Abstract
With the recent technological advancement in cultivation-independent high-throughput sequencing, metagenomes have tremendously improved our ability to characterize the genomic contents of the whole microbial communities. In this chapter, we argue the notion of pangenome can be applied beyond the available genome sequences by leveraging metagenome-assembled genomes, to form a comprehensive representation of the genetic content of a taxonomic group in a particular environment. We present the concept of the meta-pangenome, a representation of the totality of genes belonging to a species identified in multiple metagenomic samplings of a particular habitat. As an essential component in genome-centric pangenome analyses, we emphasize the importance to perform stringent quality assessment and validation to ensure the high quality of metagenomic deconvoluted genomes. This expansion from the traditional pangenome concept to the meta-pangenome overcomes many of the biases associated with whole-genome sequencing, and addresses the in vivo ecological context to further develop a systems-level understanding of microbial ecosystems.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
- Meta-pangenome
- Pan-metagenome
- Pangenome
- Metagenome
- Comparative genomics
- Metagenome-assembled genome
- Intraspecies diversity
- Metagenomic subspecies
- Community ecotype
- Habitome
1 Introduction
The first microbial genome, Haemophilus influenzae, was sequenced in 1995 (Fleischmann et al. 1995) with the second, Mycoplasma genitalium, following a few months later (Fraser et al. 1995). In analyzing the M. genitalium genome, the authors compared its sequence to that of H. influenzae, the only other available genome sequence at the time, providing insights into the ecology and evolution of these two microbes. Every subsequent genome comparison enabled the identification of shared and unique genetic characteristics between sets organisms. From this observation emerged the concept of pangenome, which describes the core (genes present in every strain of the species) and accessory (genes present in a subset of strains) genomes. Studying the similarities and differences between the genomic content of organisms can inform their evolutionary relationships, ecological roles, relationship to health, and has revolutionized our understanding of microbial diversity (Touchman 2010; Xia 2013; Hardison 2003; Miller et al. 2004; France et al. 2016).
Over the years, and with significant technological advancement, the number of available genome sequences has expanded from a few to a seemingly endless catalog. Yet this impressive collection suffers from a rather severe bias toward species and strains that are related to human health, amenable to isolation, and/or generally tractable. Metagenomics, the sequencing of whole microbial communities, is filling in these gaps by characterizing the genomes of entire populations in a community without cultivation. In this chapter, we argue the notion of pangenome can be applied beyond the available genome sequences by leveraging metagenome-assembled genomes (MAGs), to form a comprehensive representation of the genetic content of a taxonomic group in a particular environment. We present the concept of the meta-pangenome, a representation of the totality of genes belonging to a species identified in multiple metagenomic samplings of a particular habitat. This expansion from the traditional pangenome concept to the meta-pangenome overcomes many of the biases associated with whole-genome sequencing and addresses the in vivo ecological context by describing the whole genetic potential of a species in a specific environment. Further building on this new concept, one can think of the pan-metagenome as the complete genes/proteins catalog of all species found in a giving environment.
2 Metagenome Deconvolution Enables Genome-Centric Analyses of Microbial Ecosystems
An overwhelming majority of microbial species have resisted cultivation in the laboratory, largely due to strict, yet unknown, growth requirements (Bakken 1985). The cultivation of fastidious microbes requires optimal combinations of nutrients, growth temperatures, oxygen levels or even, in some cases, and the presence of key microbial partners (Amann et al. 1995; Eckburg et al. 2005). The inability to grow these organisms has undoubtedly limited our understanding of the ecology of indigenous microbial communities. State-of-the-art whole community sequencing technology via metagenomics has opened the door to in vivo studies of microbial populations and communities. By definition, metagenomic sequencing characterizes the collection of all the genetic material isolated from an environmental sample without traditional cultivation (Handelsman 2004; Iverson et al. 2012; Mackelprang et al. 2011). This has aided the development of systems-level insights into the structure and function of microbial ecosystems (Handelsman 2004; Gilbert and Dupont 2011). Advancements in sequencing technologies and throughput have, and continue to improve our ability to characterize the genomic contents of microbial communities down to the rare biosphere (Eckburg et al. 2005; Sogin et al. 2006).
Metagenomic sequencing results in a dataset of sequence reads that belong to the various species that make up the microbial community. Assembly of these datasets into stretches of contiguous DNA sequences, termed contigs, can be complicated by the presence of conserved genomic regions across species. Development of metagenomic specific short reads assembly algorithms and tools that can disentangle these similar sequences originating from different taxa has improved the quality of metagenomic assemblies (Pevzner et al. 2001), those include IDBA-UD (Peng et al. 2012), MetaVelvet (Namiki et al. 2012), SOAPdenovo (Li et al. 2010; Luo et al. 2012), ABYSS (Simpson et al. 2009), Khmer (Pell et al. 2012; Howe et al. 2012), Ray-meta (Boisvert et al. 2012), MEGAHIT (Li et al. 2015, 2016), and metaSPAdes (Nurk et al. 2017). Binning of these contigs based on genomic characteristics like GC content, tetramer frequency, sequence coverage, among others has enabled researchers to identify sets of contigs that belong to the same species. These advancements have resulted in the concept of metagenome-assembled genomes (MAGs), which represent the collection of all contigs or scaffolds from a single or closely related strains of a given species. Developments in bioinformatics tools used in assembly and binning have made the recovery of genomes from metagenomic datasets a routine analysis, including rare species and draft genomes from previously uncultivated species (Albertsen et al. 2013). Binning algorithms and tools have been reviewed previously (Sangwan et al. 2016; Breitwieser et al. 2017). For each species, the genetic contents of all strains in the population are included in a species bin, although sequencing depth, library construction methods, presence of host DNAs, and other factors may affect the metagenomic sequencing results (Zaheer et al. 2018; Pereira-Marques et al. 2019; Bowers et al. 2015).
MAGs have led to the discovery of a remarkable amount of genomic diversity and the characterization of novel bacterial membership. However, MAGs should always be used with caution for the reasons discussed above. False positives in binning, conflicted, and incomplete MAGs have been observed for a variety of different binning tools that can reduce the quality of public genome repositories if MAGs are not evaluated carefully (Shaiber and Eren 2019). Multiple studies have suggested that downstream MAGs quality assessment and validation steps are critical, and available tools published recently to serve such purpose include MetaQUAST (Koren and Phillippy 2015), CheckM (Parks et al. 2015), MAGpy (Stewart et al. 2019), Anvio (Eren et al. 2015), AMBER (Meyer et al. 2018), and DAS tool (Sieber et al. 2018). Further refinement, stringent quality assessment, extending assembly length through re-assembly after recruiting reads back to the MAGs, and genome completeness assessments are important and necessary steps to ensure the fidelity of the MAGs (Eren et al. 2015). High-quality metagenome-deconvoluted genomes are essential to perform genome-centric in vivo analyses of microbial ecosystems.
3 Metagenome-Assembled Genomes Revealed Extensive within Community Intraspecies Diversity in a Microbial Community
Microbial populations often composed of multiple strains of each species, and the resulting intraspecies diversity could have significant functional and clinical implications (Kraal et al. 2014; Greenblum et al. 2015; Oh et al. 2014). Gel microdroplet cultivation afforded nearly finished single genomes and revealed substantial intraspecies diversity within human oral and fecal microbiomes (Fitzsimons et al. 2013). Strains of dominant human skin bacterial species were shown to be heterogeneous and multiphyletic, which the authors suggested to be the result of micro-scale differences in the environment that shaped the ecology and evolution of each subpopulation (Oh et al. 2014). Another study reported extensive strain-level variation detected in the human gut microbiome using large-scale intraspecies copy number variation (Greenblum et al. 2015). This intraspecies variation is thought to be associated with obesity and inflammatory bowel disease. These studies highlight the complex relationships between within-species diversity and functional capacity, linking compositional shifts to subspecies-level variations.
Intra-species diversity obviously complicates MAGs generation, a problem that is compounded by the use of short-read sequencing technology. It is difficult to establish linkage and synteny between genotypes in a species genome. Binning strategies can separate sequences that belong to different species, but are generally not capable of distinguishing between strains of the same species in a metagenomic dataset (Huson et al. 2011). There are encouraging developments in binning algorithms recently that have addressed strain-level resolution from metagenomic short-read sequencing such as StrainPhlAn (Truong et al. 2017), ConStrains (Luo et al. 2015), MetaSNV (Costea et al. 2017), and DESMAN (Quince et al. 2017). However, the word “strain” has been used interchangeably with subspecies type, genotype, biotype, among others, in metagenome-derived strain-level resolution analyses. Although intraspecies diversity can be purged during assembly, the remainder often leads to species bins that contain composite genetic information from multiple genotypes (strains) of the species. Advancements in chromosome conformation capture (Hi-C) and long-read sequencing technologies such as PacBio SMRT sequencing and Oxford nanopore technologies could improve strain deconvolution from metagenomic data by extending the read length and assembly quality (Frank et al. 2016; Tsai et al. 2016; Belton et al. 2012). However, these technologies have not been widely adopted probably due to technical limitations.
4 A Practical Definition of Meta-Pangenome
The pangenome has been an important concept and a tool used in comparative genomics to dissect microbial diversity. A pangenome generally refers to the entire collection of genetic content from all strains of a species (Tettelin et al. 2005; Medini et al. 2005; Vernikos et al. 2015). By definition, a pangenome represents all of the genetic potentials of a species and is typically determined by homology among sets of genes belonging to multiple strains of the species in all environments the species is found. Here, we extend the pangenome concept to incorporate metagenome-derived genes and genomes. It is a natural extension as MAGs and metagenomic contigs have been used to generate species-specific gene catalog and that for all species present in a given environment (Ma et al. 2019). We introduce the term, meta-pangenome that refers to the union of genes of a species found in a habitat using both culture-independent sequencing (metagenome) and culture-based sequencing (genome) methods. In computational terms, the meta-pangenome is the entire sequence space of a species in an environment. Thus, within a sample, a metagenomic species represents known combinations of strains of a species. In this chapter, we choose to discuss the meta-pangenome in the context of a species, while the meta-pangenome paradigm can be applied to genera or broader of taxonomic groups (Lefebure and Stanhope 2007) as well as other domains of life such as fungus (McCarthy and Fitzpatrick 2019). The term “pan” itself means “whole” or “everything”, and “meta” as a prefix could mean “with”, “among”, and “beyond”. Together the words “meta-pangenome” literally mean whole genomes of a species from among samples collected in a given environment.
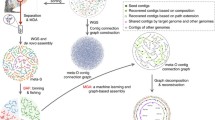
Similar to the pangenome concept, a meta-pangenome is bound to a specific species. In order to define the meta-pangenome for a species, say species A, we start from collecting all available genomes and constructing MAGs of species A from metagenomes (illustrated in Fig. 1). We then perform gene calling for these MAGs contigs after quality assessment, followed by similarity search to generate homologous gene clusters as in conventional pangenome analyses. The final step is to perform meta-pangenome size interpolation and extrapolation for species A. This procedure can then be repeated for each of the species present in a particular environment to define their meta-pangenome. Alternatively, the genetic contents characterized in all metagenomes and genomes of a habitat can be collectively pooled to generate homologous gene clusters. Taxonomic assignment of the resulting gene clusters can then be used to produce meta-pangenomes for each of the species present in the habitat.
Illustration for a workflow to generate a meta-pangenome for a species. The steps could be modified. For example, the step of gene calling could be after the step of the pooling all deconvoluted assemblies for a species. Alternatively, the genetic contents characterized in all metagenomes and genomes of a habitat can be collectively pooled to generate homologous gene clusters. Taxonomic assignment of the resulting gene clusters can then be used to produce meta-pangenomes for each of the species present in the habitat
We can then apply the concepts of core, accessory, and unique genes to the meta-pangenome framework. A species meta-pangenome core genes are those consistently present in all or almost all metagenomes in a habitat such as wastewater or the GI tract, and meta-pangenome-specific genes are only observed a single sample of the habitat. The variable or accessory meta-pangenome includes those genes only present in a subset of populations. As a metagenome can be considered a snapshot of the microbial community genetic potential at the time of collection, the core meta-pangenome can be referred as the set of genes being repeatedly observed after multiple sampling events. A closed meta-pangenome would thus refer to the case where no or very few new genes of the species are added with each additional metagenome sequenced. Conversely, a species open meta-pangenome would refer to the case where a substantive number of new genes for that species are discovered with each additional metagenome sequenced. The core meta-pangenome for a species could be quite small, or even nonexistent, if the abiotic and biotic constraints on its colonization of the environment are loose or large if these constraints are strict.
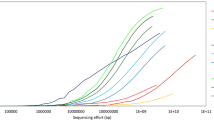
Similar to the original pangenome ecological significance (Tettelin et al. 2005), population size and niche versatility are likely to drive the size of a meta-pangenome. For example, the meta-pangenome of Gardnerella vaginalis, a highly prevalent bacterial colonizer of human vagina, is a collection of all the genes assigned to that species derived from all available vaginal metagenomes and genomes. Despite hundreds of metagenomes available containing G. vaginalis, this important species shows an open meta-pangenome (Fig. 2). On the other hand, Lactobacillus gasseri, another important and beneficial vaginal bacterial species demonstrates an essentially closed meta-pangenome such that new metagenome sequences add relatively few genes. An in-depth understanding of the genetic diversity of constituent community members and its relation to community dysbiosis will afford the development of novel strategies to evaluate and optimize prevention, diagnostics, and treatment for adverse health conditions.
Species-specific metagenome accumulation curves for the number of homologous gene clusters. Figure reproduced from Ma et al. (2019)
5 A Conceptual Framework for Microbial Comparative Genomics: Meta-Pangenome, Metagenomic Subspecies, and Pan-Metagenome
Meta-pangenome forms a practical framework that provides unprecedented insights into the genetic and functional basis underlying ecological fitness of microbial population in an environmental niche. The variable or accessory meta-pangenome of a species are the genes only present in a subset but not all of samples, which has led to the new concept of “metagenomic subspecies” (Ma et al. 2019). In essence, a metagenomic subspecies represents a slice of a species’ meta-pangenome that is commonly identified in metagenomic samplings of a habitat. This slice contains the genetic contents of a combination of strains that tend to co-occur. In theory, this co-occurrence could be driven by interactions among the strains and/or their tendency to co-colonize, termed dispersal limitations (Telford et al. 2006). Specific mechanisms that can lead to the co-existence of multiple strains in a population include frequency-dependent selection (Svensson and Connallon 2019), cross-feeding (Livingston et al. 2012; Hunt and Bonsall 2009), spatial structure (France and Forney 2019), resource partitioning (Rosenzweig et al. 1994), and interference competition (Kerr et al. 2002), among others. That said, the metagenomic subspecies concept is equivalent to a species genetic “ecotype” for an environment. Several metagenomic subspecies can exist in a given environment but cannot co-occur within a sample. The metagenomic subspecies can be determined in silico by hierarchical clustering over the data matrix such as gene prevalence or gene abundance profiles. Further development of relevant pattern recognition tools (supervised or unsupervised) as well as the approximation of the population size (number of strains) are important ongoing research developments that will contribute to this field.
The concepts of meta-pangenome and metagenomic subspecies have great value to investigate intraspecies diversity within a community and the genetic foundation underlining the functions, resilience, resistance or fitness, among others, of microbial communities. We term the entire collection of all species’ meta-pangenomes that exist in a specific environment the “pan-metagenome,” which is essentially the “habitome” that encompasses the genetic landscape of a habitat. For instance, the pan-metagenome of the human gastrointestinal (GI) tract is the collection of all genes of all species found in the human GI tract (Qin et al. 2010; Li et al. 2014), and the pan-metagenome of the human oral communities encompasses the total genetic content of all species in the human oral environment (Tierney et al. 2019). The concept of pan-metagenome is represented by extensive gene cataloging, such as those constructed for the pig (Xiao et al. 2016) or the mouse GI tract (Xiao et al. 2015). A pan-metagenome of a specific habitat, when used as a catalog of the genetic contents, has provided a comprehensive reference framework for the study of microbial communities and their interaction with the environment.
We have recently constructed a pan-metagenome for the human vaginal tract named VIRGO (the human vaginal nonredundant gene catalog) using an array of urogenital bacterial isolate genomes and vaginal metagenomes (Ma et al. 2019). VIRGO has been shown to be comprehensive and to provide an unbiased representation of the genetic diversity of each species found in the vaginal microbiome. In building VIRGO, we found that the vast majority of the genetic diversity was contributed by MAGs derived from the metagenomic datasets. In fact, the metagenomic data used to build VIRGO comprise a much larger genetic diversity (high number of nonredundant genes) than that of all combined single isolate genome sequences (Fig. 3a, b). This result indicates the importance of extending the pangenome concept beyond isolate genome sequences.
Intraspecies diversity revealed using VIRGO (human vaginal nonredundant gene catalog) of seven vaginal species including L. crispatus, L. iners, L. jensenii, L. gasseri, and G. vaginalis, A. vaginae and P. timonensis. (a) Summary of the number (N) of isolate genomes and metagenome (MG) samples with more than 80% of their average genome’s number of coding genes for a species, based on a dataset of 1507 in-house vaginal metagenomes characterized using VIRGO. (b) Boxplot of number nonredundant genes in isolate genomes versus vaginal metagenomes. (c) Heatmap of presence/absence of L. crispatus nonredundant gene profiles for 56 available isolate genomes and 413 VIRGO-characterized metagenomes that contained either high (red) or low (blue) relative abundance of the species. Hierarchical clustering of the profiles was performed using ward linkage based on their Jaccard similarity coefficient. ∗number of isolate genomes and metagenome samples. †MG: Metagenomes ∗p < 0.05, ∗∗∗p < 0.001 after correction for multiple comparisons. Figure reproduced from Ma et al. (2019)
VIRGO has afforded a different view of the vaginal microbiome, where each population is composed of complex mixtures of multiple strains, highlighting the large amount of intraspecies diversity present in these communities. We found that, in general, the majority of a species’ genes are meta-pangenomic accessory genes. For example, for Lactobacillus crispatus, the number of meta-pangenomic accessory genes is twice as many as the number of meta-pangenomic core genes (Fig. 3c). G. vaginalis demonstrated particularly high intraspecies diversity, for which the core meta-pangenome does not even exist and the majority of the genes are accessory or sample specific, suggesting that the species should be split into multiple different species within the genus Gardnerella. We further observed three distinct metagenomic subspecies of L. gasseri, among which there were two distinct types and the third being a combination of the two (Fig. 3d). This suggests that there is environmental specialized co-colonization of L. gasseri strains in the vaginal environment. Future studies are needed to reveal the linkage between specific metagenomic subspecies and pathophysiological conditions.
6 Conclusion Remarks
The field of comparative genomics has bloomed from that initial genome comparison two decades ago. Thanks to advancements in cultivation-independent whole community sequencing technology and the increased availability of metagenome-assembled genomes, we have obtained unprecedented insights into the incredible amount of diversity present within microbial populations. Intraspecies diversity exceeds that found in our current reference genome databases. The pangenome paradigm expanded to metagenome-assembled genomes and metagenomic contigs comprehensively profile microbial genetic diversity in a specific habitat. However, the incorporation of metagenome-derived genomes has to be performed carefully with stringent quality assessment to avoid spurious inflation of gene content. The meta-pangenome concept unites pangenomics and metagenomics to obtain a more compete and ecologically meaningful view of different ecosystems. Meta-pangenomes and pan-metagenomes represent a critical step in the development of a systems-level understanding of microbial ecosystems.
References
Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH (2013) Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31:533–538
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in-situ detection of individual microbial-cells without cultivation. Microbiol Rev 59:143–169
Bakken LR (1985) Separation and purification of bacteria from soil. Appl Environ Microbiol 49:1482–1487
Belton JM, McCord RP, Gibcus JH, Naumova N, Zhan Y, Dekker J (2012) Hi-C: a comprehensive technique to capture the conformation of genomes. Methods 58:268–276
Boisvert S, Raymond F, Godzaridis E, Laviolette F, Corbeil J (2012) Ray meta: scalable de novo metagenome assembly and profiling. Genome Biol 13:R122
Bowers RM, Clum A, Tice H, Lim J, Singh K, Ciobanu D, Ngan CY, Cheng JF, Tringe SG, Woyke T (2015) Impact of library preparation protocols and template quantity on the metagenomic reconstruction of a mock microbial community. BMC Genomics 16:856
Breitwieser FP, Lu J, Salzberg SL (2017) A review of methods and databases for metagenomic classification and assembly. Brief Bioinform 20(4):1125–1136
Costea PI, Munch R, Coelho LP, Paoli L, Sunagawa S, Bork P (2017) metaSNV: a tool for metagenomic strain level analysis. PLoS One 12:e0182392
Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA (2005) Diversity of the human intestinal microbial flora. Science 308:1635–1638
Eren AM, Esen OC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO (2015) Anvi’o: an advanced analysis and visualization platform for ’omics data. PeerJ 3:e1319
Fitzsimons MS, Novotny M, Lo CC, Dichosa AE, Yee-Greenbaum JL, Snook JP, Gu W, Chertkov O, Davenport KW, McMurry K et al (2013) Nearly finished genomes produced using gel microdroplet culturing reveal substantial intraspecies genomic diversity within the human microbiome. Genome Res 23:878–888
Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM et al (1995) Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496–512
France MT, Forney LJ (2019) The relationship between spatial structure and the maintenance of diversity in microbial populations. Am Nat 193:503–513
France MT, Mendes-Soares H, Forney LJ (2016) Genomic comparisons of lactobacillus crispatus and lactobacillus iners reveal potential ecological drivers of community composition in the vagina. Appl Environ Microbiol 82:7063–7073
Frank JA, Pan Y, Tooming-Klunderud A, Eijsink VGH, McHardy AC, Nederbragt AJ, Pope PB (2016) Improved metagenome assemblies and taxonomic binning using long-read circular consensus sequence data. Sci Rep 6:25373
Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, Bult CJ, Kerlavage AR, Sutton G, Kelley JM et al (1995) The minimal gene complement of mycoplasma genitalium. Science 270:397–403
Gilbert JA, Dupont CL (2011) Microbial metagenomics: beyond the genome. Annu Rev Mar Sci 3:347–371
Greenblum S, Carr R, Borenstein E (2015) Extensive strain-level copy-number variation across human gut microbiome species. Cell 160(4):583–594
Handelsman J (2004) Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev 68:669–685
Hardison RC (2003) Comparative genomics. PLoS Biol 1:E58
Howe A, Pell J, Canino-Koning R, Mackelprang R, Tringe S, Jansson J, Tiedje JM, Brown CT (2012) Illumina sequencing artifacts revealed by connectivity analysis of metagenomic datasets
Hunt JJ, Bonsall MB (2009) The effects of colonization, extinction and competition on co-existence in metacommunities. J Anim Ecol 78:866–879
Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC (2011) Integrative analysis of environmental sequences using MEGAN4. Genome Res 21:1552–1560
Iverson V, Morris RM, Frazar CD, Berthiaume CT, Morales RL, Armbrust EV (2012) Untangling genomes from metagenomes: revealing an uncultured class of marine Euryarchaeota. Science 335:587–590
Kerr B, Riley MA, Feldman MW, Bohannan BJ (2002) Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 418:171–174
Koren S, Phillippy AM (2015) One chromosome, one contig: complete microbial genomes from long-read sequencing and assembly. Curr Opin Microbiol 23:110–120
Kraal L, Abubucker S, Kota K, Fischbach MA, Mitreva M (2014) The prevalence of species and strains in the human microbiome: a resource for experimental efforts. PLoS One 9:e97279
Lefebure T, Stanhope MJ (2007) Evolution of the core and pan-genome of streptococcus: positive selection, recombination, and genome composition. Genome Biol 8:R71
Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K et al (2010) De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20:265–272
Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, Arumugam M, Kultima JR, Prifti E, Nielsen T et al (2014) An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol 32:834–841
Li D, Liu CM, Luo R, Sadakane K, Lam TW (2015) MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676
Li D, Luo R, Liu CM, Leung CM, Ting HF, Sadakane K, Yamashita H, Lam TW (2016) MEGAHIT v1.0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 102:3–11
Livingston G, Matias M, Calcagno V, Barbera C, Combe M, Leibold MA, Mouquet N (2012) Competition-colonization dynamics in experimental bacterial metacommunities. Nat Commun 3:1234
Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q et al (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 1(1):18
Luo C, Knight R, Siljander H, Knip M, Xavier RJ, Gevers D (2015) ConStrains identifies microbial strains in metagenomic datasets. Nat Biotechnol 33:1045–1052
Ma B, France M, Crabtree J, Holm J, Humphrys M, Brotman R, Ravel J (2019) VIRGO, a comprehensive non-redundant gene catalog, reveals extensive within community intraspecies diversity in the human vagina. bioRxiv
Mackelprang R, Waldrop MP, DeAngelis KM, David MM, Chavarria KL, Blazewicz SJ, Rubin EM, Jansson JK (2011) Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature 480:368–371
McCarthy CGP, Fitzpatrick DA (2019) Pan-genome analyses of model fungal species. Microb Genom 5:e000243
Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R (2005) The microbial pan-genome. Curr Opin Genet Dev 15:589–594
Meyer F, Hofmann P, Belmann P, Garrido-Oter R, Fritz A, Sczyrba A, McHardy AC (2018) AMBER: assessment of Metagenome BinnERs. Gigascience 7
Miller W, Makova KD, Nekrutenko A, Hardison RC (2004) Comparative genomics. Annu Rev Genomics Hum Genet 5:15–56
Namiki T, Hachiya T, Tanaka H, Sakakibara Y (2012) MetaVelvet: an extension of velvet assembler to de novo metagenome assembly from short sequence reads. Nucleic Acids Res 40:e155
Nurk S, Meleshko D, Korobeynikov A, Pevzner PA (2017) metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834
Oh J, Byrd AL, Deming C, Conlan S, Program NCS, Kong HH, Segre JA (2014) Biogeography and individuality shape function in the human skin metagenome. Nature 514:59–64
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW (2015) Check M: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055
Pell J, Hintze A, Canino-Koning R, Howe A, Tiedje JM, Brown CT (2012) Scaling metagenome sequence assembly with probabilistic de Bruijn graphs. Proc Natl Acad Sci U S A 109:13272–13277
Peng Y, Leung HC, Yiu SM, Chin FY (2012) IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28:1420–1428
Pereira-Marques J, Hout A, Ferreira RM, Weber M, Pinto-Ribeiro I, van Doorn LJ, Knetsch CW, Figueiredo C (2019) Impact of host DNA and sequencing depth on the taxonomic resolution of whole Metagenome sequencing for microbiome analysis. Front Microbiol 10:1277
Pevzner PA, Tang H, Waterman MS (2001) An Eulerian path approach to DNA fragment assembly. Proc Natl Acad Sci U S A 98:9748–9753
Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T et al (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65
Quince C, Delmont TO, Raguideau S, Alneberg J, Darling AE, Collins G, Eren AM (2017) DESMAN: a new tool for de novo extraction of strains from metagenomes. Genome Biol 18:181
Rosenzweig RF, Sharp RR, Treves DS, Adams J (1994) Microbial evolution in a simple unstructured environment: genetic differentiation in Escherichia coli. Genetics 137:903–917
Sangwan N, Xia F, Gilbert JA (2016) Recovering complete and draft population genomes from metagenome datasets. Microbiome 4:8
Shaiber A, Eren AM (2019) Composite metagenome-assembled genomes reduce the quality of public genome repositories. MBio 10(3):e00725–e00719
Sieber CMK, Probst AJ, Sharrar A, Thomas BC, Hess M, Tringe SG, Banfield JF (2018) Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat Microbiol 3:836–843
Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I (2009) ABySS: a parallel assembler for short read sequence data. Genome Res 19:1117–1123
Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ (2006) Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci U S A 103:12115–12120
Stewart RD, Auffret MD, Snelling TJ, Roehe R, Watson M (2019) MAGpy: a reproducible pipeline for the downstream analysis of metagenome-assembled genomes (MAGs). Bioinformatics 35:2150–2152
Svensson EI, Connallon T (2019) How frequency-dependent selection affects population fitness, maladaptation and evolutionary rescue. Evol Appl 12:1243–1258
Telford RJ, Vandvik V, Birks HJ (2006) Dispersal limitations matter for microbial morphospecies. Science 312:1015
Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS et al (2005) Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci U S A 102:13950–13955
Tierney BT, Yang Z, Luber JM, Beaudin M, Wibowo MC, Baek C, Mehlenbacher E, Patel CJ, Kostic AD (2019) The landscape of genetic content in the gut and Oral human microbiome. Cell Host Microbe 26:283–295. e288
Touchman J (2010) Comparative genomics. Nat Educ Knowl 3:13
Truong DT, Tett A, Pasolli E, Huttenhower C, Segata N (2017) Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res 27:626–638
Tsai YC, Conlan S, Deming C, Program NCS, Segre JA, Kong HH, Korlach J, Oh J (2016) Resolving the complexity of human skin metagenomes using single-molecule sequencing. MBio 7:e01948–e01915
Vernikos G, Medini D, Riley DR, Tettelin H (2015) Ten years of pan-genome analyses. Curr Opin Microbiol 23:148–154
Xia X (2013) Comparative genomics. In Briefs in Genetics. Springer, Heidelberg
Xiao L, Feng Q, Liang S, Sonne SB, Xia Z, Qiu X, Li X, Long H, Zhang J, Zhang D et al (2015) A catalog of the mouse gut metagenome. Nat Biotechnol 33:1103–1108
Xiao L, Estelle J, Kiilerich P, Ramayo-Caldas Y, Xia Z, Feng Q, Liang S, Pedersen AO, Kjeldsen NJ, Liu C et al (2016) A reference gene catalogue of the pig gut microbiome. Nat Microbiol 1:16161
Zaheer R, Noyes N, Ortega Polo R, Cook SR, Marinier E, Van Domselaar G, Belk KE, Morley PS, McAllister TA (2018) Impact of sequencing depth on the characterization of the microbiome and resistome. Sci Rep 8:5890
Acknowledgment
The author acknowledges The Gerber Foundation 2018 award.
Competing Interest Statement
The authors declare no competing financial and nonfinancial interests.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2020 The Author(s)
About this chapter
Cite this chapter
Ma, B., France, M., Ravel, J. (2020). Meta-Pangenome: At the Crossroad of Pangenomics and Metagenomics. In: Tettelin, H., Medini, D. (eds) The Pangenome. Springer, Cham. https://doi.org/10.1007/978-3-030-38281-0_9
Download citation
DOI: https://doi.org/10.1007/978-3-030-38281-0_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-38280-3
Online ISBN: 978-3-030-38281-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)