests a possible role for this change in tumorigenesis.
Similar content being viewed by others
Introduction
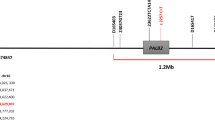
Chromosome 16q is frequently associated with loss of heterozygosity (LOH) in sporadic breast cancer, suggesting that a tumor suppressor gene is located in this region (Chen et al., 1996; Cleton-Jansen et al., 1994, Cleton-Jansen et al. 2001). A number of detailed mapping studies have identified three regions on chromosome 16q (16q22.1, 16q23.2-q24.1, and 16q24.3) with LOH (Brenner and Aldaz, 1997), of which a 650 kb region between genetic markers D16S303 and D16S3026 has been identified as the minimum LOH region at 16q24.3 (Savino et al., 1999; Whitmore et al., 1998).
During the cloning of the mouse Fanconi Anemia Group A cDNA, a novel overlapping penta zinc finger protein Zfp276 was identified (Wong et al., 2000). In this paper, we report the cloning of the human homolog of Zfp276 (ZFP276). ZFP276 maps within the region of LOH to chromosome 16q24.3 in breast tumors, suggesting that it could be a breast cancer tumor suppressor gene. We therefore performed mutation analysis of ZFP276 in 70 breast cancer patients by single strand conformation polymorphism (SSCP) analysis and subsequent sequencing.
Materials and methods
cDNA cloning
EST AA504834 was used as a probe to screen a Caco-2 cDNA library (generously provided by J. Rommens, The Hospital for Sick Children, Toronto, Canada) and a human lymphoblast cDNA library (Strathdee et al., 1992) using conditions as described (Wong et al., 2000).
Northern analysis
The human breast tumor multisample mRNA Northern blot was purchased from Biochain Institute, Inc. Single-strand antisense RNA probe corresponding to nucleotides 122–1084 of the ZFP276 cDNA was synthesized using the MAXIscript In vitro Transcription Kit (Ambion) and labeled with 32P-dUTP (Amersham). Hybridization was performed as described (Wong et al., 2000).
Specimens
Primary tumor samples from randomly selected axillary-node-negative breast cancer patients were immediately snap frozen and stored in liquid nitrogen. RNA was extracted by the guanidinium thiocyanate-cesium chloride gradient method (Chomczynski et al., 1987) Genomic DNA extracted from the peripheral blood lymphocytes of noncancer individuals were used as controls.
Mutation analysis
Ten primer pairs were used to analyze the coding region of ZFP276 (Table 1). cDNA was reverse-transcribed from 100 ng of total cellular RNA with random hexadeoxynucleotide primers and Moloney murine leukemia reverse transcriptase (MMLV-RT). The following PCR conditions were used for each primer set: 2 min at 94°C for denaturation, followed by 30 cycles of 15 s at 94°C for denaturation, 15 s at 56°C for annealing, and 20 s at 72°C for extension. The 33P-ATP-incorporated PCR products were analyzed as described (Gokgoz et al., 2001).
Results
Cloning and analysis of the human homolog for Zfp276
A Caco-2 cDNA library and a human lymphoblast cDNA library were screened with an EST clone showing significant homology to Zfp276. One and four positive clones were isolated from each library, respectively. The clone from the Caco-2 library was 2.1 kb in length, while the four clones from the human lymphoblast cDNA library were identical to each other and were 3.4 kb in length. Sequence alignment shows that the 3.4-kb clone is a longer version of the 2.1-kb clone due to a longer 3’UTR (Genbank Accession numbers AF354755 and AF354756), which probably results from the usage of two different polyadenylation sites at nucleotides 2010 and 3280 respectively.
Sequence analysis predicts an open reading frame of 1619 bp starting from nucleotide 86, which translates to a protein of 539 amino acids in length (Fig. 1a). Sequence alignment with the mouse Zfp276 protein shows that the human ZFP276 homolog is longer in the N-terminal region and has 81% amino acid identity in the region that aligns with Zfp276. All five Cys-Cys-His-His zinc finger domains are conserved, suggesting that this region is functionally significant.
a Comparison of the ZFP276 proteins: Matches between sequences are boxed and shaded, similar amino acids are shaded only. Gaps (indicated by dashes) are introduced to maintain alignment. Conserved cysteines and histidines forming the C2H2 zinc finger domains are marked with asterisks. Amino acid changes detected by SSCP analysis are indicated with arrows. b Genomic structure of ZFP276: Boxes indicate exons, hatched area represents the region that encodes for the C2H2zinc finger domain, arrow indicates the region that overlaps with FANCA gene
As with the mouse homolog, nucleotides 1332–3300 of the ZFP276 cDNA overlaps with the Fanconi Anemia Group A cDNA in a tail-to-tail manner. BLAST analysis of ZFP276 cDNA sequence with the human genome mapped the gene to contig NT_010542.13, which lies within the minimum LOH region in chromosome 16q24.3. Comparison of cDNA and genomic sequences shows that ZFP276 contains ten exons and spans approximately 17 kb of genomic DNA (Fig. 1b).
Expression and mutation analysis of ZFP276
Northern analysis of ZFP276 demonstrates that three transcripts of approximately 5.0 kb, 3.8 kb and 2.4 kb are expressed in breast tumors and in normal breast and lung tissues, and the clones we isolated probably correspond to the two shorter transcripts (Fig. 2a). Although the expression levels of ZFP276 are not significantly different between normal and tumorigenic breast tissues, but since the LOH status at 16q24.3 has not been evaluated in the tissues used for Northern analysis, it is possible that ZFP276 does not play a direct role in the tumorigenesis of these particular samples. However, the expression of ZFP276 in breast tumors suggests that it functions in breast cancer cells.
a Expression of ZFP276 in breast tumor, normal breast and lung tissues: Tissues are indicated by 1 breast tumor, moderately differentiated invasive ductal carcinoma, 2 breast tumor, poorly differentiated invasive ductal carcinoma, 3 breast tumor, moderately differentiated invasive ductal carcinoma, 4 normal breast, 5 normal lung. The same blot was hybridized with human β-actin to control for differences in lane loading. Size markers are indicated on the left of the autoradiograph. b SSCP analysis of E530D (left): Electrophoretic mobility shifts were detected in lane 1 (breast cancer patient) and lane 4 (control), as indicated by arrows. Sample 1 displayed only the altered band. Sequencing analysis of sample 1 on SSCP gel (right): The arrow indicates the base change (G to C)
We therefore investigated tumor specimens from sporadic breast cancer patients by analyzing the coding region of ZFP276 by SSCP and subsequent sequencing. Three different DNA variations were detected in this study: a silent polymorphism at amino acid position 241 (GCG→GCA); an arginine to tryptophan change (R200 W) at position 200 (CGG→TGG), and a glutamic acid to aspartic acid change (E530D) at position 530 (GAG→GAC). R200 W was found to be a common polymorphism occurring in 63 out of 82 patients (77%), of which 23 are heterozygotes (28%) and 40 (49%) are hemi/homozygotes. We were unable to calculate the allelic frequency for this variant, since we could not conclusively determine the LOH status of the hemi/homozygous samples due to unavailability of normal DNA from these patients. R200 W also occurred in similar frequency in the control individuals, as 81 of 97 controls showed this sequence variation (84%), of which 32 were heterozygotes (33%) and 49 were homozygotes (51%). In contrast, the silent polymorphism at amino acid position 241 was found only in one of 70 patients (1.4%), and this patient was hemi/homozygous both for this sequence variation and for arginine at codon 200.
E530D was present in three out of 70 patients. To determine whether this variation is a polymorphism, we analyzed the genomic DNA of 100 individuals from a noncancer population and found the E530D change in two. In the three patients with the E530D change, the wild-type band was not present in the tumor, while both controls had equal intensity of the shifted and nonshifted band in their blood DNAs (Fig. 2b). Interestingly, all three patients with the E530D change were also hemi/homozygous for arginine at codon 200.
Discussion
In this study, we cloned and evaluated ZFP276 as a potential candidate gene in sporadic breast cancer tumorigenesis. As is the case for the mouse homolog, the human ZFP276 gene also overlaps with the Fanconi Anemia Group A gene. Human ZFP276 shares a strong amino acid similarity to its mouse counterpart, especially in the zinc finger domain, suggesting that this gene is functionally important.
We have also identified three sequence alterations in ZFP276, two of which are predicted to result in amino acid changes. While the R200 W variation was common in both the cancer and control groups, the E530D variation was rare. Although the frequency of occurrence of E530D in the cancer group did not deviate significantly from that in a normal control population, it is possible that this nucleotide change may have an effect on tumorigenesis. First, the glutamic acid at position 530 is conserved in the mouse, suggesting possible functional importance. Second, tumours with this variation have only the altered band, while both controls are heterozygous for this change, suggesting that there may be LOH. However, the sample size in this study was too small for us to conclude whether E530D is associated with the occurrence of breast cancer.
It has been documented that single nucleotide polymorphisms (SNPs) can potentially affect the way genes function (Lumb and Danpure, 2000). Nonsynonymous changes (amino acid changes) may affect protein folding and thus the function of the proteins. Single base substitutions can alter or create essential sequence elements for splicing, processing, or translation of human mRNA (Shen et. al., 1999). E530D may confer an increase in cancer risk by affecting gene function. Further studies on the function of the E530D variant in a larger number of breast tumors may clarify the relationship between E530D and sporadic breast cancer.
References
Brenner AJ, Aldaz CM (1997) The genetics of sporadic breast cancer. Prog Clin Biol Res 396:63–82
Chen T, Sahin A, Aldaz CM (1996) Deletion map of chromosome 16q in ductal carcinoma in situ of the breast: refining a putative tumor suppressor gene region. Cancer Res 56:5605–5609
Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159
Cleton-Jansen AM, Moerland EW, Kuipers-Dijkshoorn NJ, Callen DF, Sutherland GR, Hansen B, Devilee P, Cornelisse CJ (1994) At least two different regions are involved in allelic imbalance on chromosome arm 16q in breast cancer. Genes Chromosomes Cancer 9:101–107
Cleton-Jansen AM, Callen DF, Seshadri R, Goldup S, McCallum B, Crawford J, Powell JA, Settasatian C, van Beerendonk H, Moerland EW, Smit VTBHM, Harris WH, Millis R, Morgan NV, Barnes D, Mathew CG, Cornelisse CJ (2001) Loss of heterozygosity mapping at chromosome arm 16q in 712 breast tumors reveals factors that influence delineation of candidate regions. Cancer Res. 61:1171–1177
Gokgoz N, Wunder JS, Mousses S, Eskandarian S, Bell RS, Andrulis IL (2001) Comparison of p53 mutations in patients with localized osteosarcoma and metastatic osteosarcoma. Cancer 92:2181–2189
Lumb NJ, Danpure CJ (2000) Functional synergism between the most common polymorphism in human alanine: glyoxylate aminotransferase and four of the most common disease-causing mutations. J Biol Chem 275:36415–36422
Savino M, d’Apolito M, Centra M, van Beerendonk HM, Cleton-Jansen AM, Whitmore SA, Crawford J, Callen DF, Zelante L, Savoia A (1999) Characterization of copine VII, a new member of the copine family, and its exclusion as a candidate in sporadic breast cancers with loss of heterozygosity at 16q24.3. Genomics 61:219–226
Shen LX, Basilion JP, Stanton VP Jr (1999) Single-nucleotide polymorphisms can cause different structural folds of mRNA. Proc Natl Acad Sci USA 6:7871–7876
Strathdee CA, Gavish H, Shannon WR, Buchwald M (1992) Cloning of cDNAs for Fanconi’s anaemia by functional complementation. Nature 356:763–767
Whitmore SA, Settasatian C, Crawford J, Lower KM, McCallum B, Seshadri R, Cornelisse CJ, Moerland EW, Cleton-Jansen AM, Tipping AJ, Mathew CG, Savnio M, Savoia A, Verlander P, Auerbach AD, Van Berkel C, Pronk JC, Doggett NA, Callen DF (1998) Characterization and screening for mutations of the growth arrest- specific 11 (GAS11) and C16orf3 genes at 16q24.3 in breast cancer. Genomics 52:325–331
Wong JC, Alon N, Norga K, Kruyt FA, Youssoufian H, Buchwald M (2000) Cloning and analysis of the mouse Fanconi anemia group A cDNA and an overlapping penta zinc finger cDNA. Genomics 67:273–283
Acknowledgements
We thank the pathologists and surgeons of Mount Sinai Hospital for specimens. This work was supported by the Canadian Institutes of Health Research (CIHR), the Lombard Insurance Chair in Pediatrics Research, and the Canadian Breast Cancer Research Initiative (ICA).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wong, J.C.Y., Gokgoz, N., Alon, N. et al. Cloning and mutation analysis of ZFP276 as a candidate tumor suppressor in breast cancer. J Hum Genet 48, 668–671 (2003). https://doi.org/10.1007/s10038-003-0088-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-003-0088-1