Abstract
Background
About 5–10% of breast cancer cases are related to genetic and hereditary factors. The application of Next Generation Sequencing (NGS) in oncology has allowed the identification of genetic variants present in several genes related to the increased risk of breast cancer. This study aimed to determine the frequency of germline genetic variants in patients with a family and/or personal history of breast cancer.
Methods
An analysis of positive reports from NGS panels was carried out in female individuals with a personal and/or family history of breast cancer, present in the database of a private laboratory in Brazil.
Results
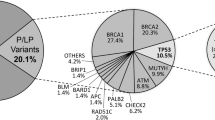
From about 2000 reports, 183 individuals presented 219 different germline genetic variants. The genes with the highest number of variants were BRCA2 (16.0%), ATM (15.0%) and BRCA1 (12.8%). Among the variants found, 78 were either pathogenic or probably pathogenic, accounting for 35% of all variants discovered. The gene with the highest proportion of pathogenic/probably pathogenic variants was TP53 (80%) and the most frequent pathogenic variant was also reported in this gene (c.1010G > A p.(Arg337His)). Furthermore, the study obtained a high proportion of variants of uncertain significance (VUS) (65%) and approximately 32% of the variants found were in genes of moderate penetrance.
Conclusions
Our results could improve the risk estimation and clinical follow-up of Brazilian patients with a history of breast cancer.


Similar content being viewed by others
Data availability
Database will be made available if required.
Code availability
Not applicable.
References
World Health Organization (2020) GLOBOCAN International Agency for Research on Cancer. https://gco.iarc.fr/today/data/factsheets/populations/900-world-fact-sheets.pdf . Accessed 12 Sept 2021
Sung H, Ferlay J, Siegel RL et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209–249. https://doi.org/10.3322/caac.2166
Instituto Nacional de Câncer José Alencar Gomes da Silva, INCA (2020) Estimativa 2020: incidência do Câncer no Brasil. https://www.inca.gov.br/sites/ufu.sti.inca.local/files/media/document/estimativa-2020-incidencia-de-cancer-no-brasil.pdf. Accessed 12 Sept 2021
Instituto Nacional de Câncer José Alencar Gomes da Silva, INCA (2021) Atlas da mortalidade. https://www.inca.gov.br/app/mortalidade. Accessed 12 Sept 2021
Silva PA, Riul SS (2011) Câncer de mama: fatores de risco e detecção precoce. Rev Bras Enferm 64:1016–1021. https://doi.org/10.1590/S0034-71672011000600005
Amendola LCB, Vieira R (2005) BRCA genes contribution in the hereditary predisposition for breast cancer. RBAC 51:325–330. https://doi.org/10.21877/2448-3877.201800615
Couch FJ, Nathanson KL, Offit K (2014) Two decades after BRCA: setting paradigms in personalized cancer care and prevention. Science 343:1466–1470. https://doi.org/10.1126/science.1251827
Harbeck N, Penault-Llorca F, Cortes J et al (2019) Breast cancer. Nat Rev Dis Primers 5:66. https://doi.org/10.1038/s41572-019-0111-2
Gomes R, Spinola PS, Brant AC et al (2021) Prevalence of germline variants in consensus moderate-to-high-risk predisposition genes to hereditary breast and ovarian câncer in BRCA1/2-negative Brazilian patients. Breast Cancer Res Treat 185:851–861. https://doi.org/10.1007/s10549-020-05985-9
Malkin D (2011) Li Fraumeni syndrome. Genes Cancer 2:475–484. https://doi.org/10.1177/1947601911413466
Umar A, Boland CR, Terdiman JP et al (2004) Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. JNCI 96:261–268. https://doi.org/10.1093/jnci/djh034
Walsh T, King MC (2007) Ten genes for inherited breast cancer. Cancer Cell 11:103–105. https://doi.org/10.1016/j.ccr.2007.01.010
Timoteo ARS, Gonçalves AEMM, Sales LAP et al (2018) A portrait of germline mutation in Brazilian at-risk for hereditary breast cancer. Breast Cancer Res Treat 172:637–646. https://doi.org/10.1007/s10549-018-4938-0
Carvalho SCS, Cury NM, Brotto DB et al (2020) Germline variants in DNA repair genes associated with hereditary breast and ovarian cancer syndrome: analysis of a 21 gene panel in the Brazilian population. BMC Med Genomics 13:1–24. https://doi.org/10.1186/s12920-019-0652-y
Instituto Nacional do Câncer José Alencar Gomes da Silva, INCA (2008) Ações de enfermagem para o controle do câncer. https://www.inca.gov.br/publicacoes/livros/acoes-de-enfermagem-para-o-controle-do-cancer. Accessed 14 Sept 2021
Barreto-Neto NJS, Pinheiro AB, Oliveira JF et al (2014) Perfil epidemiológico dos subtipos moleculares de carcinoma ductal da mama em população de pacientes em Salvador, Bahia. Rev Bras Mastologia 24:98–102
Slavin TP, Niell-Swiller M, Solomon I et al (2015) Clinical application of multigene panels: challenges of next-generation counseling and cancer risk management. Front Oncol 5:208. https://doi.org/10.3389/fonc.2015.00208
Coelho AS, Santos MAS, Caetano RI et al (2018) Predisposição hereditária ao câncer de mama e sua relação com os genes BRCA1 e BRCA2: revisão da literatura. RBAC 50:17–21. https://doi.org/10.21877/2448-3877.201800615
Nussbaum R, Mcinnes RR, Willard HF (2016) Thompson & Thompson Genética Médica. Elsevier Editora Ltda, São Paulo
Venkitaraman AR (2002) Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108:171–182. https://doi.org/10.1016/S0092-8674(02)00615-3
Rubinstein WS (2004) Hereditary breast cancer in Jews. Fam Cancer 3:249–257. https://doi.org/10.1007/s10689-004-9550-2
Karami F, Mehdipour P (2013) A comprehensive focus on global spectrum of BRCA1 and BRCA2 mutations in breast cancer. Biomed Res Int. https://doi.org/10.1155/2013/928562
Palmero EI, Schuler-Faccini L, Caleffi M et al (2008) Detection of R337H, a germline TP53 mutation predisposing to multiple cancers, in asymptomatic women participating in a breast cancer screening program in Southern Brazil. Cancer Lett 261:21–25. https://doi.org/10.1016/j.canlet.2007.10.044
Gomes R, Soares BL, Felicio OS et al (2020) Haplotypic characterization of BRCA1 c. 5266dupC, the prevailing mutation in Brazilian hereditary breast/ovarian cancer. Genet Mol Biol. https://doi.org/10.1590//1678-4685-GMB-2019-0072
Levine AJ, Momand J, Finlay CA (1991) The p53 tumour suppressor gene. Nature 6:453–466. https://doi.org/10.1038/351453a0
Pinto EM, Zambetti GP (2020) What 20 years of research has taught us about the TP53 p. R337H mutation. Cancer 126:4678–4686. https://doi.org/10.1002/cncr.33143
Assumpçãp JG, Seidinger AL, Mastellaro MJ et al (2008) Association of the germline TP53 R337H mutation with breast cancer in southern Brazil. BMC Cancer 8:1–6. https://doi.org/10.1186/1471-2407-8-357
Giacomazzi J, Graudenz MS, Osorio CA et al (2014) Prevalence of the TP53 P.R337H Mutation in Breast Cancer Patients in Brazil. PLoS ONE 9:6. https://doi.org/10.1371/journal.pone.0099893
Felix GE, Abe-Sandes C, Machado-Lopes TM et al (2014) Germline mutations in BRCA1, BRCA2, CHEK2 and TP53 in patients at high-risk for HBOC: characterizing a northeast Brazilian population. Hum Genome Var 1:1–8. https://doi.org/10.1038/hgv.2014.12
Bougeard G, Renaux-Petel M, Flaman JM et al (2015) Revisiting Li–Fraumeni syndrome from TP53 mutation carriers. J Clin Oncol 21:2345–2352. https://doi.org/10.1200/JCO.2014.59.5728
Amadou A, Achatz MIW, Hainaut P (2018) Revisiting tumor patterns and penetrance in germline TP53 mutation carriers: temporal phases of Li–Fraumeni syndrome. Curr Opin Oncol 1:23–29. https://doi.org/10.1097/CCO.0000000000000423
Achatz MI, Olivier M, Le Calvez F et al (2007) The TP53 mutation, R337H, is associated with Li-Fraumeni and Li-Fraumeni-like syndromes in Brazilian families. Cancer Lett 245:96–102. https://doi.org/10.1016/j.canlet.2005.12.039
Custódio G, Parise GA, Kiesel NF et al (2013) Impact of neonatal screening and surveillance for the TP53 R337H mutation on early detection of childhood adrenocortical tumors. J Clin Oncol 31:2619. https://doi.org/10.1200/JCO.2012.46.3711
Barbalho D, Sandoval R, Santos E et al (2021) Novel insights from the germline landscape of breast cancer in Brazil. Front Oncol 11:743231–743231. https://doi.org/10.3389/fonc.2021.743231
Sandoval RL, Polidorio N, Leite ACR et al (2022) Breast cancer phenotype associated with Li-Fraumeni syndrome: a Brazilian cohort enriched by TP53 p. R337H carriers. Front Oncol 12:826. https://doi.org/10.3389/fonc.2022.836937
Guindalini RSC, Viana DV, Kitajima JPFW et al (2022) Detection of germline variants in Brazilian breast cancer patients using multigene panel testing. Sci Rep 12:1–12. https://doi.org/10.1038/s41598-022-07383-1
Thompson D, Duedal S, Kirner J et al (2005) Cancer risks and mortality in heterozygous ATM mutation carriers. JNCI 97:813–822. https://doi.org/10.1093/jnci/dji141
Couch FJ, Hart SN, Sharma P et al (2015) Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J Clin Oncol 33:304. https://doi.org/10.1200/JCO.2014.57.1414
Cybulski C, Wokolorczyk D, Jakubowska A et al (2011) Risk of breast cancer in women with a CHEK2 mutation with and without a family history of breast cancer. J Clin Oncol 29:3747–3752. https://doi.org/10.1200/JCO.2010.34.0778
Rahman N, Seal S, Thompson D et al (2007) PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet 39:165–167. https://doi.org/10.1038/ng1959
Heikkinen T, Karkkainen H, Aaltonen K et al (2009) The breast cancer susceptibility mutation PALB2 1592delT is associated with an aggressive tumor phenotype. Clin Cancer Res 15:3214–3222. https://doi.org/10.1158/1078-0432.CCR-08-3128
Lynch HT, Boland CR, Gong G et al (2006) Phenotypic and genotypic heterogeneity in the Lynch syndrome: diagnostic, surveillance and management implications. Eur J Hum Genet 14:390–402. https://doi.org/10.1038/sj.ejhg.5201584
Richards S, Aziz N, Bale S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405–423. https://doi.org/10.1038/gim.2015.30
Kurian AW, Hare EE, Mills MA et al (2014) Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J Clin Oncol 32:2001. https://doi.org/10.1200/JCO.2013.53.6607
Tung N, Battelli C, Allen B et al (2015) Frequency of mutations in individuals with breast cancer referred for BRCA 1 and BRCA 2 testing using next-generation sequencing with a 25-gene panel. Cancer 121:25–33. https://doi.org/10.1002/cncr.29010
Goldgar DE, Easton DF, Graham BB et al (2008) Genetic evidence and integration of various data sources for classifying uncertain variants into a single model. Hum Mutat 29:1265–1272. https://doi.org/10.1002/humu.20897
Li MM, Datto M, Duncavage EJ et al (2017) Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn 19:4–23. https://doi.org/10.1016/j.jmoldx.2016.10.002
Robson ME, Storm CD, Weitzel J, Wollins DS, Ofitt K (2010) American Society of Clinical Oncology policy statement update: genetic and genomic testing for cancer susceptibility. J Clin Oncol 33:3660–3667. https://doi.org/10.1200/JCO.2009.27.0660
Carnevali I, Riva C, Chiaravalli AM et al (2019) Inherited cancer syndromes in 220 Italian ovarian cancer patients. Cancer Genet 237:55–62. https://doi.org/10.1016/j.cancergen.2019.06.005
Funding
KBG is grateful to Conselho Nacional de Desenvolvimento Científico e Tecnológico—CNPq for the research fellowship.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by JZP, JGC, MSV, BMV, PZO, CLM, CLVC, KBG The first draft of the manuscript was written by JZP, CLVC, KBG. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest.
Ethical approval
The study was approved by the Ethics Comitte of Federal University of Minas Gerais (CAAE- 54070121.0.0000.5149).
Consent for publication
Not applicable.
Consent to participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pereira, J.Z., Carneiro, J.G., Vieira, M.S. et al. Frequency of germline genetic variants in women with a personal or family history of breast cancer from Brazil. Mol Biol Rep 49, 9509–9520 (2022). https://doi.org/10.1007/s11033-022-07840-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07840-0