Abstract
Restriction–modification (R-M) systems form a large superfamily constituting bacterial innate immunity mechanism. The restriction endonucleases (REases) are very diverse in subunit structure, DNA recognition, co-factor requirement, and mechanism of action. Among the different catalytic motifs, HNH active sites containing REases are the second largest group distinguished by the presence of the ββα-metal finger fold. KpnI is the first member of the HNH-family REases whose homologs are present in many bacteria of Enterobacteriaceae having varied degrees of sequence similarity between them. Considering that the homologs with a high similarity may have retained KpnI-like properties, while those with a low similarity could be different, we have characterized a distant KpnI homolog present in a pathogenic Klebsiella pneumoniae NTUH K2044. A comparison of the properties of KpnI and KpnK revealed that despite their similarity and the HNH motif, these two enzymes have different properties viz oligomerization, cleavage pattern, metal ion requirement, recognition sequence, and sequence specificity. Unlike KpnI, KpnK is a monomer in solution, nicks double-stranded DNA, recognizes degenerate sequence, and catalyses the degradation of DNA into smaller products after the initial cleavage at preferred sites. Due to several distinctive properties, it can be classified as a variant of the Type IIS enzyme having nicking endonuclease activity.
Key points
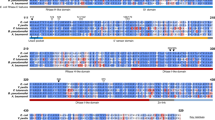
• KpnK is a distant homolog of KpnI and belongs to the ββα-metal finger superfamily.
• Both KpnI and KpnK have widespread occurrence in K. pneumoniae strains.
• KpnK is a Type IIS restriction endonuclease with a single-strand nicking property.






Similar content being viewed by others
Data availability
Most data generated or analysed during this study are included in this manuscript (and its ‘Supplementary information’ files).
References
Basak S, Nagaraja V (2001) A versatile in vivo footprinting technique using 1,10-phenanthroline-copper complex to study important cellular processes. Nucleic Acids Res 29(21):E105–E115. https://doi.org/10.1093/nar/29.21.e105
Bheemanaik S, Chandrashekaran S, Nagaraja V, Rao DN (2003) Kinetic and catalytic properties of dimeric KpnI DNA methyltransferase. J Biol Chem 278(10):7863–7874. https://doi.org/10.1074/jbc.M211458200
Chak KF, Kuo WS, Lu FM, James R (1991) Cloning and characterization of the ColE7 plasmid. J Gen Microbiol 137(1):91–100. https://doi.org/10.1099/00221287-137-1-91
Chan S-H, Opitz L, Higgins L, O’loane D, Xu S-Y (2010) Cofactor requirement of HpyAV restriction endonuclease. PloS One 5(2):e9071. https://doi.org/10.1371/journal.pone.0009071
Chandrashekaran S, Saravanan M, Radha DR, Nagaraja V (2004) Ca(2+)-mediated site-specific DNA cleavage and suppression of promiscuous activity of KpnI restriction endonuclease. J Biol Chem 279(48):49736–49740. https://doi.org/10.1074/jbc.M409483200
Chatterjee DK, Hammond AW, Blakesley RW, Adams SM, Gerard GF (1991) Genetic organization of the KpnI restriction–modification system. Nucleic Acids Res 19(23):6505–6509. https://doi.org/10.1093/nar/19.23.6505
Gralla JD (1985) Rapid “footprinting” on supercoiled DNA. Proc Natl Acad Sci U S A 82(10):3078–3081. https://doi.org/10.1073/pnas.82.10.3078
Hensley P, Nardone G, Chirikjian JG, Wastney ME (1990) The time-resolved kinetics of superhelical DNA cleavage by BamHI restriction endonuclease. J Biol Chem 265(25):15300–15307. https://doi.org/10.1016/S0021-9258(18)77256-6
Ibryashkina EM, Zakharova MV, Baskunov VB, Bogdanova ES, Nagornykh MO, Den’mukhamedov MM, Melnik BS, Kolinski A, Gront D, Feder M, Solonin AS, Bujnicki JM (2007) Type II restriction endonuclease R.Eco29kI is a member of the GIY-YIG nuclease superfamily. BMC Struct Biol 7:48. https://doi.org/10.1186/1472-6807-7-48
Kisiala M, Copelas A, Czapinska H, Xu S-Y, Bochtler M (2018) Crystal structure of the modification-dependent SRA-HNH endonuclease TagI. Nucleic Acids Res 46(19):10489–10503. https://doi.org/10.1093/nar/gky781
Kumar S, Bangru S, Kumar R, Rao DN (2020) Promiscuous DNA cleavage by HpyAII endonuclease is modulated by the HNH catalytic residues. Biosci Rep 40(9):1–14. https://doi.org/10.1042/BSR20201633
Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R (2019) The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47(W1):W636–W641. https://doi.org/10.1093/nar/gkz268
Maxam AM, Gilbert W (1980) Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol 65(1):499–560. https://doi.org/10.1016/s0076-6879(80)65059-9
Miyazono K, Watanabe M, Kosinski J, Ishikawa K, Kamo M, Sawasaki T, Nagata K, Bujnicki JM, Endo Y, Tanokura M, Kobayashi I (2007) Novel protein fold discovered in the PabI family of restriction enzymes. Nucleic Acids Res 35(6):1908–1918. https://doi.org/10.1093/nar/gkm091
Mizuuchi K, Kemper B, Hays J, Weisberg RA (1982) T4 endonuclease VII cleaves holliday structures. Cell 29(2):357–365. https://doi.org/10.1016/0092-8674(82)90152-0
Nagamalleswari E, Rao S, Vasu K, Nagaraja V (2017) Restriction endonuclease triggered bacterial apoptosis as a mechanism for long time survival. Nucleic Acids Res 45(14):8423–8434. https://doi.org/10.1093/nar/gkx576
Nishigaki K, Kaneko Y, Wakuda H, Husimi Y, Tanaka T (1985) Type II restriction endonucleases cleave single-stranded DNAs in general. Nucleic Acids Res 13(16):5747–5760. https://doi.org/10.1093/nar/13.16.5747
Orlowski J, Bujnicki JM (2008) Structural and evolutionary classification of Type II restriction enzymes based on theoretical and experimental analyses. Nucleic Acids Res 36(11):3552–3569. https://doi.org/10.1093/nar/gkn175
Pingoud A, Jeltsch A (1997) Recognition and cleavage of DNA by type-II restriction endonucleases. Eur J Biochem 246(1):1–22. https://doi.org/10.1111/j.1432-1033.1997.t01-6-00001.x
Pingoud A, Jeltsch A (2001) Structure and function of type II restriction endonucleases. Nucleic Acids Res 29(18):3705–3727. https://doi.org/10.1093/nar/29.18.3705
Pingoud A, Fuxreiter M, Pingoud V, Wende W (2005) Type II restriction endonucleases: structure and mechanism. Cell Mol Life Sci 62(6):685–707. https://doi.org/10.1007/s00018-004-4513-1
Pingoud V, Wende W, Friedhoff P, Reuter M, Alves J, Jeltsch A, Mones L, Fuxreiter M, Pingoud A (2009) On the Divalent metal ion dependence of DNA cleavage by restriction endonucleases of the EcoRI Family. J Mol Biol 393(1):140–160. https://doi.org/10.1016/j.jmb.2009.08.011
Pommer AJ, Cal S, Keeble AH, Walker D, Evans SJ, Kühlmann UC, Cooper A, Connolly BA, Hemmings AM, Moore GR, James R, Kleanthous C (2001) Mechanism and cleavage specificity of the H-N-H endonuclease colicin E9. J Mol Biol 314(4):735–749. https://doi.org/10.1006/jmbi.2001.5189
Robert X, Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42(Web Server issue), W320–4. https://doi.org/10.1093/nar/gku316
Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev SK, Dryden DTF, Dybvig K, Firman K, Gromova ES, Gumport RI, Halford SE, Hattman S, Heitman J, Hornby DP, Janulaitis A, Jeltsch A, Josephsen J, Kiss A, Klaenhammer TR, Kobayashi I, Kong H, Krüger DH, Lacks S, Marinus MG, Miyahara M, Morgan RD, Murray NE, Nagaraja V, Piekarowicz A, Pingoud A, Raleigh E, Rao DN, Reich N, Repin VE, Selker EU, Shaw PC, Stein DC, Stoddard BL, Szybalski W, Trautner TA, Van Etten JL, Vitor JM, Wilson GG, Xu SY (2003) A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res 31(7):1805–1812. https://doi.org/10.1093/nar/gkg274
Roberts RJ, Vincze T, Posfai J, Macelis D (2015) REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res 43(Database issue):D298-9. https://doi.org/10.1093/nar/gku1046
Roberts RJ, Vincze T, Posfai J, Macelis D (2023) REBASE: a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res 51:D629–D630. https://doi.org/10.1093/nar/gkac975
Sapranauskas R, Sasnauskas G, Lagunavicius A, Vilkaitis G, Lubys A, Siksnys V (2000) Novel subtype of type IIs restriction enzymes. BfiI endonuclease exhibits similarities to the EDTA-resistant nuclease Nuc of Salmonella typhimurium. J Biol Chem 275(40):30878–30885. https://doi.org/10.1074/jbc.M003350200
Saravanan M, Bujnicki JM, Cymerman IA, Rao DN, Nagaraja V (2004) Type II restriction endonuclease R.KpnI is a member of the HNH nuclease superfamily. Nucleic Acids Res 32(20):6129–6135. https://doi.org/10.1093/nar/gkh951
Saravanan M, Vasu K, Ghosh S, Nagaraja V (2007a) Dual role for Zn2+ in maintaining structural integrity and inducing DNA sequence specificity in a promiscuous endonuclease. J Biol Chem 282(44):32320–32326. https://doi.org/10.1074/jbc.M705927200
Saravanan M, Vasu K, Kanakaraj R, Rao DN, Nagaraja V (2007b) R.KpnI, an HNH superfamily REase, exhibits differential discrimination at non-canonical sequences in the presence of Ca2+ and Mg2+. Nucleic Acids Res 35(8):2777–2786. https://doi.org/10.1093/nar/gkm114
Stoddard BL (2005) Homing endonuclease structure and function. Q Rev Biophys 38(1):49–95. https://doi.org/10.1017/S0033583505004063
Szybalski W, Kim SC, Hasan N, Podhajskab AJ (1991) Class-IIS restriction enzymes-a review. Gene 100:31–59. https://doi.org/10.1016/0378-1119(91)90345-C
Vasu K, Nagaraja V (2013) Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol Mol Biol Rev 77(1):53–72. https://doi.org/10.1128/MMBR.00044-12
Vasu K, Saravanan M, Nagaraja V (2011) Endonuclease active site plasticity allows DNA cleavage with diverse alkaline Earth and transition metal ions. ACS Chem Biol 6(9):934–942. https://doi.org/10.1021/cb200107y
Vasu K, Nagamalleswari E, Nagaraja V (2012) Promiscuous restriction is a cellular defense strategy that confers fitness advantage to bacteria. Proc Natl Acad Sci U S A 109(20):E1287–E1293. https://doi.org/10.1073/pnas.1119226109
Walker DC, Georgiou T, Pommer AJ, Walker D, Moore GR, Kleanthous C, James R (2002) Mutagenic scan of the H-N-H motif of colicin E9: implications for the mechanistic enzymology of colicins, homing enzymes and apoptotic endonucleases. Nucleic Acids Res 30(14):3225–3234. https://doi.org/10.1093/nar/gkf420
Wilson GG, Murray NE (1991) Restriction and modification systems. Annu Rev Genet 25:585–627. https://doi.org/10.1146/annurev.ge.25.120191.003101
Wright DJ, Jack WE, Modrich P (1999) The kinetic mechanism of EcoRI endonuclease. J Biol Chem 274(45):31896–31902. https://doi.org/10.1074/jbc.274.45.31896
Wu KM, Li NH, Yan JJ, Tsao N, Liao TL, Tsai HC, Fung CP, Chen HJ, Liu YM, Wang JT, Fang CT, Chang SC, Shu HY, Liu TT, Chen YT, Shiau YR, Lauderdale TL, Su IJ, Kirby R, Tsai SF (2009) Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J Bacteriol 191(14):4492–4501. https://doi.org/10.1128/JB.00315-09
Xu S, Gupta YK (2013) Natural zinc ribbon HNH endonucleases and engineered zinc finger nicking endonuclease. Nucleic Acids Res 41(1):378–390. https://doi.org/10.1093/nar/gks1043
Zhang Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. https://doi.org/10.1186/1471-2105-9-40
Zhang L, Radziwon A, Reha-Krantz LJ (2014) Targeted mutagenesis of a specific gene in yeast. Methods Mol Biol 1163:109–129. https://doi.org/10.1007/978-1-4939-0799-1_8
Zhang L, Huang Y, Xu D, Yang L, Qian K, Chang G, Gong Y, Zhou X, Ma K (2016) Biochemical characterization of a thermostable HNH endonuclease from deep-sea thermophilic bacteriophage GVE2. Appl Microbiol Biotechnol 100(18):8003–8012. https://doi.org/10.1007/s00253-016-7568-7
Zhang L, Xu D, Huang Y, Zhu X, Rui M, Wan T, Zheng X, Shen Y, Chen X, Ma K, Gong Y (2017) Structural and functional characterization of deep-sea thermophilic bacteriophage GVE2 HNH endonuclease. Sci Rep 7(January):1–13. https://doi.org/10.1038/srep42542
Acknowledgements
We thank Dr. Vijaya Srinivasan from CSIR-IMTECH for providing the K. pneumoniae NTUH strain. Debarghya Ghose is acknowledged for the initial cloning of KpnK, and Iqball Faheem is acknowledged for his help in size exclusion chromatography. We acknowledge the central facility of phosphor imaging at the Indian Institute of Science, supported by the Department of Biotechnology, Government of India, and VN laboratory members for their valuable suggestions and critical inputs. MS acknowledges the University Grants Commission, Government of India, for financial assistance through Junior Research Fellowship (JRF) and Senior Research Fellowship (SRF).
Funding
This research was supported by grants to V.N. from the Science and Engineering Research Board, Department of Science and Technology (CRG/2019/000077), and Department of Biotechnology (BT/PR41348/MED/29/1536/2020), Government of India.
Author information
Authors and Affiliations
Contributions
MS, BE and SME: performed the experiments. EN, SME, and BE: prepared constructs used in the study. ISR: performed bioinformatic analyses. MS and VN: analysed the data and wrote the manuscript. VN: designed the experiments, supervision, and resources. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not involve any human participants or animal experiments.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Santoshi, M., Engleng, B., Eligar, S.M. et al. Identification and characterization of a new HNH restriction endonuclease with unusual properties. Appl Microbiol Biotechnol 107, 6263–6275 (2023). https://doi.org/10.1007/s00253-023-12717-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-023-12717-8