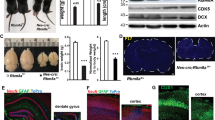
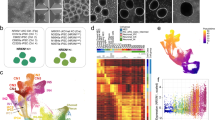
Abstract
Tourette Syndrome (TS) is a neuropsychiatric disorder thought to involve a reduction of basal ganglia (BG) interneurons and malfunctioning of the BG circuitry. However, whether interneurons fail to develop or are lost postnatally remains unknown. To investigate the pathophysiology of early development in TS, induced pluripotent stem cell (iPSC)-derived BG organoids from TS patients and healthy controls were compared on multiple levels of measurement and analysis. BG organoids from TS individuals manifested an impaired medial ganglionic eminence fate and a decreased differentiation of cholinergic and GABAergic interneurons. Transcriptome analyses revealed organoid mispatterning in TS, with a preference for dorsolateral at the expense of ventromedial fates. Our results point to altered expression of GLI transcription factors downstream of the Sonic Hedgehog signaling pathway with cilia disruption at the earliest stages of BG organoid differentiation as a potential mechanism for the BG mispatterning in TS. This study uncovers early neurodevelopmental underpinnings of TS neuropathological deficits using organoids as a model system.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Eblen F, Graybiel AM. Highly restricted origin of prefrontal cortical inputs to striosomes in the macaque monkey. J Neurosci. 1995;15:5999–6013.
Wang Z, Maia TV, Marsh R, Colibazzi T, Gerber A, Peterson BS. The neural circuits that generate tics in Tourette’s syndrome. Am J Psychiatry. 2011;168:1326–37.
Albin RL, Young AB, Penney JB. The functional anatomy of basal ganglia disorders. TINS. 1995;18:63–4.
Bloch MH, Leckman JF, Zhu H, Peterson BS. Caudate volumes in childhood predict symptom severity in adults with Tourette syndrome. Neurology. 2005;65:1253–8.
Peterson BS, Thomas P, Kane MJ, Scahill L, Zhang H, Bronen R, et al. Basal Ganglia volumes in patients with Gilles de la Tourette syndrome. Arch Gen Psychiatry. 2003;60:415–24.
Kalanithi PS, Zheng W, Kataoka Y, DiFiglia M, Grantz H, Saper CB, et al. Altered parvalbumin-positive neuron distribution in basal ganglia of individuals with Tourette syndrome. Proc Natl Acad Sci USA. 2005;102:13307–12.
Kataoka Y, Kalanithi PS, Grantz H, Schwartz ML, Saper C, Leckman JF, et al. Decreased number of parvalbumin and cholinergic interneurons in the striatum of individuals with Tourette Syndrome. J Comp Neurol. 2010;518:277–91.
Lennington JB, Coppola G, Kataoka-Sasaki Y, Fernandez TV, Palejev D, Li Y, et al. Transcriptome analysis of the human striatum in Tourette Syndrome. Biol Psychiatry. 2016;79:372–82.
Kawaguchi Y. Physiological, morphological and histochemical characterization of three classess of interneurons in rat neostriatum. JNeurosci. 1993;13:4908–23.
Mariani J, Simonini MV, Palejev D, Tomasini L, Coppola G, Szekely AM, et al. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109:12770–5.
Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Brauninger M, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci USA. 2015;112:15672–7.
Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–9.
Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min Yang S, Berger DR, et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53.
Marton RM, Pasca SP. Organoid and assembloid technologies for investigating cellular crosstalk in human brain development and disease. Trends Cell Biol. 2020;30:133–43.
Lim L, Mi D, Llorca A, Marin O. Development and functional diversification of cortical interneurons. Neuron. 2018;100:294–313.
Hoch RV, Clarke JA, Rubenstein JL. Fgf signaling controls the telencephalic distribution of Fgf-expressing progenitors generated in the rostral patterning center. Neural Dev. 2015;10:8.
Bloch MH, Peterson BS, Scahill L, Otka J, Katsovich L, Zhang H, et al. Adulthood outcome of tic and obsessive-compulsive symptom severity in children with Tourette syndrome. Arch Pediatr Adolesc Med. 2006;160:65–9.
Park IH, Lerou PH, Zhao R, Huo H, Daley GQ. Generation of human-induced pluripotent stem cells. Nat Protoc. 2008;3:1180–6.
Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8:409–12.
Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, et al. FOXG1-dependent dysregulation of GABA/Glutamate neuron differentiation in autism spectrum disorders. Cell 2015;162:375–90.
Lopez-Coviella I, Berse B, Krauss R, Thies RS, Blusztajn JK. Induction and maintenance of the neuronal cholinergic phenotype in the central nervous system by BMP-9. Science. 2000;289:313–6.
Lopez-Coviella I, Follettie MT, Mellott TJ, Kovacheva VP, Slack BE, Diesl V, et al. Bone morphogenetic protein 9 induces the transcriptome of basal forebrain cholinergic neurons. Proc Natl Acad Sci USA. 2005;102:6984–9.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39:D712–7.
Li M, Santpere G, Imamura Kawasawa Y, Evgrafov OV, Gulden FO, Pochareddy S, et al. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science. 2018;362.
Shi Y, Wang M, Mi D, Lu T, Wang B, Dong H, et al. Mouse and human share conserved transcriptional programs for interneuron development. Science. 2021;374:eabj6641.
Nobrega-Pereira S, Kessaris N, Du T, Kimura S, Anderson SA, Marin O. Postmitotic Nkx2-1 controls the migration of telencephalic interneurons by direct repression of guidance receptors. Neuron. 2008;59:733–45.
Sussel L, Marin O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126:3359–70.
Corbin JG, Rutlin M, Gaiano N, Fishell G. Combinatorial function of the homeodomain proteins Nkx2.1 and Gsh2 in ventral telencephalic patterning. Development. 2003;130:4895–906.
Magno L, Barry C, Schmidt-Hieber C, Theodotou P, Hausser M, Kessaris N. NKX2-1 is required in the embryonic septum for cholinergic system development, learning, and memory. Cell Rep. 2017;20:1572–84.
Silberberg SN, Taher L, Lindtner S, Sandberg M, Nord AS, Vogt D, et al. Subpallial enhancer transgenic lines: a data and tool resource to study transcriptional regulation of GABAergic cell fate. Neuron. 2016;92:59–74.
Luccardini C, Hennekinne L, Viou L, Yanagida M, Murakami F, Kessaris N, et al. N-cadherin sustains motility and polarity of future cortical interneurons during tangential migration. J Neurosci. 2013;33:18149–60.
Rataj-Baniowska M, Niewiadomska-Cimicka A, Paschaki M, Szyszka-Niagolov M, Carramolino L, Torres M, et al. Retinoic acid receptor beta controls development of striatonigral projection neurons through FGF-dependent and Meis1-dependent mechanisms. J Neurosci. 2015;35:14467–75.
Long JE, Cobos I, Potter GB, Rubenstein JL. Dlx1&2 and Mash1 transcription factors control MGE and CGE patterning and differentiation through parallel and overlapping pathways. Cereb Cortex. 2009;19:i96–106.
Sandberg M, Flandin P, Silberberg S, Su-Feher L, Price JD, Hu JS, et al. Transcriptional networks controlled by NKX2-1 in the development of Forebrain GABAergic neurons. Neuron 2016;91:1260–75.
Scott BB, Lois C. Generation of tissue-specific transgenic birds with lentiviral vectors. Proc Natl Acad Sci USA. 2005;102:16443–7.
Bassett AS, Scherer SW. Copy number variation in Tourette Syndrome. Neuron 2017;94:1041–3.
Huang AY, Yu D, Davis LK, Sul JH, Tsetsos F, Ramensky V, et al. Rare copy number variants in NRXN1 and CNTN6 increase risk for Tourette Syndrome. Neuron. 2017;94:1101–11 e7.
Qin S, Madhavan M, Waclaw RR, Nakafuku M, Campbell K. Characterization of a new Gsx2-cre line in the developing mouse telencephalon. Genesis. 2016;54:542–9.
Yun K, Potter S, Rubenstein JL. Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development. 2001;128:193–205.
Xu Q, Tam M, Anderson SA. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J Comp Neurol. 2008;506:16–29.
Manabe T, Tatsumi K, Inoue M, Makinodan M, Yamauchi T, Makinodan E, et al. L3/Lhx8 is a pivotal factor for cholinergic differentiation of murine embryonic stem cells. Cell Death Differ. 2007;14:1080–5.
Zhao Y, Marin O, Hermesz E, Powell A, Flames N, Palkovits M, et al. The LIM-homeobox gene Lhx8 is required for the development of many cholinergic neurons in the mouse forebrain. Proc Natl Acad Sci USA. 2003;100:9005–10.
Murdoch JN, Copp AJ. The relationship between sonic Hedgehog signaling, cilia, and neural tube defects. Birth Defects Res A Clin Mol Teratol. 2010;88:633–52.
Andreu-Cervera A, Anselme I, Karam A, Laclef C, Catala M, Schneider-Maunoury S. The ciliopathy gene Ftm/Rpgrip1l controls mouse forebrain patterning via region-specific modulation of Hedgehog/Gli signaling. J Neurosci. 2019;39:2398–415.
Andreu-Cervera A, Catala M, Schneider-Maunoury S. Cilia, ciliopathies and hedgehog-related forebrain developmental disorders. Neurobiol Dis. 2021;150:105236.
Park SM, Jang HJ, Lee JH. Roles of primary cilia in the developing brain. Front Cell Neurosci. 2019;13:218.
Backman M, Machon O, Mygland L, van den Bout CJ, Zhong W, Taketo MM, et al. Effects of canonical Wnt signaling on dorso-ventral specification of the mouse telencephalon. Dev Biol. 2005;279:155–68.
Arai Y, Cwetsch AW, Coppola E, Cipriani S, Nishihara H, Kanki H, et al. Evolutionary gain of Dbx1 expression drives subplate identity in the cerebral cortex. Cell Rep. 2019;29:645–58 e5.
Thomsen MS, Routhe LJ, Moos T. The vascular basement membrane in the healthy and pathological brain. J Cereb Blood Flow Metab. 2017;37:3300–17.
Hartwig C, Veske A, Krejcova S, Rosenberger G, Finckh U. Plexin B3 promotes neurite outgrowth, interacts homophilically, and interacts with Rin. BMC Neurosci. 2005;6:53.
Zhu B, Chen C, Xue G, Moyzis RK, Dong Q, Chen C. et al. The SEMA5A gene is associated with hippocampal volume, and their interaction is associated with performance on Raven’s Progressive Matrices. NeuroImage. 2014;88:181–7.
Bloch MH, Leckman JF. Clinical course of Tourette Syndrome. J Psychosom Res. 2009;67:497–501.
Leckman JF, Peterson B. The pathogenesis of Tourette’s syndrome: role od epigenetic factors active in early CNS development. Biol Psychiatry. 1993;34:425–7.
Peterson B, Riddle MA, Cohen DJ, Katz LD, Smith JC, Hardin MT, et al. Reduced basal ganglia volumes in Tourette’s syndrome using three-dimensional reconstruction techniques from magnetic resonance images. Neurology. 1993;43:941–9.
Heinz A, Knable MB, Wolf SS, Jones DW, Gorey JG, Hyde TM. et al. Tourette’s syndrome: [I-123]beta-CIT SPECT correlates of vocal tic severity. Neurology. 1998;51:1069–74.
Muller-Vahl KR, Meyer GJ, Knapp WH, Emrich HM, Gielow P, Brucke T, et al. Serotonin transporter binding in Tourette Syndrome. Neurosci Lett. 2005;385:120–5.
Wong DF, Brasic JR, Singer HS, Schretlen DJ, Kuwabara H, Zhou Y, et al. Mechanisms of dopaminergic and serotonergic neurotransmission in Tourette Syndrome: clues from an in vivo neurochemistry study with PET. Neuropsychopharmacology. 2008;33:1239–51.
Muller-Vahl KR, Szejko N, Wilke F, Jakubovski E, Geworski L, Bengel F, et al. Serotonin transporter binding is increased in Tourette Syndrome with obsessive compulsive disorder. Sci Rep. 2019;9:972.
Rallu M, Corbin JG, Fishell G. Parsing the prosencephalon. Nat Rev Neurosci. 2002;3:943–51.
Wen X, Lai CK, Evangelista M, Hongo JA, de Sauvage FJ, Scales SJ. Kinetics of hedgehog-dependent full-length Gli3 accumulation in primary cilia and subsequent degradation. Mol Cell Biol. 2010;30:1910–22.
Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, Yoder BK. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53.
Kim J, Kato M, Beachy PA. Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc Natl Acad Sci USA. 2009;106:21666–71.
Li J, Wang C, Wu C, Cao T, Xu G, Meng Q, et al. PKA-mediated Gli2 and Gli3 phosphorylation is inhibited by Hedgehog signaling in cilia and reduced in Talpid3 mutant. Dev Biol. 2017;429:147–57.
Lee B, Panda S, Lee HY. Primary ciliary deficits in the dentate gyrus of fragile X syndrome. Stem Cell Rep. 2020;15:454–66.
Acknowledgements
We wish to thank the participants who donated samples and time to our study. We want to thank Jeremy Schreiner and Livia Tomasini for technical assistance and Scott Norton for guidance in performing the transcriptome analyses. We thank Michael Higley and Riccardo Parra for use of the dual photon microscope. We are grateful to Drs. Christopher Pittenger and Nenad Sestan for comments and suggestions on an earlier version of this work. We acknowledge the Yale Center for Clinical Investigation for clinical support in obtaining the biopsy specimens, the Yale Stem Cell Center for the generation of the iPSC lines, and the Yale Center for Genome Analysis for library preparation and sequencing. We thank Dr. Pamela Ventola, Dr. Katarzyna Chawarska and Dr. Kevin Pelphrey for help with recruitment of control subjects.
Funding
The recruitment and production of iPSC lines for control subjects were supported by the following grants: MH087879, MH089176, and MH109648 from the National Institutes of Health, and by the Simons Foundation. The recruitment and production of iPSC lines for TS subjects were supported by MH118453 from the National Institutes of Health, by the NARSAD- Brain and Behavior Research Fund and by the Tourette Association of America.
Author information
Authors and Affiliations
Contributions
FMV conceived the study, designed and supervise experiments; JFL, RAK, AL-W, MHB, helped recruit patients and obtained clinical data; AS evaluated donor subjects and obtained skin biopsies; MVB contributed to the experimental design, cultured primary cells, performed reprogramming, developed the BG organoid protocol, generated organoid preps, processed them for all assays and performed and analyzed all experiments; JM oversaw organoid protocol development and optimization; YK performed the RNA-seq bioinformatic analyses; MVB, JM, YK and FMV generated display items and wrote the manuscript; all authors provided edits and comments on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Brady, M.V., Mariani, J., Koca, Y. et al. Mispatterning and interneuron deficit in Tourette Syndrome basal ganglia organoids. Mol Psychiatry 27, 5007–5019 (2022). https://doi.org/10.1038/s41380-022-01880-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-022-01880-5