Abstract
Background
MIKC type MADS-box transcription factors are one of the largest gene families and play a pivotal role in flowering time and flower development. Chimonanthus salicifolius belongs to the family Calycanthaceae and has a unique flowering time and flowering morphology compared to other Chimonanthus species, but the research on MIKC type MADS-box gene family of C. salicifolius has not been reported.
Objective
Identification, comprehensive bioinformatic analysis, the expression pattern of MIKC-type MADS-box gene family from different tissues of C. salicifolius.
Methods
Genome-wide investigation and expression pattern under different tissues of the MIKC-type MADS-box gene family in C. salicifolius, and their phylogenetic relationships, evolutionary characteristics, gene structure, motif distribution, promoter cis-acting element were performed.
Results
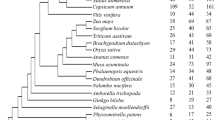
A total of 29 MIKC-type MADS-box genes were identified from the whole genome sequencing. Interspecies synteny analysis revealed more significant collinearity between C. salicifolius and the magnoliids species compared to eudicots and monocots. MIKC-type MADS-box genes from the same subfamily share similar distribution patterns, gene structure, and expression patterns. Compared with Arabidopsis thaliana, Nymphaea colorata, and Chimonanthus praecox, the FLC genes were absent in C. salicifolius, while the AGL6 subfamily was expanded in C. salicifolius. The selectively expanded promoter (AGL6) and lack of repressor (FLC) genes may explain the earlier flowering in C. salicifolius. The loss of the AP3 homologous gene in C. salicifolius is probably the primary cause of the morphological distinction between C. salicifolius and C. praecox. The csAGL6a gene is specifically expressed in the flowering process and indicates the potential function of promoting flowering.
Conclusion
This study offers a genome-wide identification and expression profiling of the MIKC-types MADS-box genes in the C. salicifolius, and establishes the foundation for screening flowering development genes and understanding the potential function of the MIKC-types MADS-box genes in the C. salicifolius.









Similar content being viewed by others
Availability of data and materials
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Code availability
Not applicable.
References
Alvarez-Buylla ER, Pelaz S, Liljegren SJ, Gold SE, Burgeff C, Ditta GS, Yanofsky MF (2000) An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc Natl Acad Sci 97:5328–5333
Amasino RM (2005) Vernalization and flowering time. Curr Opin Biotechnol 16:154–158
Anderson JT, Willis JH, Mitchell-Olds T (2011) Evolutionary genetics of plant adaptation. Trends Genet 27:258–266
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202-208
Cabin Z, Derieg NJ, Garton A, Ngo T, Quezada A, Gasseholm C, Simon M, Hodges SA (2022) Non-pollinator selection for a floral homeotic mutant conferring loss of nectar reward in Aquilegia coerulea. Curr Biol 32:1332.e1335-1341.e1335
Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4:10
Carlsbecker A, Tandre K, Johanson U, Englund M, Engstrom P (2004) The MADS-box gene DAL1 is a potential mediator of the juvenile-to-adult transition in Norway spruce (Picea abies). Plant J 40:546–557
Chang YY, Chiu YF, Wu JW, Yang CH (2009) Four orchid (Oncidium Gower Ramsey) AP1/AGL9-like MADS-box genes show novel expression patterns and cause different effects on floral transition and formation in Arabidopsis thaliana. Plant Cell Physiol 50:1425–1438
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13:1194–1202
Cho LH, Yoon J, An G (2017) The control of flowering time by environmental factors. Plant J 90:708–719
Cui L, Wall PK, Leebens-Mack JH, Lindsay BG, Soltis DE, Doyle JJ, Soltis PS, Carlson JE, Arumuganathan K, Barakat A, Albert VA, Ma H, dePamphilis CW (2006) Widespread genome duplications throughout the history of flowering plants. Genome Res 16:738–749
Cyranoski D (2018) The big push for Chinese medicine. Nature 561:448–450
Dong X, Deng H, Ma W, Zhou Q, Liu Z (2021) Genome-wide identification of the MADS-box transcription factor family in autotetraploid cultivated alfalfa (Medicago sativa L.) and expression analysis under abiotic stress. BMC Genom 22:603
Ernst J, Bar-Joseph Z (2006) STEM: a tool for the analysis of short time series gene expression data. BMC Bioinform 7:191
Ernst J, Nau GJ, Bar-Joseph Z (2005) Clustering short time series gene expression data. Bioinformatics 21(Suppl 1):i159-168
Fan J, Li W, Dong X, Guo W, Shu H (2007) Ectopic expression of a hyacinth AGL6 homolog caused earlier flowering and homeotic conversion in Arabidopsis. Sci China, Ser C Life Sci 50:676–689
Ge H, Shi YN, Zhang MX, Li X, Yin XR, Chen KS (2021) The MADS-Box transcription factor EjAGL65 controls Loquat flesh lignification via direct transcriptional inhibition of EjMYB8. Front Plant Sci 12:652959
Gramzow L, Theissen G (2010) A hitchhiker’s guide to the MADS world of plants. Genome Biol 11:1–11
Hemming MN, Trevaskis B (2011) Make hay when the sun shines: the role of MADS-box genes in temperature-dependant seasonal flowering responses. Plant Sci 180:447–453
Henschel K, Kofuji R, Hasebe M, Saedler H, Münster T, Theißen G (2002) Two ancient classes of MIKC-type MADS-box genes are present in the Moss Physcomitrella patens. Mol Biol Evol 19:801–814
Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35:W585-587
Hsing-Fun H, Chih-Hsiang H, Lu-Tung C, Chang-Hsien Y (2003) Ectopic expression of an orchid (Oncidium Gower Ramsey) AGL6-like gene promotes flowering by activating flowering time genes in Arabidopsis thaliana. Plant Cell Physiol 44:783–794
Hsu HF, Chen WH, Shen YH, Hsu WH, Mao WT, Yang CH (2021) Multifunctional evolution of B and AGL6 MADS box genes in orchids. Nat Commun 12:902
Huang B, Huang Z, Ma R, Ramakrishnan M, Chen J, Zhang Z, Yrjala K (2021) Genome-wide identification and expression analysis of LBD transcription factor genes in Moso bamboo (Phyllostachys edulis). BMC Plant Biol 21:296
Kanno A, Nakada M, Akita Y, Hirai M (2007) Class B gene expression and the modified ABC model in nongrass monocots. Sci World J 7:268–279
Katoh K, Rozewicki J, Yamada KD (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20:1160–1166
Kaufmann K, Melzer R, Theißen G (2005) MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347:183–198
Kim S, Koh J, Yoo MJ, Kong H, Hu Y, Ma H, Soltis PS, Soltis DE (2005) Expression of floral MADS-box genes in basal angiosperms: implications for the evolution of floral regulators. Plant J 43:724–744
Kuan J, Saier MH (1993) The Mitochondria1 carrier family of transport proteins: structural, functional, and evolutionary relationships. Crit Rev Biochem Mol Biol 28:209–233
Kumar S, Suleski M, Craig JM, Kasprowicz AE, Sanderford M, Li M, Stecher G, Hedges SB (2022) TimeTree 5: an expanded resource for species divergence times. Mol Biol Evol 39(8):msac174
Letunic I, Khedkar S, Bork P (2021) SMART: recent updates, new developments and status in 2020. Nucleic Acids Res 49:D458–D460
Li S-L, Zou Z-R (2018) Research progress on flavonoids and coumarins from Chimonanthus plants and its pharmacological activities. In: Chinese Traditional and Herbal Drugs, pp 3425–3430
Li B-J, Zheng B-Q, Wang J-Y, Tsai W-C, Lu H-C, Zou L-H, Wan X, Zhang D-Y, Qiao H-J, Liu Z-J, Wang Y (2020) New insight into the molecular mechanism of colour differentiation among floral segments in orchids. Commun Biol 3:1–13
Liang Y, Liu Q, Wang X, Huang C, Xu G, Hey S, Lin HY, Li C, Xu D, Wu L, Wang C, Wu W, Xia J, Han X, Lu S, Lai J, Song W, Schnable PS, Tian F (2019) ZmMADS69 functions as a flowering activator through the ZmRap2.7-ZCN8 regulatory module and contributes to maize flowering time adaptation. New Phytol 221:2335–2347
Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, Thanki N, Yamashita RA, Yang M, Zhang D, Zheng C, Lanczycki CJ, Marchler-Bauer A (2020) CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res 48:D265–D268
Lv Q, Qiu J, Liu J, Li Z, Zhang W, Wang Q, Fang J, Pan J, Chen Z, Cheng W, Barker MS, Huang X, Wei X, Cheng K (2020) The Chimonanthus salicifolius genome provides insight into magnoliid evolution and flavonoid biosynthesis. Plant J 103:1910–1923
Morel P, Chambrier P, Boltz V, Chamot S, Rozier F, Rodrigues Bento S, Trehin C, Monniaux M, Zethof J, Vandenbussche M (2019) Divergent functional diversification patterns in the SEP/AGL6/AP1 MADS-Box transcription factor superclade. Plant Cell 31:3033–3056
Ning K, Han Y, Chen Z, Luo C, Wang S, Zhang W, Li L, Zhang X, Fan S, Wang Q (2019) Genome-wide analysis of MADS-box family genes during flower development in lettuce. Plant, Cell Environ 42:1868–1881
Ohmori S, Kimizu M, Sugita M, Miyao A, Hirochika H, Uchida E, Nagato Y, Yoshida H (2009) MOSAIC FLORAL ORGANS1, anAGL6-Like MADS box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 21:3008–3025
Parenicova L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, Angenent GC, Colombo L (2003) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15:1538–1551
Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD (2018) HMMER web server: 2018 update. Nucleic Acids Res 46:W200–W204
Ren Z, Yu D, Yang Z, Li C, Qanmber G, Li Y, Li J, Liu Z, Lu L, Wang L, Zhang H, Chen Q, Li F, Yang Z (2017) Genome-wide identification of the MIKC-Type MADS-Box gene family in Gossypium hirsutum L. unravels their roles in flowering. Front Plant Sci 8:384
Rijpkema AS, Royaert S, Zethof J, van der Weerden G, Gerats T, Vandenbussche M (2006) Analysis of the Petunia TM6 MADS box gene reveals functional divergence within the DEF/AP3 lineage. Plant Cell 18:1819–1832
Ruelens P, de Maagd RA, Proost S, Theissen G, Geuten K, Kaufmann K (2013) FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nat Commun 4:2280
Schilling S, Kennedy A, Pan S, Jermiin LS, Melzer R (2020) Genome-wide analysis of MIKC-type MADS-box genes in wheat: pervasive duplications, functional conservation and putative neofunctionalization. New Phytol 225:511–529
Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ (2006) The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133:251–261
Shang J, Tian J, Cheng H, Yan Q, Li L, Jamal A, Xu Z, Xiang L, Saski CA, Jin S, Zhao K, Liu X, Chen L (2020) The chromosome-level wintersweet (Chimonanthus praecox) genome provides insights into floral scent biosynthesis and flowering in winter. Genome Biol 21:200
Shen G, Yang CH, Shen CY, Huang KS (2019) Origination and selection of ABCDE and AGL6 subfamily MADS-box genes in gymnosperms and angiosperms. Biol Res 52:25
Staedler YM, Weston PH, Endress PK (2007) Floral Phyllotaxis and floral architecture in Calycanthaceae (Laurales). Int J Plant Sci 168:285–306
Stone R (2008) Lifting the veil on traditional Chinese medicine. Science 319:709–710
Theißen G (2001) Development of floral organ identity: stories from the MADS house. Curr Opin Plant Biol 4:75–85
Theißen G, Kim JT, Saedler H (1996) Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J Mol Evol 43:484–516
Tuan PA, Bai S, Saito T, Ito A, Moriguchi T (2017) Dormancy-associated MADS-Box (DAM) and the abscisic acid pathway regulate pear endodormancy through a feedback mechanism. Plant Cell Physiol 58:1378–1390
Vachon G, Engelhorn J, Carles CC (2018) Interactions between transcription factors and chromatin regulators in the control of flower development. J Exp Bot 69:2461–2471
Wang B-G, Zhang Q, Wang L-G, Duan K, Pan A-H, Tang X-M, Sui S-Z, Li M-Y (2011) The AGL6-like Gene CpAGL6, a potential regulator of floral time and organ identity in Wintersweet (Chimonanthus praecox). J Plant Growth Regul 30:343–352
Wang K-W, Li D, Wu B, Cao X-J (2016) New cytotoxic dimeric and trimeric coumarins from Chimonanthus salicifolius. Phytochem Lett 16:115–120
Wang P, Wang S, Chen Y, Xu X, Guang X, Zhang Y (2019) Genome-wide analysis of the MADS-box gene family in watermelon. Comput Biol Chem 80:341–350
Waterhouse A, Procter J, Martin DA, Barton GJ (2005) Jalview: visualization and analysis of molecular sequences, alignments, and structures. BMC Bioinform 6:1–1
Wen H-P, Fei J, Ji J (2013) Study on antidiarrheal effect of Chimonanthus salicifolius. Chin Arch Trad Chin Med, pp 182–183
WenBo W, Xiangfeng H, Xiaoying L, Wenhe W (2022) Transcriptome profiling during double-flower development provides insight into stamen petaloid in cultivated Lilium. Ornam Plant Res 2:1–11
WenChieh T, HongHwa C (2007) The orchid MADS-box genes controlling floral morphogenesis. In: Orchid Biotechnology, pp 163–183
Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, DF H (1999) Protein identification and analysis tools on the ExPASy server. Methods Mol Biol 112:531–552
Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM (1990) The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346:35–39
Yoo SK, Wu X, Lee JS, Ahn JH (2011) AGAMOUS-LIKE 6 is a floral promoter that negatively regulates the FLC/MAF clade genes and positively regulates FT in Arabidopsis. Plant J 65:62–76
Zhang Q, Wang B-G, Duan K, Wang L-G, Wang M, Tang X-M, Pan A-H, Sui S-Z, Wang G-D (2011) The paleoAP3-type gene CpAP3, an ancestral B-class gene from the basal angiosperm Chimonanthus praecox, can affect stamen and petal development in higher eudicots. Dev Genes Evol 221:83–93
Zhang Y, Tang D, Lin X, Ding M, Tong Z (2018) Genome-wide identification of MADS-box family genes in moso bamboo (Phyllostachys edulis) and a functional analysis of PeMADS5 in flowering. BMC Plant Biol 18:176
Zhang L, Chen F, Zhang X, Li Z, Zhao Y, Lohaus R, Chang X, Dong W, Ho SYW, Liu X, Song A, Chen J, Guo W, Wang Z, Zhuang Y, Wang H, Chen X, Hu J, Liu Y, Qin Y, Wang K, Dong S, Liu Y, Zhang S, Yu X, Wu Q, Wang L, Yan X, Jiao Y, Kong H, Zhou X, Yu C, Chen Y, Li F, Wang J, Chen W, Chen X, Jia Q, Zhang C, Jiang Y, Zhang W, Liu G, Fu J, Chen F, Ma H, Van de Peer Y, Tang H (2020) The water lily genome and the early evolution of flowering plants. Nature 577:79–84
Zheng Q, Si-Jie Z, You-Quan J, Xin-Feng Z (2019) Transcriptome sequencing of leaves and flowers and screening and expression of differential genes in aroma synthesis in Chimonanthus salicifolius. J Agric Biotechnol 27:844–855
Zhou S, Renner SS, Wen J (2006) Molecular phylogeny and intra- and intercontinental biogeography of Calycanthaceae. Mol Phylogenet Evol 39:1–15
Zhou X-L, Yu Y, Zhou C, Li S, Zhu Y-G, Xiao-Ling Y (2014) Anti-depression effect of Chimonanthus salicifolicus essential oil in chronic stressed rats. J Med Plants Res 8:430–435
Acknowledgements
The authors would like to thank Dr. Jin-Peng Wan (XTBG Tropical Botanical Garden, Chinese Academy of Sciences) and Yang Yang (Zhejiang A&F University) for linguistic assistance during preparation of this manuscript.
Funding
This study was supported by the Research Development Fund of Zhejiang A&F University (2019FR028) and Open Fund of Zhejiang Provincial Key Laboratory of Germplasm Innovation and Utilization for Garden Plants (2020E10013-K202104) and The key research and development program of Zhejiang Province (2021C02071).
Author information
Authors and Affiliations
Contributions
G-FF: Conceptualization Methodology, Data curation, Writing—original draft. Y-LY: Methodology, Writing—review and editing. J-GG: Software, Validation. FQ, XZ, Z-SW and W-YG: Investigation. DB ang Y-LY: Supervision. Z-HB: Project administration. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gui, FF., Jiang, GG., Bin Dong et al. Genome-wide identification and analysis of MIKC-type MADS-box genes expression in Chimonanthus salicifolius. Genes Genom 45, 1127–1141 (2023). https://doi.org/10.1007/s13258-023-01420-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-023-01420-7