Corrigendum: Global Transcriptomic Analysis and Function Identification of Malolactic Enzyme Pathway of Lactobacillus paracasei L9 in Response to Bile Stress
- 1Beijing Advanced Innovation Center for Food Nutrition and Human Health, College of Food Science and Nutritional Engineering, China Agricultural University, Beijing, China
- 2Key Laboratory of Functional Dairy, Co-constructed by the Ministry of Education and Beijing Municipality, China Agricultural University, Beijing, China
Tolerance to bile stress is crucial for Lactobacillus paracasei to survive in the intestinal tract and exert beneficial actions. In this work, global transcriptomic analysis revealed that 104 genes were significantly changed (log2FoldChange > 1.5, P < 0.05) in detected transcripts of L. paracasei L9 when exposed to 0.13% Ox-bile. The different expressed genes involved in various biological processes, including carbon source utilization, amino acids and peptide metabolism processes, transmembrane transport, transcription factors, and membrane proteins. It is noteworthy that gene mleS encoding malolactic enzyme (MLE) was 2.60-fold up-regulated. Meanwhile, L-malic acid was proved to enhance bile tolerance, which could be attributed to the intracellular alkalinization caused by MLE pathway. In addition, membrane vesicles were observed under bile stress, suggesting a disturbance in membrane charge without L-malic acid. Then, genetic and physiological experiments revealed that MLE pathway enhanced the bile tolerance by maintaining a membrane balance in L. paracasei L9, which will provide new insight into the molecular basis of MLE pathway involved in bile stress response in Lactic acid bacteria.
Introduction
Lactobacillus paracasei is a common constituent of inhabitants in the human and animal gut, and is extensively used in the food industry as a starter culture for dairy fermentation (Sisto and Lavermicocca, 2012; Di Renzo et al., 2018; Stefanovic et al., 2018). Furthermore, some strains of L. paracasei have attracted additional interest as health-promotion probiotics (De Vrese and Schrezenmeir, 2008). To exert their beneficial effects, high survival rate and stable colonization in the human gastrointestinal tract (GIT) are crucial for L. paracasei strains. However, during digestion, L. paracasei suffers from various challenges in the GIT, such as acidic pH in the stomach and bile salts in the intestine (Corcoran et al., 2008; Bove et al., 2013). Bile salts are known as detergent-like biological compounds with antimicrobial activity, which are capable of disrupting the lipid bilayer structure of cellular membranes. Damage due to bile salts includes protein misfolding and denaturation, DNA or RNA damage, and intracellular acidification (Begley et al., 2005). Therefore, tolerance to bile stress is essential for L. paracasei to survive and colonize in the GIT.
A number of studies have explored the mechanism of bile resistance in members of genus Lactobacillus, such as Lactobacillus plantarum (Hamon et al., 2011b), Lactobacillus rhamnosus (Koskenniemi et al., 2011), and Lactobacillus casei (Hamon et al., 2011a; Alcantara and Zuniga, 2012). Generally, bile salts hydrolases (BSHs) contribute to bile tolerance by hydrolyzing the protonated conjugated bile salts into unconjugated counterparts (Smet et al., 1995). On the other hand, efflux pumps are a common mechanism in lactobacilli, and this could extrude the bile salts from the cell, such as the MDR transporters in L. acidophilus (Pfeiler and Klaenhammer, 2009). In addition, changes in fatty acid metabolism have been described under bile exposure, which could alter the lipid composition of bacterial membranes to reduce bile diffusion (Taranto et al., 2003). Probiotic bacterial species differ from each other in their resistances to bile salts, and even within one species, the strain-specific variation in bile salt tolerance is remarkable. For example, the gene encoding glucosamine-6-phosphate deaminase (NagB) involved in N-acetylneuraminate degradation was 7.54-fold up-regulated in detected transcripts in L. casei BL23. However, this gene was down-regulated in other L. casei strains under bile stress (Hamon et al., 2011a; Alcantara and Zuniga, 2012).
Lactobacillus paracasei L9 (CGMCC No. 9800) originated from healthy human intestine (Yang et al., 2015). Previous research has revealed the health-promoting effects of this strain, such as regulating host immunity (Yang et al., 2015) and preventing intestinal damage (Pan et al., 2014). In this study, RNA-Seq was performed to analyze the global bile stress response and resistance mechanisms in L. paracasei L9. Transcriptomic data revealed that differentially expressed genes (DEGs) participate in various biological processes, such as carbohydrate and amino acid metabolism, transcriptional regulation, fatty acid biosynthesis, and general stress response. It is worth noting that there was an increase in detected transcripts for malolactic enzyme (MLE) pathway genes. The MLE pathway is a secondary fermentation during wine making, in which the dicarboxylic acid L-malate is converted into L-lactate and carbon dioxide by the MLE (Caspritz and Radler, 1983). The MLE pathway enhanced bacterial viability under environmental stress conditions in Streptococcus mutans, such as low pH (Lemme et al., 2010), oxidative stress and starvation (Sheng et al., 2010). In the present work, physiological and genetic experiments were performed to demonstrate that MLE enhanced the bile tolerance by maintaining a membrane balance in L. paracasei L9.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
Bacterial strains and plasmids used in this study are listed in the Supplementary Table S1. L. paracasei L9 was cultured in de Man-Rogosa-Sharpe (MRS) medium anaerobically at 37°C. Growth assays were carried out in chemically defined medium (CDM; Zhou, 2010). CDMM was CDM supplemented with 2.5 mg/mL L-malic acid. Escherichia coli was grown aerobically at 37°C in Luria-Bertani (LB) medium with shaking at 200 rpm. When required, media were supplemented with antibiotics at the following concentrations: 10 μg/mL erythromycin for L. paracasei L9, 100 μg/mL ampicillin for E. coli DH5α. Ox-bile (Sigma, St. Louis, MO, United States) was added at different concentration as needed. To investigate the influence of L-malic acid on the growth of cells under bile stress, overnight cultures of L. paracasei strains were 1% inoculated in CDM and CDMM containing 0.2% Ox-bile. And the OD600 was determined at 2 h intervals. All results were obtained from at least three independent experiments.
Transcriptomics
Overnight cultures of L9 were inoculated at 1% in 50 mL MRS with or without 0.13% Ox-bile, and then incubated anaerobically at 37°C. Cells were harvested when cultures reached mid-exponential phase (OD600 of 0.6, 3–4 × 107 cfu/mL). Three independent biological replicates were performed in this study. Total RNA isolation was performed with TRIzol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. For RNA sample preparation, 3 μg RNA per sample was used as input material. Sequencing libraries were generated using NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (NEB, United States) following the manufacturer’s recommendations and index codes were added to attribute sequences. Library fragments were then purified using AMPure XP system (Beckman Coulter, Beverly, MA, United States), in order to preferentially select cDNA fragments of 150–200 bp in length. Size-selected and adaptor-ligated cDNA was then treated with 3 μl USER Enzyme (NEB, United States) at 37°C for 15 min followed by 5 min at 95°C. PCR was then performed using Phusion High-Fidelity DNA polymerase with Universal PCR primers and Index (X) Primer. Finally, products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq platform and paired-end reads were generated.
Raw data (raw reads) of fastq format were firstly processed through in-house perl scripts. All sequenced reads were aligned to the genome of L. paracasei L9 with GenBank Accession No. NZ_CP012148.1. HTSeq v0.6.1 was used to count the reads numbers mapped to each gene. Differential expression analysis of two groups was performed using the DESeq R package (1.18.0). The resulting P-values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P-value < 0.05 found by DESeq were assigned as differentially expressed.
DNA Manipulation Techniques
Chromosomal DNA from L. paracasei was extracted using TIANamp Bacteria DNA Kit according to the manufacturer’s instructions (Tiangen, Beijing, China). Lysis of L. paracasei was performed by adding 30 mg/mL lysozyme dissolved in TES buffer (50 mM Tris-HCl, 1 mM EDTA, 25% sucrose; pH 8.0), and incubating the suspension at 37°C for 1 h. Miniprep plasmid isolation from E. coli was performed using the Plasmid Mini Kit I (OMEGA Bio-tek, Inc., Doraville, GA, United States). PCR was carried out using Q5®High-Fidelity DNA Polymerase (NEB, United States). Primers used in PCR reactions are listed in Supplementary Table S2. Restriction endonuclease digestions were conducted according to the supplier’s instructions (Takara, Dalian, China). DNA ligation was performed using the T4 DNA Ligation Kit (Thermo Fisher Scientific, Beijing, China). Plasmids were introduced into E. coli DH5α using standard heat shock transformation (Tiangen, Beijing, China). Plasmids were introduced into L. paracasei L9 by electroporation as described previously (Aukrust et al., 1995). DNA sequencing was performed with the BigDye Terminator cycle sequencing kit (Sangon, Beijing, China) and the results were further analyzed with the DNAMAN software package (Version 6.0.3, Lynnon Biosoft, Canada).
Insertional Inactivation of the Gene mleS
In order to study the role of MLE pathway in bile stress, a mleS mutant of L. paracasei L9 was obtained by a single crossover homologous recombination as shown in Supplementary Figure S3. A 650-bp internal region of the mleS gene (300–950 bp) was chosen as a homologous sequence and amplified using the primers pair LPL9_M0797F and LPL9_M0797R with flanking XbaI and EcoRI sites, respectively. The resulting PCR product was restriction enzyme digested and ligated into the corresponding restriction sites of the suicide plasmid pUC19EM (Yang et al., 2017). The recombinant plasmid, designated pUCmleS, was then introduced into L. paracasei L9 by electroporation, and the recombinant strain were cultivated on MRS solid medium containing erythromycin. As the pUCmleS could not replicate in L. paracasei L9, the erythromycin selection pressure resulted the integration of the plasmid into the mleS gene region of the L. paracasei L9 genome. The resulting mutant was designated L9mleS-. To confirm the integration of pUCmleS into the correct genome locus, PCR was performed with forward primer EM-F and reverse primer 0797D-R, which were designed according to the DNA sequence of the erythromycin resistance gene (GenBank Accession No. KM017875.1) and the downstream sequence of LPL9_0797, respectively.
Field-Emission Scanning Electron Microscopy
Cells from L. paracasei L9 in CDM with or without 0.2% Ox-bile and cells in CDMM with 0.2% Ox-bile were collected at 12 h and fixed with 2.5% glutaraldehyde in phosphate buffer solution (PBS) at 4°C. Samples were dehydrated in a graded ethanol series, and then transferred into the chamber of the critical point dryer (CPD 030 critical point dryer; Bal-Tec) to replace the ethanol with liquid CO2. Glass samples were sputter-coated with platinum layer to be imaged by a field-emission scanning electron microscopy (FESEM, Hitachi SU8010, Japan) with an acceleration voltage of 5 kV.
Hydrophobicity Assays
The microbial adhesion to solvents (MATS) method was employed for the evaluation of the hydrophobic/hydrophilic character of the cell surface of strains of L. paracasei (Bellon-Fontaine et al., 1996). Cells from L. paracasei L9 and L9mleS- in CDMM were harvested at 12 h by centrifugation (7000 g, 10 min, 4°C), washed twice with 150 mM NaCl and resuspended in the same solution to an optical density of 0.8 at 400 nm, Then 2.4 ml of this bacterial suspension was mixed with 0.4 ml xylene and vibrated on a vortex-type agitator for 60 s. After standing for 15 min, the under layer sample (1 ml) was removed from the aqueous phase and the OD400 was measured. The formula H % = [(A0 – A) /A0] × 100 was used to calculate the bacterial surface hydrophobicity (H %), where A0 and A represent the absorbance of the aqueous phase before and after xylene extraction with xylene respectively. All results were obtained from at least three independent experiments. Unpaired t-tests were used to evaluate the data.
Zeta Potential
The procedure for preparation of samples was described previously (Giaouris et al., 2009). Cells from L. paracasei L9 and L9mleS- in CDMM were harvested at 12 h by centrifugation (7000 g, 10 min, 4°C), washed twice with 1.5 mM NaCl and resuspended in the same solution at an OD400 of 0.6. The electrophoretic mobility was measured with a Nano Particle Analyzer (Nano Particle Analyzer SZ-100, HORIBA Scientific, Japan). All results were obtained from at least three independent experiments. One-way analysis of variance (ANOVA) tests were used to evaluate the data.
Results
Global Transcriptomic Analysis of the Bile Stress Response in L. paracasei L9
Lactobacillus paracasei L9 was initially cultivated with different Ox-bile concentrations (0.1, 0.13, 0.15 and 0.2%, w/v). The growth rate was reduced by approximately 50% in the presence of 0.13% (w/v) Ox-bile (Supplementary Figure S1). Therefore, 0.13% bile salts was used for further studies of bile stress response in L. paracasei L9. The Illumina Hiseq platform was used to investigate transcriptome level changes in L. paracasei L9 under bile salts stress. All the raw data could be downloaded from GEO with the Accession No. GSE108713. Correlation of gene expression level between three independent biological replicates was shown by the Pearson correlation coefficient (Supplementary Figure S2). The number of raw and clean reads used in this study is listed in the Supplementary Table S3, and the information of reads mapping to the reference genome is shown in the Supplementary Table S4. Data analysis showed a total of 1184 differential expressed genes comprising 587 up-regulated and 597 down-regulated genes (Supplementary Table S5). The GOseq R package (Release2.12) was used to assign the gene ontology (GO) terms to the DEGs. Genes involved in the response to bile stress were selected for further analysis according to following standards: (1) log2FoldChange > 1.5; (2) statistically significant level P < 0.05. Finally, the transcription of 104 genes was detected to be related to bile stress, which comprised 58 up-regulated genes and 46 down-regulated genes (Supplementary Table S6). The putative functions of these genes were classified in different categories grouped by GO (Figure 1).
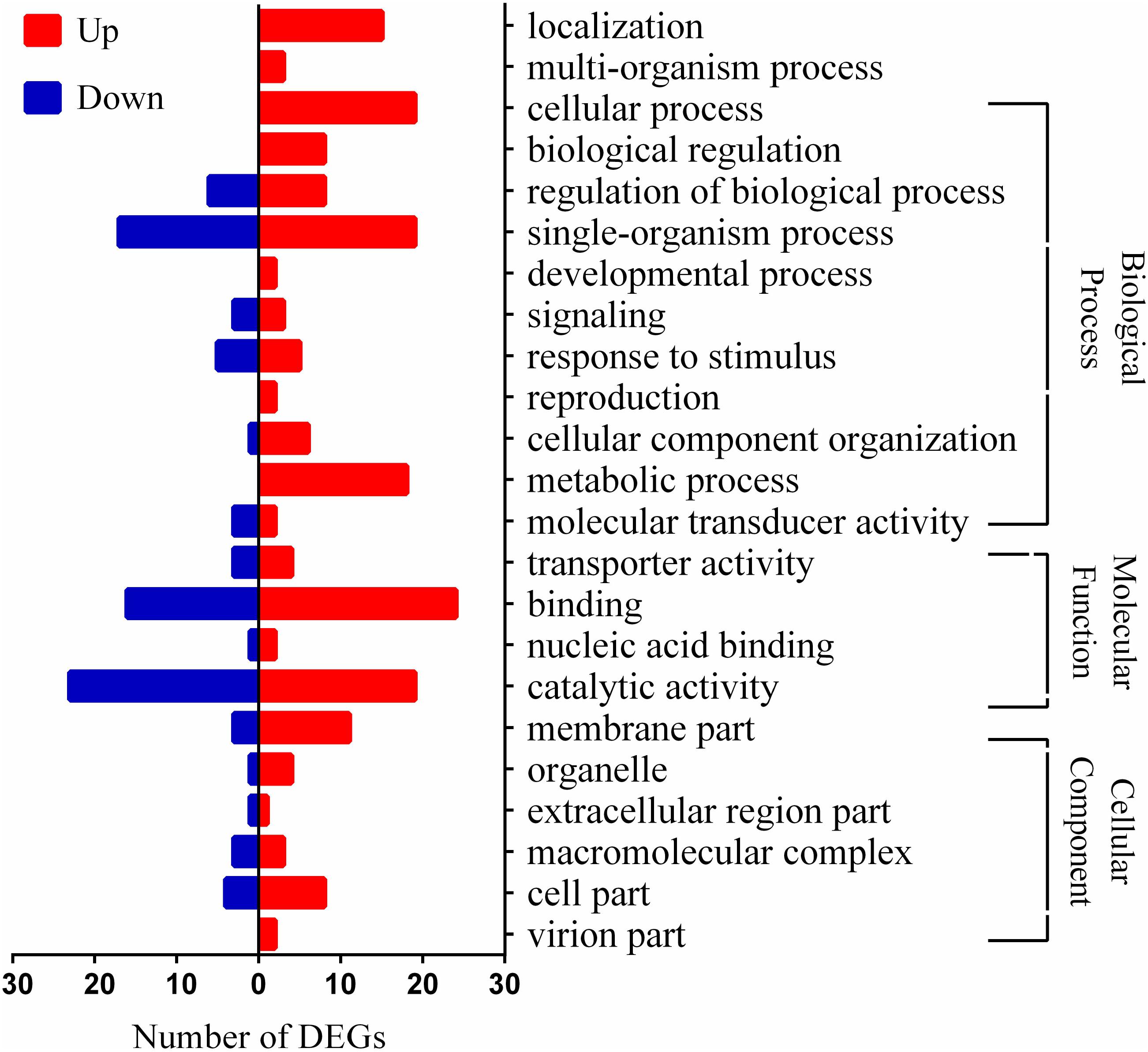
FIGURE 1. Distribution of differentially expressed genes (DEGs) in Lactobacillus paracasei L9 under bile stress. Bars indicate up (red) and down (blue) regulated genes (log2FoldChange > 1.5) in the presence of 0.13% Ox-bile.
Bile Stress Expands Carbon Source Utilization Profile and Changes Carbohydrate Metabolism Pattern in L. paracasei L9
Genes LPL9_0432 and LPL9_0433 involved in a mannose/fructose-inducible phosphotransferase system (PTS) were 2.98 and 7.29-fold up-regulated, respectively, after exposure to bile. Meanwhile, the downstream gene (LPL9_0434) encoding the beta-fructosidase was up-regulated 2.18-fold. This beta-fructosidase can degrade the fructose polymer levan to fructose which can be transported through the mannose/fructose PTS system (Mazé et al., 2004; Monedero et al., 2007). In addition, the gene (LPL9_2760) encoding alpha-glucosidase was 3.21-fold up-regulated. Alpha-glucosidase usually extracellularly breaks down starch and disaccharides to glucose. The gene (LPL9_1931) encoding PTS system cellobiose-specific IIB component was 3.28-fold increased after exposure to bile salts. Up-regulation of cellobiose-specific EIIA transporter was also detected in L. johnsonii under bile stress (Lee et al., 2012). Some oligosaccharides such as fructo-oligosaccharides are considered as prebiotics because they are non-digestible food ingredients, but they may be selective substrates for potentially probiotics in the colon. The presence of bile could function as a signal for gut entry and stimulate L. paracasei L9 to get better adapt to the available carbon sources in the intestinal environment.
As for glucose degradation, only the gene (LPL9_1849) encoding phosphoglycerate mutase was 3.83-fold up-regulated. However, D-xylulose-5P phosphoketolase (LPL9_0177) in the hexose monophophate pathway (HMP) was 3.12-fold down-regulated at the transcriptome level. This enzyme catalyzes the conversion of xylulose-5P to acetyl-P and glyceraldehyde-3-phosphate (GAP) (Mozzi and Vignolo, 2010). Another critical enzyme in HMP, glucose-6-P dehydrogenase (LPL9_0242) was also decreased 1.52-fold under bile stress. HMP, which was repressed in response to bile in L. paracasei L9, mainly generates reducing power and metabolic intermediates for biosynthetic processes. These results suggest that the demand of reducing power and metabolic intermediates might be reduced as the growth of bacteria was slowed down under bile salt stress. In addition, gene (LPL9_1666) encoding a zinc-containing alcohol dehydrogenase was 2.93-fold up-regulated. In lactic acid bacteria, alcohol dehydrogenase is involved in the mixed-acid fermentation which facilitates the conversion from acetaldehyde to ethanol with the reduction of NAD+ to NADH (Mozzi and Vignolo, 2010). It has been reported that ethanol was produced at a higher level by the bile-resistant derivative of B. animalis IPLA 4549 (Ruas-Madiedo et al., 2005). In the mixed-acid fermentation, the formation of formic acid from pyruvate instead of lactic acid could yield an extra ATP, and subsequently enable the cell to recycle NAD+ through the generate of ethanol (Ballongue, 1998). Taken together, L9 may shift its carbohydrate metabolism to mixed-acid fermentation and then generate extra energy to enhance bile salt tolerance under bile stress.
Effect of Bile Stress on Amino Acids and Peptide Metabolism Processes
The opp operon encoding an ABC oligopeptide transport system (oppA, oppB, oppC, oppD, oppF) was approximately threefold up-regulated. Opp transporters are membrane-associated five-protein complexes (oppABCDF) of the ATP-binding cassette involved in uptake of di- and tripeptides (Higgins, 1992). Homologous overexpression of oppA has been shown to increase the tolerance to Ox-bile in L. salivarius Ren and confer specific resistance to taurine-conjugated bile salts, suggesting the envelope-located protein OppA may be capable of binding taurine-conjugated bile salts and then reducing the entry of these toxic compounds (Wang et al., 2015). Gene encoding a putative peptide ABC transporter was also strongly up-regulated under salt stress in L. paracasei ATCC 334 (Palud et al., 2018). In addition, the branched-chain amino acid (BCAA) aminotransferase (LPL9_2126), which participated in the last step of the synthesis of L-Leucine, L-Valine and L-Isoleucine, was increased 5.56-fold at transcriptional level. BCAA could form high hydrophobic structure in proteins which may resist bile salt, and similar results have also been reported in Bifidobacterium longum (Sánchez et al., 2005).
Several genes involved in the biosynthesis of L-lysine were up-regulated (LPL9_0091, LPL9_0086, LPL9_1309), among which the gene encoding succinyl-diaminopimelate desuccinylase (LPL9_1309, 4.69-fold) was the most significant. Meanwhile, LysX (LPL9_2970) and LysY (LPL9_2971) belonging to the putative lysine ABC transport system were up-regulated 2.06 and 2.97-fold, respectively. The lysX-lysY system is involved in transporting lysine and is necessary for phosphatidylglycerol (PG) lysinylation (Rodionov et al., 2003; Maloney et al., 2011), which has been found to play a role in the resistance to cationic antimicrobial peptides (CAMPs) (Peschel et al., 2001). Meanwhile, genes encoding the biosynthesis of lysine and proteins related to lysinylation were up-regulated under bile stress in Lactobacillus rhamnosus GG. This modulation of bacterial surface may improve the resistance to bile by altering the surface charge to repulse cationic bile compounds (Koskenniemi et al., 2011).
Effect of Bile Stress on Transmembrane Transport in L. paracasei L9
Genes (LPL9_1281, LPL9_1282) encoding a multidrug ABC transport system were all up-regulated approximately 3.5-fold. Genes LPL9_1667 and LPL9_1668 encoding an uncharacterized ABC transport system were up-regulated about 15-fold in the bile stress response. It has been reported that multidrug transporters could export a wide range of diverse cytotoxic drugs including bile salts through cell membranes (Piddock, 2006; Whitehead et al., 2008; Pfeiler and Klaenhammer, 2009). In addition, gene (LPL9_1477) encoding a FtsE family cell division transporter was decreased 3.46-fold. FtsE is a cytoplasmic ATPase locating to the septal ring, and plays a role in cell division together with the integral membrane protein FtsX (Schmidt et al., 2004). The upstream gene (LPL9_1476) encoding a periplasmic-binding component of an ABC transporter was also 3.28-fold repressed, which probably functions related to FtsX. Recent research showed that FtsEX may be an important regulator of peptidoglycan hydrolases at the division site (Yang et al., 2011) and cells would divide poorly in the absence of FtsEX, resulting in increased cell length (Arends et al., 2009).
Analysis of Transcription Factors Involved in Bile Stress
A TetR family transcription factor (LPL9_0057) was 8.02-fold up-regulated, and the gene (LPL9_0056) in the same operon encoding a mycobacterial membrane protein large (MMPL) family transporter was 5.05-fold up-regulated in response to bile salt. In another operon, a TetR regulator (LPL9_1280) was 4.39-fold up-regulated and the downstream genes (LPL9_1281, LPL9_1282) encoding a multidrug ABC transporter system were also 3.95 and 3.69-fold up-regulated, respectively. The MMPL family transporters and multidrug transporters of the resistance-nodulation-cell division (RND) transporters superfamily, are usually regulated by TetR family members (Goldberg et al., 1999) and play a role in antibiotic resistance (Cuthbertson and Nodwell, 2013). The genes breA and breB encoding efflux pumps of the RND superfamily, were tightly controlled by the TetR family regulator BreR, and were also induced in the presence of cholate, deoxycholate, or chenodeoxycholate in Vibrio cholerae (Cerda-Maira et al., 2008). Another regulator (LPL9_0083) belonging to the Xre family was 4.39-fold up-regulated in detected transcripts. In Lactobacillus acidophilus, inactivation of the gene encoding a XRE family-like protein resulted in a more sensitive phenotype under low pH stress (Azcarate-Peril et al., 2004). Because the protonated form of bile salts could lead to intracellular acidification in a manner similar to organic acids (Smet et al., 1995). It is speculated that this Xre family regulator may be involved in bile resistance by overcoming intracellular acidification caused by bile salt in L. paracasei L9.
Response of Membrane Proteins in Bile Stress
Three genes encoding membrane proteins were up-regulated in response to bile stress, which are located in the same operon. Gene LPL9_0968 encoding a PspC-domain containing protein was 9.68-fold up-regulated, gene LPL9_0969 encoding a hypothetical protein was 10.34-fold up-regulated and gene LPL9_0970 encoding a phage holin family protein was 23.76-fold up-regulated. The PspC (phage shock protein C) is involved in extra cytoplasmic stress such as filamentous phage infection, mislocalization of some envelope proteins, extremes of temperature, osmolarity or ethanol concentration (Model et al., 1997). In Lactobacillus reuteri, a homolog of the phage shock transcriptional regulator PspC was reported to be related to bile stress (Whitehead et al., 2008). These membrane proteins may protect cells from environmental stress by maintaining the integrity of the cytoplasmic membrane (Darwin, 2005).
In addition, the gene (LPL9_1521) encoding a hemolysin III protein was 3.80-fold up-regulated in response to bile stress in L9. A hemolysin-like protein TlyC1 was 10-fold up-regulated upon Ox-bile treatment in B. longum BBMN68. Heterologous expression of tlyC1 in L. lactis conferred 45-fold higher resistance to sodium taurocholate and sodium taurodeoxycholate (Liu et al., 2014). Therefore, the hemolysin protein may function as a barrier to protect L. paracasei L9 from bile toxicity. A gene encoding the L-Ala-D/L-Glu epimerase was increased 4.04-fold in detected transcripts, which was involved in peptidoglycan turnover and recycling (Reith and Mayer, 2011). Peptidoglycan can be degraded to the dipeptide L-Ala-D-Glu and then L-Ala-D/L-Glu epimerase could convert L-Ala-D-Glu to L-Ala-L-Glu. Only the latter can be further hydrolyzed with dipeptidases (Schmidt et al., 2001). As bile salt could cause cell wall damage (Begley et al., 2005), this epimerase may increase survival under bile stress conditions through recycling the dipeptide or amino acid from peptidoglycan degradation in L. paracasei L9.
L-Malic Acid Increased the Bile Tolerance of L9
Genes encoding MLE (mleS, LPL9_0797) and putative L-malate transporter MleT (mleT, LPL9_0798) were 2.60 and 3.26-fold up-regulated, respectively, in response to bile in L. paracasei L9. To investigate whether L-malic acid could enhance the bile resistance, L. paracasei was cultured in CDM with or without L-malic acid supplement. Growth of L. paracasei L9 was not affected by the addition of L-malic acid when cultured in the absence of Ox-bile. However, under 0.2% Ox-bile, the OD600 of L. paracasei L9 reached the maximum about 1.0 at 14 h in CDM, while the OD600 was 2.0 at the corresponding time in CDMM (Figure 2). These results demonstrated that L-malic acid in CDMM participated in the resistance to bile stress in L. paracasei L9.
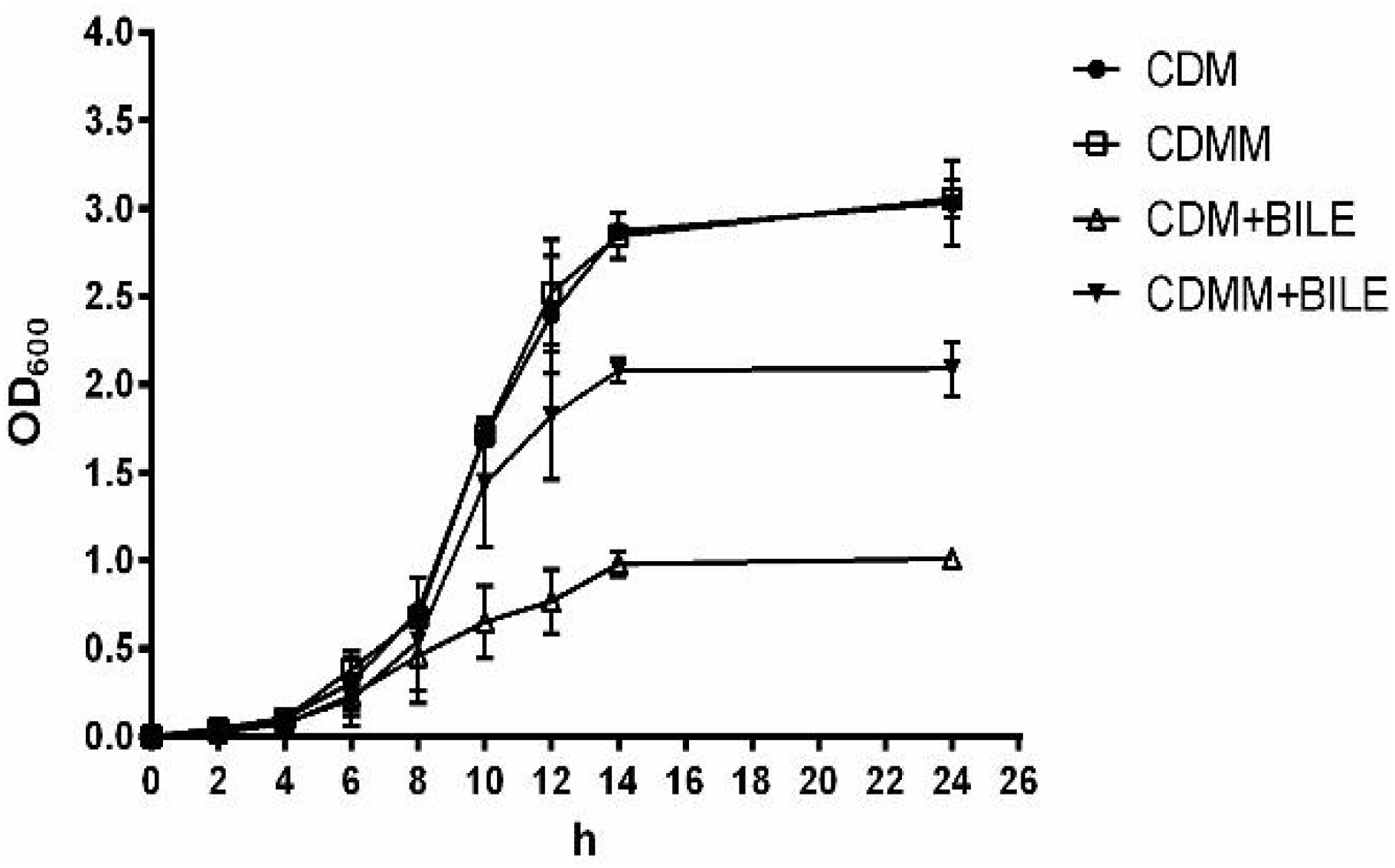
FIGURE 2. Growth of L. paracasei L9 in CDM (chemically defined medium) and CDMM (CDM with 2.5 mg/mL L-malic acid). Ox-bile was added at 0.2% final concentration. All results were obtained from at least three independent experiments. Error bars correspond to the standard error (SD).
The Morphology of L. paracasei L9 in the Presence of Bile Salts
Under 0.2% bile stress, the morphology of L. paracasei L9 was observed using FESEM. Compared with L. paracasei L9 in CDMM+Bile, bacteria grown in CDM+Bile displayed a more rough and shrunken appearance (Figures 3E,F). Without L-malic acid, the length of a proportion of cells were increased to various degrees (Figure 3A), suggesting disruption in the cell division. In addition, most of the cells were not completely separated, and had a tendency to twist and clump together (Figure 3A), which may attribute to a change in cell surface properties. Membrane vesicles (MVs) were observed on the surface of cells under bile stress (Figures 3C,E). For L. paracasei L9 grown with L-malic acid, cells were dispersive and rod-shaped with little variation in the length. No twist or MV structures were observed in cells cultured in the medium supplemented with L-malic acid (Figures 3B,D,F). The morphology of bacteria grown in CDM without bile (Figure 3G) was more similar to bacteria grown in CDMM+Bile (Figure 3F). These experiments showed that L-malic acid could maintain the integrity of the cell membrane and enhance survival of L. paracasei L9 under bile stress.
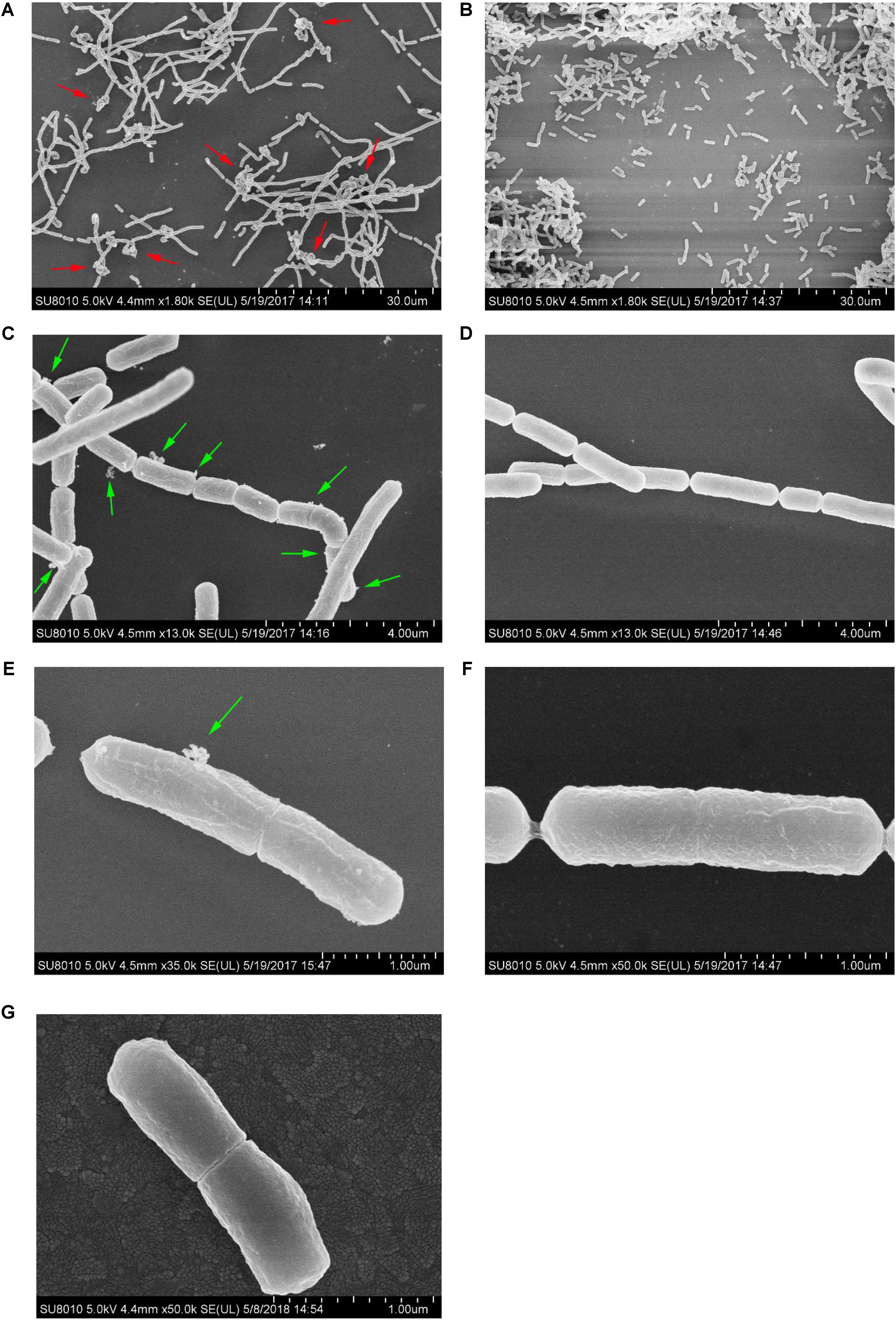
FIGURE 3. Field-emission scanning electron microscopy examination of cells of L. paracasei L9 treated with 0.2% Ox-bile. For three groups, the left row (A,C,E) was bacteria from CDM+Bile and the right (B,D,F) was from CDMM+Bile. (G) Shows bacteria from CDM without bile salt. The twist structure (red arrows) and MVs (green arrows) are indicated.
Malolactic Enzyme (MLE) Pathway of L. paracasei L9 in Bile Stress Response
To investigate if the MLE pathway is involved in the increase of bile resistance in L. paracasei L9, the L9mleS- mutant was constructed using the suicide plasmid pUC19EM. When chromosomal DNA from the L9mleS- mutant was used as template for PCR, the expected 1.5-kb product was obtained, and sequencing revealed amplification of the expected fragments of the genes Emr and mleS, confirming the correct integration of pUCmleS into the chromosome of L. paracasei L9 by a single crossover homologous recombination event. The growth assay of L9mleS- showed that growth of L9mleS- was not affected by addition of L-malic acid when cultured without Ox-bile (Figure 4). However, the enhancement of bile tolerance in CDMM was abolished in L9mleS-, and both groups finally reached a maximal OD600 of 1.0 in CDM or CDMM under bile stress. These results demonstrated that L-malic acid is metabolized through the MLE pathway to participate in bile resistance in L. paracasei L9.
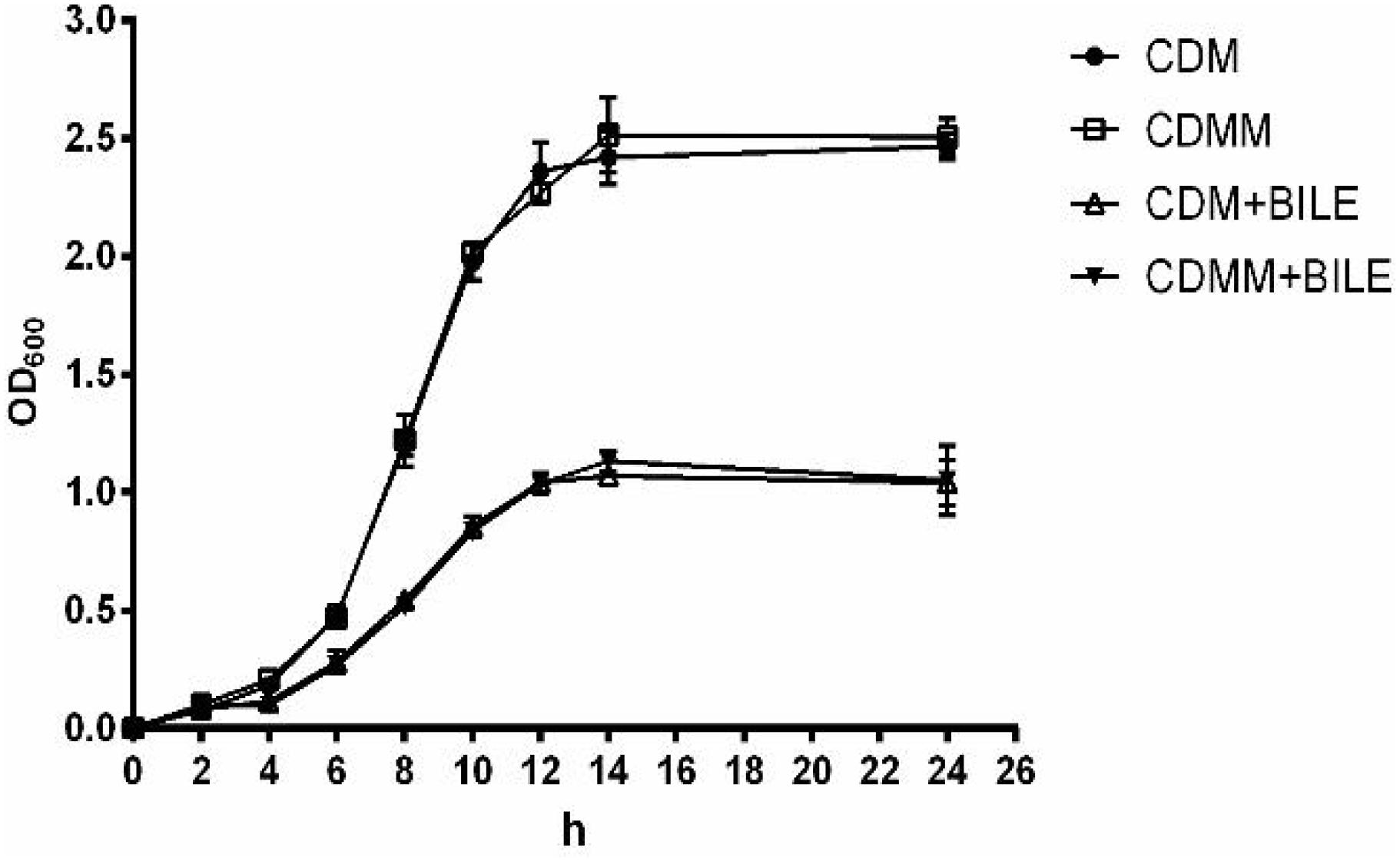
FIGURE 4. Growth of L9mleS- in CDM and CDMM. Ox-bile was added at 0.2%. All results were obtained from at least three independent experiments. Error bars correspond to the standard error (SD).
Alteration of Cell Surface Properties in Response to Bile Stress
In the growth assays experiment, autoaggregation of L9mleS- was observed at the bottom of the tube in the CDMM under 0.2% Ox-bile (Supplementary Figure S4). Therefore the hydrophobicity and zeta potential of L. paracasei strains were tested to investigate the surface properties. As shown in Figure 5, there is no significant difference in hydrophobicity between L9 and L9mleS- cultured in CDMM without Ox-bile. However, the hydrophobicity of L9mleS- was about threefold higher than that of L9 when cultured in CDMM under bile stress, which may lead to the enhancement of the autoaggregation of L9mleS-. In addition, there was a slight difference in the zeta potential between L9 and L9mleS- cultured in CDMM (Figure 6), but the L9mleS- strain had a significantly more negative zeta potential under bile stress. These results suggest inactivation of the MLE pathway could lead to changes in cell surface properties, which may be responsible for the decreased bile resistance in L9mleS-.
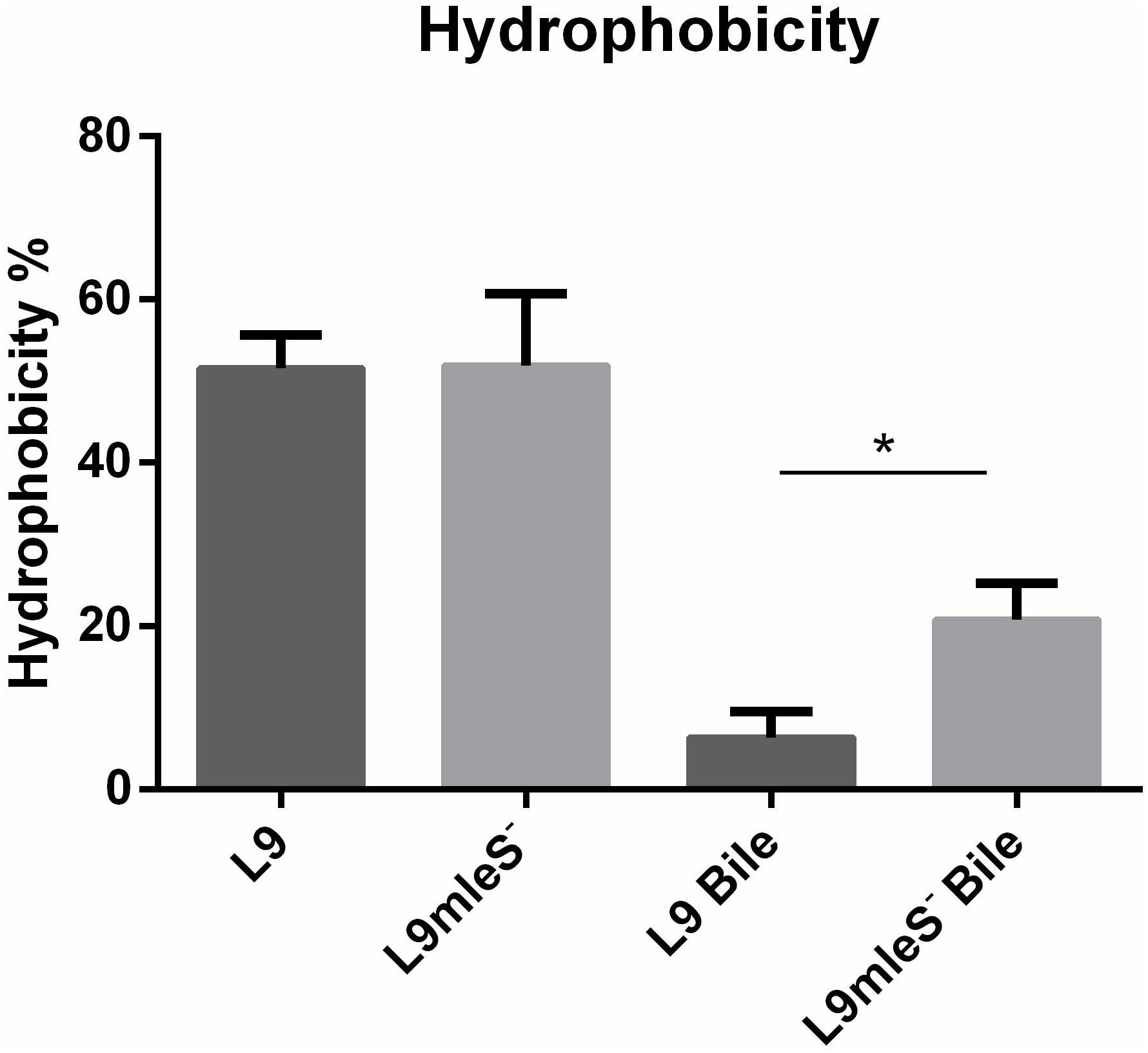
FIGURE 5. Hydrophobicity of L. paracasei L9 and L9mleS- cultured in CDMM with or without 0.2% Ox-bile. All results were obtained from at least three independent experiments. An unpaired t-test was used to evaluate the data, ∗P < 0.05.
Discussion
Under bile salt stress, metabolism changes at the glycolytic level which lead to enhanced energy production could be crucial for bacteria to resist bile salt. In Lactobacillus delbrueckii subsp. lactis, key enzymes in central glycolysis were over-expressed under bile stress, resulting in increased glucose utilization and lactic acid generation (Burns et al., 2010). While the bifid shunt was enhanced in B. animalis subsp. lactis and B. longum BBMN68 in response to bile (Sánchez et al., 2007; An et al., 2014). In this study, the carbohydrate metabolism may be shifted into the mixed-acid fermentation in L. paracasei L9. So, different mechanism about metabolic shifts in carbohydrate was employed to enhance energy production to resist bile stress in the different strains. As the bacterial cell membrane is the first physical target of bile salt (Begley et al., 2005), membrane proteins were often induced to resist bile stress in a variety of mechanisms, such as excreting bile salts (Ruiz et al., 2012) and acting as a barrier to protect the bacteria from bile toxicity (Liu et al., 2014). The operon (LPL9_0968-LPL9_0970) encoding membrane proteins showed the highest increase at the transcriptome level in L. paracasei L9, which has not been reported to be involved in the bile stress in other bacteria. Therefore, further research on these membrane proteins are needed to explain their roles in bile salt resistance.
A previous report indicated that the protonated form of bile salts could result in intracellular acidification in a way similar to organic acids (Smet et al., 1995). Generally, the genes encoding F1F0-ATPase would be up-regulated, which could maintain the internal pH under bile stress (Senior, 1990). In addition, the bile salt hydrolases (BSHs) can hydrolyze the protonated conjugated bile salts into unconjugated counterparts, and these unconjugated bile salts would prevent the pH-dropping by recapturing and exporting the co-transported proton (Smet et al., 1995). However, genes encoding BSHs do not exist in the genome of L. paracasei L9 and the transcription level of genes encoding F1F0-ATPase did not change under bile stress. It is noteworthy that the transcriptomic data showed that the MLE pathway was involved in bile tolerance, and this was further confirmed by genetic and physical evidence. In this pathway, L-malate acid is converted to L-lactate and carbon dioxide (CO2) by the MLE. As L-lactate (pKa = 3.85) has lower acidity than L-malate (pKa = 3.4), this decarboxylation contributes to alkalinization of the cytoplasm (Lemme et al., 2010). In addition, CO2 produced by MLE diffuses out of the cell or partially be used by carbonic anhydrase to form bicarbonate, which could further increase the intracellular pH (Papadimitriou et al., 2016). It has been reported that L-malate acid increased the survival of L. casei ATCC 334 under acidic conditions (Broadbent et al., 2010).
There are two gene clusters related to the metabolism of L-malate in L. paracasei L9. The MLE pathway consists of the gene mleS (LPL9_0797) encoding the MLE and gene mleT (LPL9_0798) encoding the putative L-malate transporter. The malic enzyme (ME) pathway in L. paracasei L9 consists of two diverging operons, maePE (LPL9_2996, LPL9_2997) and maeKR (LPL9_2998, LPL9_2999). In L. casei BL23, growth could not be sustained by utilization of L-malate through MLE as most of lactic acid bacteria cannot metabolize lactate into the gluconeogenic pathway. In contrast, the gene maeE encodes a ME converting L-malate into pyruvate, which could sustain the growth of strains using L-malate as a sole carbon source. However, the ME pathway undergoes carbon catabolite repression in L. casei BL23 (Landete et al., 2013). In this study, it is speculated that the ME pathway is repressed by glucose in the presence of culture medium. Therefore, L-malate enhances bile tolerance mainly through the MLE pathway rather than the ME pathway in L. paracasei L9.
Membrane vesicles are lumen-containing spheres formed from bulging or blebbing of the lipid-bilayer, which range in size from 20 to 500 nm in diameter, and can be found in eukaryotes, archaea, and bacteria (Deatherage and Cookson, 2012). Under different environmental stresses, formation of MVs is a common phenomenon in Gram-negative bacteria (Baumgarten et al., 2012). A molecular model of MVs formation was proposed whereby changes of electric charge on the lipopolysaccharide (LPS) caused charge-to-charge repulsion and membrane instability, and subsequently resulted in outward membrane blebbing and trapping of periplasmic components within MVs (Kadurugamuwa and Beveridge, 1995). In particular, MVs have also been observed in Gram-positive bacteria including lactic acid bacteria. MVs were observed in L. plantarum (Bron et al., 2004) and B. animalis ssp. lactis (Ruiz et al., 2007) under bile stress. Deformation or disruption of the cell wall was crucial for MVs to release (Brown et al., 2015), and MVs could occur when the cell wall undergoes thinning during daughter cell budding (Kopecka et al., 2000). In addition, cells with MVs were found to have a more hydrophobic surface in Pseudomonas putida (Baumgarten et al., 2012). Bile salts could disturb membrane integrity, which causes the leakage of protons and further dissipates the transmembrane pH and electrical potential (Kurdi et al., 2006). Based on these studies, we speculate bile salts stress could result in charge repulsions between phospholipid molecules and the bulge of membrane to generate MVs. Then the damaged cell wall may facilitate the release of MVs, which was verified by the higher hydrophobicity in L9mleS- grown in CDMM under bile stress (Figure 6). Uptake of L- malic acid needs to be driven by a proton gradient (ΔpH) or an electrical potential (ΔΨ) (Salema et al., 1994). When bile stress disrupts the ΔΨ, internalization of L-malic acid results in the translocation of net negative charges across the membrane, which in turn creates a ΔΨ to recover the disturbance in the membrane and the zeta potential of cells could then return to a less negative level (Figure 7). This speculation could be verified by finding that the zeta potential of L9mleS- grown in CDMM under bile stress was more negative than that of L. paracasei L9 (Figure 5). Taking all the evidence together, it is coucluded that MLE enhanced the bile tolerance not only through increasing the intracellular pH, but also by maintaining a membrane balance in L. paracasei L9.
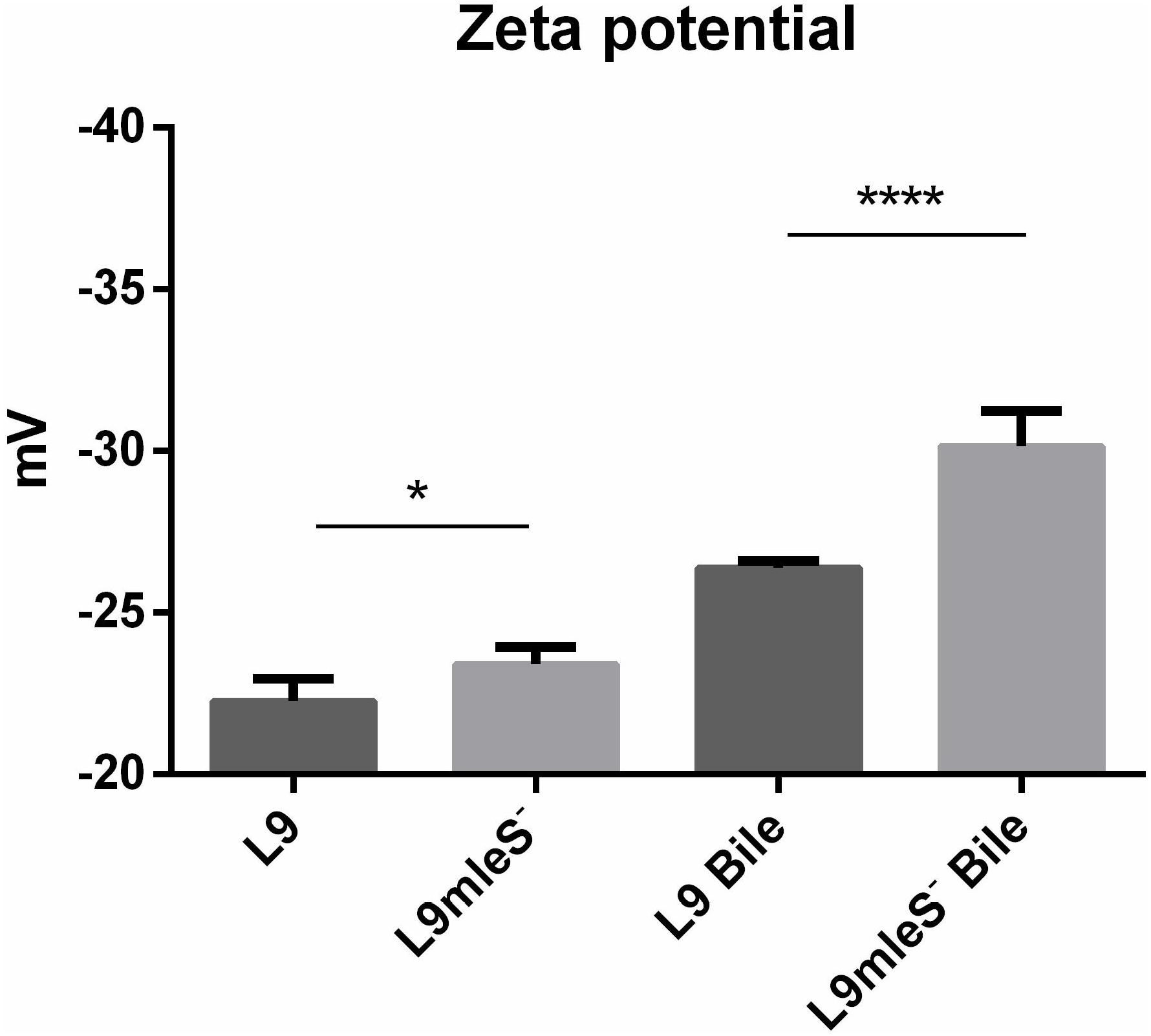
FIGURE 6. Zeta potential of L. paracasei L9 and L9mleS- cultured in CDMM with or without 0.2% Ox-bile. All results were obtained from at least three independent experiments. One-way analysis of variance (ANOVA) test was used to evaluate the data, ∗P < 0.05, ∗∗∗∗P < 0.0001.
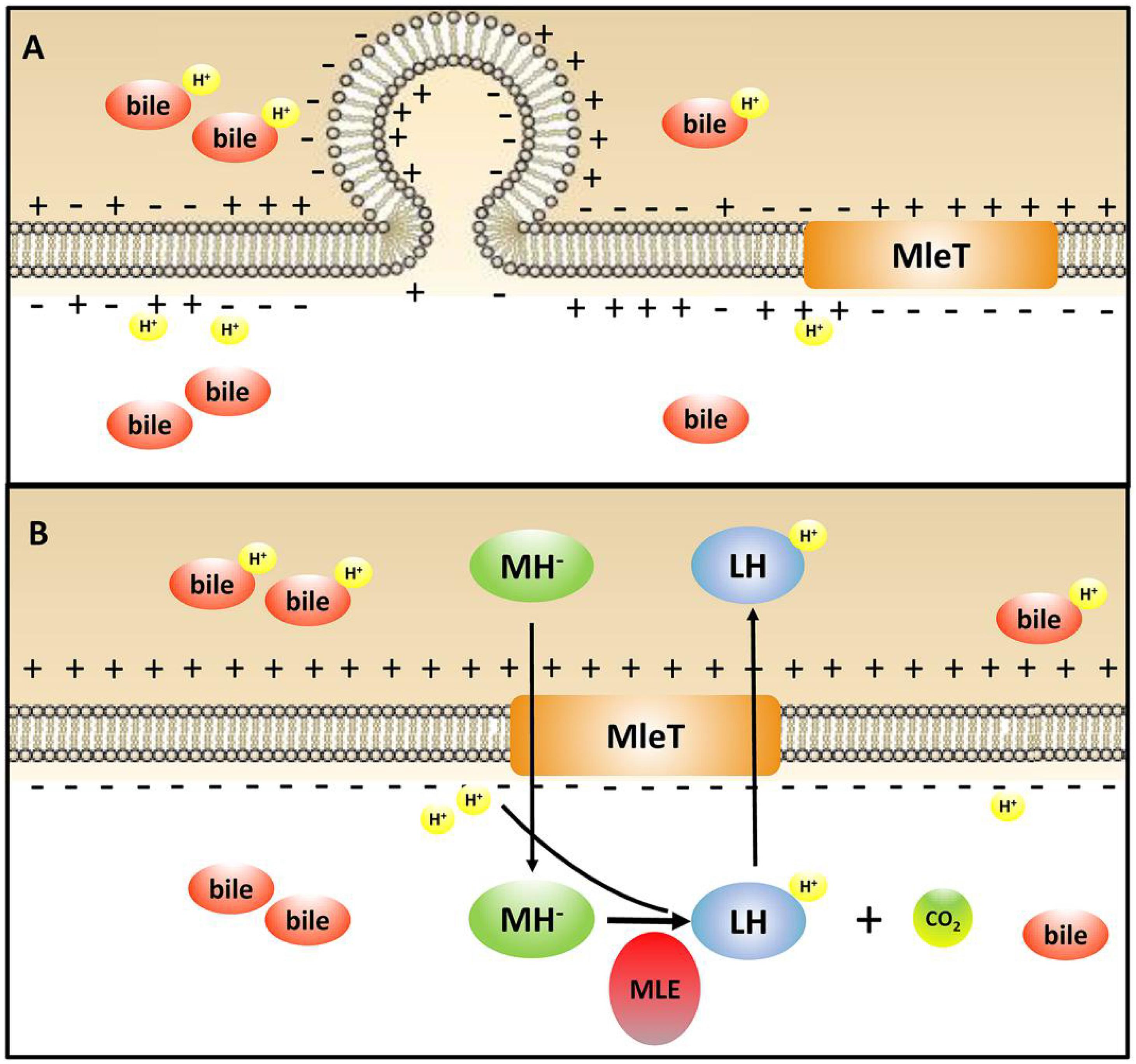
FIGURE 7. Illustration of the mechanism of MLE pathway involved in the bile stress response in L. paracasei L9. MH-, monoanionic L-malate; LH, L-lactic acid; MLE, malolactic enzyme. (A) L. paracasei L9 under bile stress without L-malic acid; (B) L. paracasei L9 under bile stress with L-malic acid.
Author Contributions
YH and XM designed the study. XM and PZ performed the experiments. XM analyzed and evaluated the data. XM and YH wrote the manuscript. YH, GW, and ZZ revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Contract No. 21676294).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01978/full#supplementary-material
References
Alcantara, C., and Zuniga, M. (2012). Proteomic and transcriptomic analysis of the response to bile stress of Lactobacillus casei BL23. Microbiology 158, 1206–1218. doi: 10.1099/mic.0.055657-0
An, H., Douillard, F. P., Wang, G., Zhai, Z., Yang, J., Song, S., et al. (2014). Integrated transcriptomic and proteomic analysis of the bile stress response in a centenarian-originated probiotic Bifidobacterium longum BBMN68. Mol. Cell. Proteomics 13, 2558–2572. doi: 10.1074/mcp.M114.039156
Arends, S. R., Kustusch, R. J., and Weiss, D. S. (2009). ATP-binding site lesions in FtsE impair cell division. J. Bacteriol. 191, 3772–3784. doi: 10.1128/JB.00179-09
Aukrust, T. W., Brurberg, M. B., and Nes, I. F. (1995). “Transformation of Lactobacillus by electroporation. Electroporation,” in Electroporation Protocols for Microorganisms. Methods in Molecular Biology™, Vol. 47, ed. J. A. Nickoloff (New York, NY: Humana Press), 201–208. doi: 10.1385/0-89603-310-4:201
Azcarate-Peril, M. A., Altermann, E., Hoover-Fitzula, R. L., Cano, R. J., and Klaenhammer, T. R. (2004). Identification and inactivation of genetic loci involved with Lactobacillus acidophilus acid tolerance. Appl. Environ. Microbiol. 70, 5315–5322. doi: 10.1128/AEM.70.9.5315-5322.2004
Ballongue, J. (1998). Bifidobacteria and Probiotic Action. Lactic Acid Bacteria: Microbiology and Functional Aspects, 3rd Edn. New York, NY: Marcel Dekker Inc.
Baumgarten, T., Sperling, S., Seifert, J., von Bergen, M., Steiniger, F., Wick, L. Y., et al. (2012). Membrane vesicle formation as a multiple-stress response mechanism enhances Pseudomonas putida DOT-T1E cell surface hydrophobicity and biofilm formation. Appl. Environ. Microbiol. 78, 6217–6224. doi: 10.1128/AEM.01525-12
Begley, M., Gahan, C. G., and Hill, C. (2005). The interaction between bacteria and bile. FEMS Microbiol. Rev. 29, 625–651. doi: 10.1016/j.femsre.2004.09.003
Bellon-Fontaine, M. N., Rault, J., and Van Oss, C. J. (1996). Microbial adhesion to solvents: a novel method to determine the electron-donor/electron-acceptor or Lewis acid-base properties of microbial cells. Colloid Surf. B 7, 47–53. doi: 10.1016/0927-7765(96)01272-6
Bove, P., Russo, P., Capozzi, V., Gallone, A., Spano, G., and Fiocco, D. (2013). Lactobacillus plantarum passage through an oro-gastro-intestinal tract simulator: carrier matrix effect and transcriptional analysis of genes associated to stress and probiosis. Microbiol. Res. 168, 351–359. doi: 10.1016/j.micres.2013.01.004
Broadbent, J. R., Larsen, R. L., Deibel, V., and Steele, J. L. (2010). Physiological and transcriptional response of Lactobacillus casei ATCC 334 to acid stress. J. Bacteriol. 192, 2445–2458. doi: 10.1128/JB.01618-09
Bron, P. A., Marco, M., Hoffer, S. M., Van Mullekom, E., De Vos, W. M., and Kleerebezem, M. (2004). Genetic characterization of the bile salt response in Lactobacillus plantarum and analysis of responsive promoters in vitro and in situ in the gastrointestinal tract. J. Bacteriol. 186, 7829–7835. doi: 10.1128/JB.186.23.7829-7835.2004
Brown, L., Wolf, J. M., Prados-Rosales, R., and Casadevall, A. (2015). Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 13, 620–630. doi: 10.1038/nrmicro3480
Burns, P., Sanchez, B., Vinderola, G., Ruas-Madiedo, P., Ruiz, L., Margolles, A., et al. (2010). Inside the adaptation process of Lactobacillus delbrueckii subsp. lactis to bile. Int. J. Food Microbiol. 142, 132–141. doi: 10.1016/j.ijfoodmicro.2010.06.013
Caspritz, G., and Radler, F. (1983). Malolactic enzyme of Lactobacillus plantarum. Purification, properties, and distribution among bacteria. J. Biol. Chem. 258, 4907–4910.
Cerda-Maira, F. A., Ringelberg, C. S., and Taylor, R. K. (2008). The bile response repressor BreR regulates expression of the Vibrio cholerae breAB efflux system operon. J. Bacteriol. 190, 7441–7452. doi: 10.1128/JB.00584-08
Corcoran, B. M., Stanton, C., Fitzgerald, G., and Ross, R. P. (2008). Life under stress: the probiotic stress response and how it may be manipulated. Curr. Pharm. Des. 14, 1382–1399. doi: 10.2174/138161208784480225
Cuthbertson, L., and Nodwell, J. R. (2013). The TetR family of regulators. Microbiol. Mol. Biol. R. 77, 440–475. doi: 10.1128/MMBR.00018-13
Darwin, A. J. (2005). The phage-shock-protein response. Mol. Microbiol. 57, 621–628. doi: 10.1111/j.1365-2958.2005.04694.x
De Vrese, M., and Schrezenmeir, J. (2008). Probiotics, prebiotics, and synbiotics. Adv. Biochem. Eng. 111, 1–66. doi: 10.1007/10_2008_097
Deatherage, B. L., and Cookson, B. T. (2012). Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect. Immun. 80, 1948–1957. doi: 10.1128/IAI.06014-11
Di Renzo, T., Reale, A., Boscaino, F., and Messia, M. C. (2018). Flavoring production in Kamut®, quinoa and wheat doughs fermented by Lactobacillus paracasei, Lactobacillus plantarum, and Lactobacillus brevis: a SPME-GC/MS Study. Front. Microbiol. 9:429. doi: 10.3389/fmicb.2018.00429
Giaouris, E., Chapot-Chartier, M. P., and Briandet, R. (2009). Surface physicochemical analysis of natural Lactococcus lactis strains reveals the existence of hydrophobic and low charged strains with altered adhesive properties. Int. J. Food Microbiol. 131, 2–9. doi: 10.1016/j.ijfoodmicro.2008.09.006
Goldberg, M., Pribyl, T., Juhnke, S., and Nies, D. H. (1999). Energetics and topology of CzcA, a cation/proton antiporter of the resistance-nodulation-cell division protein family. J. Biol. Chem. 274, 26065–26070. doi: 10.1074/jbc.274.37.26065
Hamon, E., Horvatovich, P., Bisch, M., Bringel, F., Marchioni, E., Aoudé-Werner, D., et al. (2011a). Investigation of biomarkers of bile tolerance in Lactobacillus casei using comparative proteomics. J. Proteome Res. 11, 109–118. doi: 10.1021/pr200828t
Hamon, E., Horvatovich, P., Izquierdo, E., Bringel, F., Marchioni, E., Aoudé-Werner, D., et al. (2011b). Comparative proteomic analysis of Lactobacillus plantarum for the identification of key proteins in bile tolerance. BMC Microbiol. 11:63. doi: 10.1186/1471-2180-11-63
Higgins, C. F. (1992). ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8, 67–113. doi: 10.1146/annurev.cb.08.110192.000435
Kadurugamuwa, J. L., and Beveridge, T. J. (1995). Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J. Bacteriol. 177, 3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995
Kopecka, M., Yamaguchi, M., Gabriel, M., Takeo, K., and Svoboda, A. (2000). Morphological transitions during the cell division cycle of Cryptococcus neoformans as revealed by transmission electron microscopy of ultrathin sections and freeze-substitution. Scr. Med. 73, 369–380.
Koskenniemi, K., Laakso, K., Koponen, J., Kankainen, M., Greco, D., Auvinen, P., et al. (2011). Proteomics and transcriptomics characterization of bile stress response in probiotic Lactobacillus rhamnosus GG. Mol. Cell. Proteomics 10, M110–M002741. doi: 10.1074/mcp.M110.002741
Kurdi, P., Kawanishi, K., Mizutani, K., and Yokota, A. (2006). Mechanism of growth inhibition by free bile acids in lactobacilli and Bifidobacteria. J. Bacteriol. 188, 1979–1986. doi: 10.1128/JB.188.5.1979-1986.2006
Landete, J. M., Ferrer, S., Monedero, V., and Zúñiga, M. (2013). Malic enzyme and malolactic enzyme pathways are functionally linked but independently regulated in Lactobacillus casei BL23. Appl. Environ. Microbiol. 79, 5509–5518. doi: 10.1128/AEM.01177-13
Lee, J. Y., Pajarillo, E. A. B., Kim, M. J., Chae, J. P., and Kang, D. K. (2012). Proteomic and transcriptional analysis of Lactobacillus johnsonii PF01 during bile salt exposure by iTRAQ shotgun proteomics and quantitative RT-PCR. J. Proteome Res. 12, 432–443. doi: 10.1021/pr300794y
Lemme, A., Sztajer, H., and Wagner-Döbler, I. (2010). Characterization of mleR, a positive regulator of malolactic fermentation and part of the acid tolerance response in Streptococcus mutans. BMC Microbiol. 10:58. doi: 10.1186/1471-2180-10-58
Liu, Y., An, H., Zhang, J., Zhou, H., Ren, F., and Hao, Y. (2014). Functional role of tlyC1 encoding a hemolysin-like protein from Bifidobacterium longum BBMN68 in bile tolerance. FEMS Microbiol. Lett. 360, 167–173. doi: 10.1111/1574-6968.12601
Maloney, E., Lun, S., Stankowska, D., Guo, H., Rajagoapalan, M., Bishai, W. R., et al. (2011). Alterations in phospholipid catabolism in Mycobacterium tuberculosis lysX mutant. Front. Microbiol. 2:19. doi: 10.3389/fmicb.2011.00019
Mazé, A., Boël, G., Poncet, S., Mijakovic, I., Le Breton, Y., Benachour, A., et al. (2004). The Lactobacillus casei ptsHI47T mutation causes overexpression of a LevR-regulated but RpoN-independent operon encoding a mannose class phosphotransferase system. J. Bacteriol. 186, 4543–4555. doi: 10.1128/JB.186.14.4543-4555.2004
Model, P., Jovanovic, G., and Dworkin, J. (1997). The Escherichia coli phage-shock-protein (psp) operon. Mol. Microbiol. 24, 255–261. doi: 10.1046/j.1365-2958.1997.3481712.x
Monedero, V., Mazé, A., Boël, G., Zúñiga, M., Beaufils, S., Hartke, A., et al. (2007). The phosphotransferase system of Lactobacillus casei: regulation of carbon metabolism and connection to cold shock response. J. Mol. Microbiol. Biotechnol. 12, 20–32. doi: 10.1159/000096456
Mozzi, F., and Vignolo, G. M. (eds). (2010). Biotechnology of Lactic Acid Bacteria: Novel Applications. Hoboken, NJ: John Wiley and Sons, Inc. doi: 10.1002/9780813820866
Palud, A., Scornec, H., Cavin, J. F., and Licandro, H. (2018). New genes involved in mild stress response identified by transposon mutagenesis in Lactobacillus paracasei. Front. Microbiol. 9:535. doi: 10.3389/fmicb.2018.00535
Pan, T., Guo, H. Y., Zhang, H., Liu, A. P., Wang, X. X., and Ren, F. Z. (2014). Oral administration of Lactobacillus paracasei alleviates clinical symptoms of colitis induced by dextran sulphate sodium salt in BALB/c mice. Benef. Microbes 5, 315–322. doi: 10.3920/BM2013.0041
Papadimitriou, K., Alegría,Á., Bron, P. A., De Angelis, M., Gobbetti, M., Kleerebezem, M., et al. (2016). Stress physiology of lactic acid bacteria. Microbiol. Mol. Biol. R. 80, 837–890. doi: 10.1128/MMBR.00076-15
Peschel, A., Jack, R. W., Otto, M., Collins, L. V., Staubitz, P., Nicholson, G., et al. (2001). Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. Tohoku J. Exp. Med. 193, 1067–1076. doi: 10.1084/jem.193.9.1067
Pfeiler, E. A., and Klaenhammer, T. R. (2009). Role of transporter proteins in bile tolerance of Lactobacillus acidophilus. Appl. Environ. Microbiol. 75, 6013–6016. doi: 10.1128/AEM.00495-09
Piddock, L. J. (2006). Multidrug-resistance efflux pumps? Not just for resistance. Nat. Rev. Microbiol. 4, 629–636. doi: 10.1038/nrmicro1464
Reith, J., and Mayer, C. (2011). Peptidoglycan turnover and recycling in Gram-positive bacteria. Appl. Microbiol. Biotechnol. 92, 1–11. doi: 10.1007/s00253-011-3486-x
Rodionov, D. A., Vitreschak, A. G., Mironov, A. A., and Gelfand, M. S. (2003). Regulation of lysine biosynthesis and transport genes in bacteria: yet another RNA riboswitch? Nucleic Acids Res. 31, 6748–6757.
Ruas-Madiedo, P., Hernández-Barranco, A., Margolles, A., and Clara, G. (2005). A bile salt-resistant derivative of Bifidobacterium animalis has an altered fermentation pattern when grown on glucose and maltose. Appl. Environ. Microbiol. 71, 6564–6570. doi: 10.1128/AEM.71.11.6564-6570.2005
Ruiz, L., Sánchez, B., Ruas-Madiedo, P., De Los Reyes-Gavilán, C. G., and Margolles, A. (2007). Cell envelope changes in Bifidobacterium animalis ssp. lactis as a response to bile. FEMS Microbiol. Lett. 274, 316–322. doi: 10.1111/j.1574-6968.2007.00854.x
Ruiz, L., Zomer, A., O’Connell-Motherway, M., van Sinderen, D., and Margolles, A. (2012). Discovering novel bile protection systems in Bifidobacterium breve UCC2003 through functional genomics. Appl. Environ. Microbiol. 78, 1123–1131. doi: 10.1128/AEM.06060-11
Salema, M., Poolman, B., Lolkema, J. S., Dias, M. C. L., and Konings, W. N. (1994). Uniport of monoanionic L-malate in membrane vesicles from Leuconostoc oenos. FEBS J. 225, 289–295. doi: 10.1111/j.1432-1033.1994.00289.x
Sánchez, B., Champomier-Vergès, M. C., Anglade, P., Baraige, F., Clara, G., Margolles, A., et al. (2005). Proteomic analysis of global changes in protein expression during bile salt exposure of Bifidobacterium longum NCIMB 8809. J. Bacteriol. 187, 5799–5808. doi: 10.1128/JB.187.16.5799-5808.2005
Sánchez, B., Champomier-Vergés, M.-C., Stuer-Lauridsen, B., Ruas-Madiedo, P., Anglade, P., Baraige, F., et al. (2007). Adaptation and response of Bifidobacterium animalis subsp. lactis to Bile: a proteomic and physiological approach. Appl. Environ. Microbiol. 73, 6757–6767. doi: 10.1128/AEM.00637-07
Schmidt, D. M., Hubbard, B. K., and Gerlt, J. A. (2001). Evolution of enzymatic activities in the enolase superfamily: functional assignment of unknown proteins in Bacillus subtilis and Escherichia coli as L-Ala-D/L-Glu epimerases. Biochemistry 40, 15707–15715. doi: 10.1021/bi011640x
Schmidt, K. L., Peterson, N. D., Kustusch, R. J., Wissel, M. C., Graham, B., Phillips, G. J., et al. (2004). A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J. Bacteriol. 186, 785–793. doi: 10.1128/JB.186.3.785-793.2004
Senior, A. E. (1990). The proton-translocating ATPase of Escherichia coli. Annu. Rev. Biophys. Biophys. Chem. 19, 7–41. doi: 10.1146/annurev.bb.19.060190.000255
Sheng, J., Baldeck, J. D., Nguyen, P. T., Quivey, R. G., and Marquis, R. E. (2010). Alkali production associated with malolactic fermentation by oral streptococci and protection against acid, oxidative, or starvation damage. Can. J. Microbiol. 56, 539–547. doi: 10.1139/w10-039
Sisto, A., and Lavermicocca, P. (2012). Suitability of a probiotic Lactobacillus paracasei strain as a starter culture in olive fermentation and development of the innovative patented product “probiotic table olives”. Front. Microbiol. 3:174. doi: 10.3389/fmicb.2012.00174
Smet, I., Hoorde, L. V., Woestyne, M., Christiaens, H., and Verstraete, W. (1995). Significance of bile salt hydrolytic activities of lactobacilli. J. Appl. Microbiol. 79, 292–301. doi: 10.1111/j.1365-2672.1995.tb03140.x
Stefanovic, E., Kilcawley, K. N., Roces, C., Rea, M., O’ Sullivan, M., Sheehan, D. J., et al. (2018). Evaluation of the potential of Lactobacillus paracasei adjuncts for flavour compounds development and diversification in short-aged Cheddar cheese. Front. Microbiol. 9:1506. doi: 10.3389/fmicb.2018.01506
Taranto, M. P., Fernandez Murga, M. L., Lorca, G., and Valdez, G. D. (2003). Bile salts and cholesterol induce changes in the lipid cell membrane of Lactobacillus reuteri. J. Appl. Microbiol. 95, 86–91. doi: 10.1046/j.1365-2672.2003.01962.x
Wang, G., Li, D., Ma, X., An, H., Zhai, Z., Ren, F., et al. (2015). Functional role of oppA encoding an oligopeptide-binding protein from Lactobacillus salivarius Ren in bile tolerance. J. Ind. Microbiol. Biotechnol. 42, 1167–1174. doi: 10.1007/s10295-015-1634-5
Whitehead, K., Versalovic, J., Roos, S., and Britton, R. A. (2008). Genomic and genetic characterization of the bile stress response of probiotic Lactobacillus reuteri ATCC 55730. Appl. Environ. Microbiol. 74, 1812–1819. doi: 10.1128/AEM.02259-07
Yang, D. C., Peters, N. T., Parzych, K. R., Uehara, T., Markovski, M., and Bernhardt, T. G. (2011). An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc. Natl. Acad. Sci. U.S.A. 108, E1052–E1060. doi: 10.1073/pnas.1107780108
Yang, J., Ren, F. Z., Zhang, H., Jiang, L., Hao, Y., and Luo, X. (2015). Induction of regulatory dendritic cells by Lactobacillus paracasei L9 prevents allergic sensitization to bovine beta-lactoglobulin in mice. J. Microbiol. Biotechnol. 25, 1687–1696. doi: 10.4014/jmb.1503.03022
Yang, Y., Yin, J., Liu, J., Xu, Q., Lan, T., Ren, F., et al. (2017). The copper homeostasis transcription factor CopR is involved in H2O2 stress in Lactobacillus plantarum CAUH2. Front. Microbiol. 8:2015. doi: 10.3389/fmicb.2017.02015
Keywords: Lactobacillus paracasei L9, bile stress, RNA-seq, malolactic enzyme pathway, alkalinization, membrane vesicles, membrane balance
Citation: Ma X, Wang G, Zhai Z, Zhou P and Hao Y (2018) Global Transcriptomic Analysis and Function Identification of Malolactic Enzyme Pathway of Lactobacillus paracasei L9 in Response to Bile Stress. Front. Microbiol. 9:1978. doi: 10.3389/fmicb.2018.01978
Received: 07 March 2018; Accepted: 06 August 2018;
Published: 23 August 2018.
Edited by:
Vittorio Capozzi, University of Foggia, ItalyReviewed by:
Tamara Smokvina, Danone, FranceHelene Licandro, Université Bourgogne Franche-Comté, France
Copyright © 2018 Ma, Wang, Zhai, Zhou and Hao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanling Hao, haoyl@cau.edu.cn
 Xiayin Ma1,2
Xiayin Ma1,2 Guohong Wang
Guohong Wang Yanling Hao
Yanling Hao