Expression and protease characterization of a conserved protein YgjD in Vibrio harveyi
- Published
- Accepted
- Received
- Academic Editor
- Mohammed Gagaoua
- Subject Areas
- Agricultural Science, Biochemistry, Bioengineering, Biotechnology, Molecular Biology
- Keywords
- Vibrio harveyi, Protease activity, Site-directed mutagenesis, Cell growth
- Copyright
- © 2020 Zhang et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2020. Expression and protease characterization of a conserved protein YgjD in Vibrio harveyi. PeerJ 8:e9061 https://doi.org/10.7717/peerj.9061
Abstract
The glycopeptidase GCP and its homologue proteins are conserved and essential for survival of bacteria. The ygjD gene (Glycopeptidase homologue) was cloned from Vibrio harveyi strain SF-1. The gene consisted of 1,017 bp, which encodes a 338 amino acid polypeptide. The nucleotide sequence similarity of the ygjD gene with that of V. harveyi FDAARGOS 107 was 95%. The ygjD gene also showed similarities of 68%, 67% and 50% with those of Salmonella enterica, Escherichia coli and Bacillus cereus. The ygjD gene was expressed in E. coli BL21 (DE3) and the recombinant YgjD was purified by Ni2+ affinity chromatography column. The purified YgjD showed a specific 37 kDa band on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and exhibited protease activities of 59,000 units/mg, 53,700 units/mg and 8,100 units/mg, respectively, on N-Acetyl-L-tyrosine ethyl ester monohydrate (ATEE), N-Benzoyl-L-tyrosine ethyl ester (BTEE) and N-Benzoyl-DL-arginine-4-nitroanilide hydrochloride (BAPNA) substrates. When the conserved amino acids of His111, Glu113 and His115 in the YgjD were replaced with alanine, respectively, the protease activities of the mutants were partly decreased. The two conserved His111 and His115 of YgjD were mutated and the protein lost the protease activity, which implied that the two amino acid played very important roles in maintaining its protease activity. The addition of the purified YgjD to the culture medium of V. harveyi strain SF-1 can effectively promote the bacteria growth. These results indicated that the protease activities may be involved in the survival of bacteria.
Introduction
As one of the top 10 essential proteins (Galperin, 2004), the GCP proteins are described as putative glycoprotease, which were first isolated from Mannheimia haemolitica. The GCP and its homologue proteins are conserved in eukaryotes, bacteria and archaea. These proteins were found to be essential for survival both of Gram-positive and Gram-negative bacteria such as Staphylococcus aureus (Zheng et al., 2005), Bacillus subtilis (Hunt, 2006; Kobayashi et al., 2003), Mycoplasm genetalium (Glass et al., 2006), Francisella novicida (Gallagher et al., 2007), Pseudomonas aeruginosa (Liberati et al., 2006) and Escherichia coli (Fabrizio et al., 1998). GCP is also important for the growth of Saccharomyces cerevisiae (Downey et al., 2006; Hofmann et al., 2006) and embryo development (Haussuehl et al., 2009) in Arabidopsis thaliana.
It has been reported that the GCP homologues are also involved in cell wall biosynthesis, cell division, cell autolysis and antibiotics resistances (Zheng et al., 2007; Bergmiller et al., 2011), and the depletion of protein-coding genes can lead to significant alterations in cell morphology and ultra structure in prokaryotes (Bergmiller et al., 2011; Boyle-Vavra, Yin & Daum, 2006; Nichols et al., 2006). The GCP homolog of Synechocystis sp. is also related with its salt tolerance, pigment and cyanophycin production (Karandashova et al., 2002; Zuther, Schubert & Hagemann, 1998). The ygjD gene of E. coli was previously recognized to consist the rpsU-dnaG-rpoD macromolecular-synthesis operon (Nesin et al., 1987), and involved in the survival by hydrolyzing toxic glycation proteins in E. coli (Katz et al., 2010). YgjD has recently been reported to be necessary for the synthesis of universal tRNA N6-threonylcarbamoyladenosine (t6A) (Hashimoto et al., 2011; Thiaville et al., 2015). YgjD could interact with YjeE and YeaZ to form an YgjD-YeaZ-YjeE complex which determined the function of t6A in E. coli. It also revealed that the ATPase activity of YjeE is regulated by YgjD-YeaZ heterodimer. Deletion of YgjD, YeaZ or YjeE leads to the loss of the t6A biosynthesis (Zhang et al., 2015). The GCP of S. aureus is able to regulate the murein hydrolase activities, which further implied the GCP may have multiple function and regulate expression of genes involved in some critical pathways for bacteria survival.
V. harveyi is one of the important pathogens of aquatic animals, which poses a serious threat to marine aquaculture (Alvarez, 1998; Wei et al., 2019). Our earlier work revealed that V. harveyi could enter into viable but nonculturable (VBNC) state and resuscitated V. harveyi cells were found to retain their pathogenicity (Sun et al., 2008). We also found that the purified recombinant YeaZ could improve the resuscitation of the VBNC cells of V. harveyi to culturable state (Li et al., 2017). YeaZ also shared some identities with YgjD proteins of Vibrio spp. Although the YgjD and YeaZ proteins are inextricably linked, the exact biological functions of the GCP in the bacteria remain unclear. In this study, the ygjD gene was cloned and its enzyme activities were evaluated. The promoting effect on cell growth and resuscitation-promoting activities of the purified YgjD were also studied. The underlying mechanisms of resuscitation for bacteria also were explored in this study.
Material and Methods
Bacteria strains and plasmids
The main bacteria strains and plasmids are listed in Table 1. The V. harveyi strain SF-1 was isolated from diseased sea perch (Lateolabrax japonicus) in Qingdao, P. R. China (He, Chen & Li, 2011; Wang et al., 2002), and confirmed by 16S rDNA sequence analysis (Accession Number: SUB6973071). The V. harveyi cells were cultured on Zobell’s 2216 E medium at 30 °C. E. coil cells were cultured on Luria-Bertani (LB) medium at 37 °C. All chemical reagents were analytical grade.
| Strains or plasmids | Characteristics | Sources |
|---|---|---|
| Strains | ||
| Vibrio harveyi strain SF-1 | Wild-type | Laboratory collection |
| E. coli DH5 α | deoR, recA1, endA1, hsdR17(rk−, mk+), phoA, supE44, λ−, thi−1, gyrA96, relA1 | Tiangen, China |
| E. coli BL21 (DE3) | E. coli B, F−−, dcm, ompT, hsdS(rB−,mB−), gal, λ(DE3) | Tiangen, China |
| Arctic-Express-pET-28a (+)-ygjD | Kanr, ygjD containing pET-28a (+) | This work |
| Plasmids | ||
| PEASY-T1 Vector | Kanr, Ampr, 3.928 kb, high-copy-number cloning vector | TransGen, China |
| pET-28a (+) | f1 origin; Kanr; PT7 | Zoonbio |
| PEASY-T1-ygjD | Ampr, PEASY-T1 with an 1117 bp fragment containing ygjD gene of SF-1 | This work |
| pET-28a (+)-ygjD | Kanr, pET-28a (+) with an 1017 bp fragment containing ygjD gene of SF-1 | This work |
Cloning and bioinformatics analysis of ygjD gene of V. harveyi
A pair of primers (forward primer 5′-AATTTTAATCCGATCAAC-3′, reverse primer 5′-TATTTCAAACCTTCAGTAGA-3′) was designed according to the ygjD gene of V. harveyi and other Vibrio spp. on NCBI database (http://blast.ncbi.nlm.nih.gov/). The ygjD gene was amplified by PCR from chromosomal DNA of V. harveyi strain SF-1. The reaction conditions were as follows: denaturation at 94 °C for 5 min, 30 cycles of denaturation at 94 °C for 60 s, annealing at 42 °C for 1 min, extension at 72 °C for 1.5 min, and a final extension at 72 °C for 10 min. The PCR products were checked by 1.0% agarose gel eletrophoresis and further purified using a DNA purification Kit (Tiangen, Beijing, China). The gene was cloned into PEASY-T1 vector (TransGEN, Beijing, China) and sequenced by Sangon Biotech (Shanghai) Co., Ltd., China. The ygjD gene sequence was subjected to similarity alignment analysis on the NCBI. The gene similarities were analyzed with DNAMAN software (version 5.1; Lynnon Biosoft, Quebec, Canada). A phylogenic tree was constructed using MEGA 5.0 by the neighbor joining method with 1,000 replicates.
Expression and purification of the recombinant YgjD
The ygjD gene was amplified by PCR from the PEASY-T1-ygjD with a pair of primers (forward primer 5′-GGAGTCTAGAATGCGCATTATTGGTA-3′ with Xba I restriction site, reverse primer 5′-CGAATGCGGTTAGCGCGAGCTCCGGC-3′ with Xho I restriction site). The PCR products were also detected with the same method above described. The amplified sequence was inserted into pET-28a (+). The recombinant pET-28a (+)-ygjD was then transformed into E. coli BL21 (DE3). The E. coli containing pET-28a (+)-ygjD was inoculated into 5 mL of LB liquid medium containing kanamycin (0.05 mg/mL), and cultured overnight at 37 °C with shaking. The culture was then added to 500 mL of the same medium and kept at 37 °C for further 3 h, and then induced with isopropyl-β-D-thiogalactopyranoside (IPTG) at 37 °C for 4 h. The bacteria cells were then collected by centrifugation at 10,000 g for 15 min at 4 °C. The cell pellet was resuspended in 20 mL of lysate (pH 8.0, 20 mM Tris-HCl) containing phenylmethanesulfonyl fluoride (PMSF) and protease inhibitor, sonicated (work 4 sec, interval 8 sec, 20 min total) at 400 W by Ultrasonic Processor (SCIENTZ-950E; Ningbo Xinzhi Biotechnology Co., Ltd, Ningbo, China) in an ice bath. The lysate was centrifuged at 9,000 g for 20 min at 4 °C and the precipitate was collected. The inclusion bodies were washed 3 times with inclusion body washing solution (20 mM Tris, 1 mM ethylene diamine tetraacetic acid (EDTA), 2 mM Urea, 1 M NaCl, 1% Triton X-100, pH 8.0), and then were dissolved in a certain ratio with a lysis buffer (20 mM Tris, 5 mM dithiothreitol, 8 mM Urea, pH 8.0). Centrifuged at 15,000 g for 15 min and the inclusion bodies were filtered through a 0.22 µm sterile filter (Millipore, USA). The recombinant protein was purified using a Ni2+ affinity chromatography as described (Mukhija & Erni, 1996). The inclusion bodies solutions were loaded into the pre-equilibrium Ni-ID Sepharose CL-6B column, and eluted with Ni-IDA Binding-Buffer at 1.0 mL/min until the absorbance at 280 nm achieved the stable baseline. The targeted protein was eluted with Ni-IDA Elution-Buffer and collected for determining the concentration of protein by Bradford assay (Bradford, 1976). The purified protein was dialyzed with deionized water for analysis by 12% SDS-PAGE (Laemmli, 1970), and protease activity determination.
Construction, expression and purification of the YgjD mutants
The ygjD gene of V. harvey i strain SF-1 was analyzed and compared with the other bacteria. The amino acids of the conserved “HXEXH” sequence of the proteases were selected and replaced with other amino acids (Table 2). These mutant gene sequences were synthesized by General Biosystems (Anhui) Co., Ltd., China. The expressing plasmids pET-28a (+) containing mutated ygjD gene was constructed and transformed to E. coli BL21 (DE3). The recombinant YgjD mutants were expressed by IPTG induction and purified with the abovementioned methods.
| Mutant proteins | Mutations | Oligo nucleotides (5′–3′)* |
|---|---|---|
| H111A | His111→Ala | CTGTTCACGCTATGGAAGGTCACCTA |
| E113A | Glu113→Ala | CTGTTCACCATATGGCAGGTCACCTACT |
| H115A | His115→Ala | CACCATATGGAAGGTGCCCTACTGGCGCCTAT |
| H111A+H115A | His111→Ala | TTCACGCTATGGAAGGTGCCCTACTGGCGCC |
| His115→Ala |
Protein electrophoresis and Western blotting
The purified proteins were detected with western blot method (Das et al., 2018). The purified proteins were cut off acrylamide gels (SDS-PAGE) and transferred on the nitrocellulose membrane after electrophoresis. The membrane was then incubated with rabbit anti-6 ×His tag antibody (Genscript, Nanjing, China) at 37 °C for 2 h. Horseradish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin (BoShiDe Biotechnology, Wuhan, China) was used as the secondary antibody. The hybrid membrane was washed with TBST buffer (pH 7.4), and detection was carried out with the ECL Western Blotting Detection kit (SW2010, Solarbio, Beijing).
Proteolytic activity determination of the purified YgjD
Amidase activity was determined with BAPNA substrate (Erlanger, Kokowsky & Cohen, 1961). Briefly, 0.1 mL of protein solution were mixed with 0.5 mL of BAPNA solution (1 mmol/L), incubated at 37 °C for 30 min, and added 30 µL of 30% acetic acid solution to stop the reaction. The optical densities were detected at a wavelength of 410 nm and the activity was calculated (Erlanger, Kokowsky & Cohen, 1961). 50 µL of protein solution were added into 0.7 mL of ATEE solution (1 mmol/L) or BTEE solution (1 mmol/L), and incubated at 37 °C for 30 min. 30 µL of 30% acetic acid solution were used to stop the reaction. The absorbance at 256 nm were measured and the esterase activities were calculated as described (Hummel, 1959; Schwert & Takenaka, 1955).
Protease characterization of the recombinant YgjD under different conditions
According to the above described method, the esterase activity of the YgjD also was determined with ATEE substrate at various temperatures (4 °C, 20 °C, 30 °C, 40 °C, 50 °C, 60 °C, 70 °C, 80 °C). The optimal pH and pH stability of the enzyme activities were determined with Na2HPO4/C6H8O7 buffer (pH 2.0–8.0) and Na2HPO4/KH2PO4 buffer (pH 8.0–10.0), respectively. To determine the effect of metal ions on enzyme activities of the recombinant YgjD, different amounts of Mn2+, Ca2+, Zn2+, Co2+, Mg2+ and Cu2+ were added to the mixtures of recombinant YgjD and ATEE solution at 37 °C for 30 min, and the protease activities were determined. Enzyme inhibitors such as DTT, PMSF, EDTA and ethylene glycol tetraacetic acid (EGTA) also were added into the mixtures of recombinant YgjD and ATEE solution at the same conditions, and then dialyzed with deionized water for the enzyme activities determination.
Effects of the recombinant YgjD proteins on the growth of V. harveyi strain SF-1
The V. harvey cells were cultured at 30 °C to reach stationary phase, and inoculated into 5 mL of 2216 E broth with addition of the purified wild-type and mutant YgjD proteins, respectively. The bacteria mixtures were then incubated at the same temperature for 72 h. The optical density of bacteria was determined at a wavelength of 600 nm, and the growth curve was drawn for evaluating the effects of the recombinant proteins on the growth of V. harveyi.
Recovery effect of the YgjD on VBNC cells of V. harveyi strain SF-1
The V. harveyi was cultured in 2216 E broth at 30 °C, 200 rpm overnight. 45 µL of H2O2 was added to the bacteria solution with a final concentration of 50 mM. The bacteria solution was kept at 30 °C. The viable counts were determined with plate counting method (Kogure, Simidu & Taga, 1979). The bacteria cells were considered to enter into the uncultivable state when the viable cells cannot be detected (Baffone et al., 2006).
5 mL of the VBNC cells were taken out from the VBNC cell microcosm of V. harveyi. The cells were collected with centrifugation. The collected cells were washed with sterile physiological saline. They were inoculated into 5 mL of sterile physiological saline. 500 µL of the purified YgjD (0.2 mg/mL) were added to 4.5 mL of the VBNC cells to determine its promoting effect on VBNC cells. A boiled protein solution was used as a control. The mixed solution was kept under the same condition. The culturable cell counts were determined by plate count method.
Statistical analysis
All the measurements were performed in three separate replicates. Statistical analysis of the mean of the results were performed with SPSS software (SPSS, Chicago, IL). The graphs were constructed using Origin 95 software (OriginLab Corp., USA).
Results
Cloning and analysis of ygjD gene from V. harveyi strain SF-1
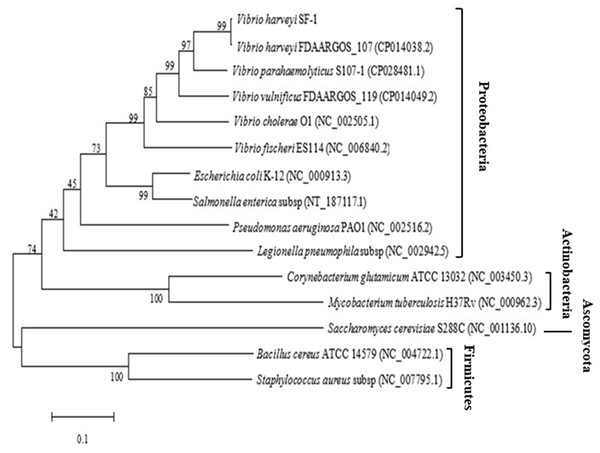
The ygjD gene of V. harveyi strain SF-1 was composed of 1,017 bp with a stop codon, which encoded a polypeptide of 338 amino acids. The sequence was submitted to NCBI, and an accession was obtained as MN699966. The phylogenic tree from 15 strains were established based on the ygjD gene (part segment) as shown in Fig. 1. Its nucleotide sequence showed 95% similarity with that of V. harveyi FDAARGOS 107. The nucleotide sequence similarities with V. cholerae, V. vulnificus, V. parahaemolyticus were 75%–83%, respectively. The gene also showed 68%, 67% and 50% similarities with the ygjDs of S. enterica, E. coli and B. cereus, respectively. According to the amino acid sequence alignment analysis, the amino acid sequence in this study showed 100% similarity with other V. harveyi strains. Meanwhile, the amino acid similarities with V. cholerae, V. vulnificus and V. parahaemolyticus also reached 89%, 96% and 99%, respectively. However, the similarities with S. enterica, E. coli and B. cereus were 77%, 76% and 43%, respectively. Further it showed 31% similarity with eukaryotic organism S. cerevisiae. It was clear from the phylogenetic tree that the cloned ygjD gene was more conservative in Vibrio sp, and existed in many microorganisms. These results implied ygjD gene played an important roles in microbes survival (Zheng et al., 2007; Bergmiller et al., 2011).
Figure 1: Phylogenetic tree based on the multiple sequence alignment of ygjD genes from different strains.
Expression, purification and enzyme activity analysis of the recombinant YgjD
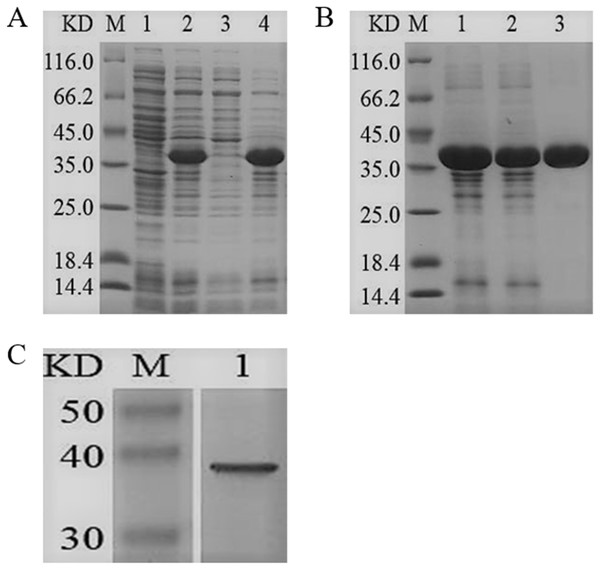
The ygjD gene was cloned into pET-28a (+) and the recombined plasmid transformed into E. coli BL21 (DE3) for the expression of YgjD protein. The amount of expressed protein purified with Ni2+ affinity chromatography column reached at 3 mg. The molecular weight showed a 37 kDa band on SDS-PAGE, which corresponded to the molecular weight of fusion protein (Fig. 2B). The Western blotting also showed a 37 kDa specific band (Fig. 2C), further indicating the protein was correctly expressed and induced by IPTG.
Figure 2: SDS-PAGE and Western blotting analysis of the purified recombinant YgjD.
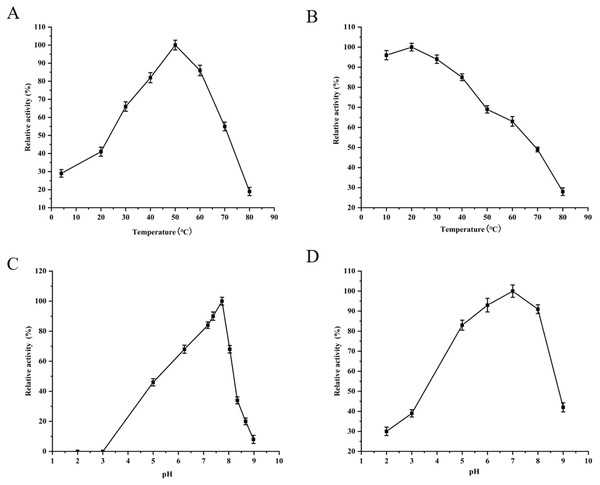
M denotes the middle-ranged protein marker. (A) When the recombination strain was induced for 4 h by IPTG at 37 °C, 20 µL of the culture supernatant was collected. The culture supernatant mixed with the 6× loading buffer were boiled and then determined by SDS-PAGE as shown in band 2 with the un-induced culture supernatant as control (band 1); When the induced cells were cracked by ultrasonic, the supernatant and the precipitation were analyzed by SDS-PAGE (band 3 and 4). (B) The solubilized inclusion body and dialysis solution were measured by SDS-PAGE (band 1 and 2); The purified YgjD by Ni-ID Sepharose CL-6B column was determined shown in band 3. (C) Western blot analysis of the purified YgjD is shown in band 1.The purified YgjD showed protease activities with ATEE, BTEE and BAPNA substrates, the specific activities were of 59,000 units/mg, 53,700 units/mg and 8100 units/mg, respectively. The protein showed maximum activity at 50 °C and pH 7.7 (Fig. 3). As shown in Table 3, Zn2+increased the protease activity of the YgjD. Cu2+, Mn2+, Ca2+ and Co2+ partly inhibited the enzyme activities. PMSF, EDTA and EGTA all had different degrees of inhibitory effect on the protease activity.
Figure 3: Optimum determination value and effects of temperature and pH on the protease activity of YgjD.
(A) Esterase activity of the YgjD with ATEE substrate at various temperatures; (B) esterase activity of the YgjD treated at different temperatures; (C) esterase activity of the YgjD with ATEE substrate at various pH; (D) esterase activity of the YgjD treated at different pH.Mutation analysis of the enzyme active site of the YgjD
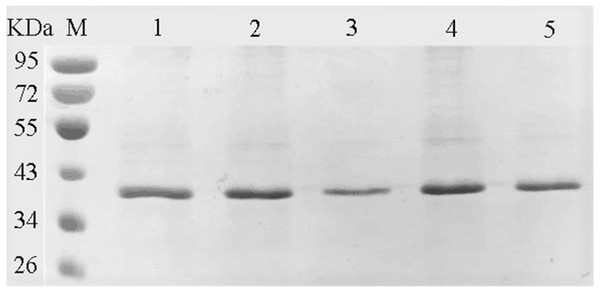
The amino acids of the conserved sequence “HXEXH” of the YgjD were separately substituted with alanine. The purified YgjD mutants showed specific bands of the same size with the wild-type protein on SDS-PAGE (Fig. 4). The protease activities of the YgjD mutant were decreased at different degrees (Table 4). When ATEE, BTEE, BAPNA was used as substrate, the protease activities of protein with a single amino acid mutation (H111A, E113A and H115A) decreased significantly. The protease activities with the H111A mutant nearly inactivated with BAPNA substrate, the protease activities with the E113A mutant also inactivated with BTEE substrate, the protease activities with the H115A mutant nearly inactivated with ATEE and BTEE substrates, while the YgjD mutant with His111+His115(H111A+H115A) lost its activity completely. These results revealed that the two amino acid site (H111A+H115A) played the most important roles in maintaining its protease activity.
| Chemical reagents | Concentrations (mM) | Specific activity (u/mg) | Relative activity (%) | Concentrations (mM) | Specific activity (u/mg) | Relative activity (%) |
|---|---|---|---|---|---|---|
| Normal saline | / | 4500 ± 200 | 100.00 | / | 4580 ± 350 | 100.00 |
| Zn2+ | 0.1 | 7020 ± 600 | 155.55 ± 4.23 | 1 | 10850 ± 1100 | 237.05 ± 101 |
| Mg2+ | 0.1 | 4580 ± 300 | 101.85 ± 2.46 | 1 | 4440 ± 290 | 97.05 ± 2.13 |
| Co2+ | 0.1 | 4220 ± 600 | 93.82 ± 2.85 | 1 | 3520 ± 140 | 77.06 ± 1.18 |
| Ca2+ | 0.1 | 3990 ± 150 | 88.75 ± 3.84 | 1 | 3870 ± 230 | 84.71 ± 2.95 |
| Cu2+ | 0.1 | 2650 ± 100 | 59.25 ± 1.34 | 1 | 3630 ± 410 | 79.41 ± 5.23 |
| Mn2+ | 0.1 | 2330 ± 240 | 51.85 ± 2.17 | 1 | 1670 ± 120 | 36.47 ± 1.66 |
| EDTA | 0.1 | 3300 ± 320 | 73.46 ± 1.91 | 1 | 2540 ± 370 | 55.59 ± 3.41 |
| EGTA | 0.1 | 2330 ± 170 | 51.85 ± 1.85 | 1 | 3090 ± 270 | 67.64 ± 2.04 |
| DTT | 0.1 | 4110 ± 1160 | 91.36 ± 5.98 | 1 | 4440 ± 180 | 97.05 ± 6.44 |
| PMSF | 0.1 | 2160 ± 260 | 48.15 ± 1.37 | 1 | 1690 ± 160 | 37.06 ± 0.78 |
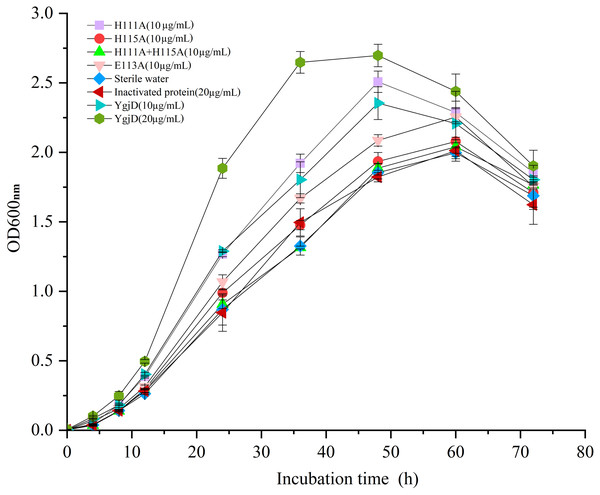
Effects of the purified YgjD on growth of V. harveyi strain SF-1
As shown in Fig. 5, the different growth curve of V. harveyi strain SF-1 were determined by addition of the purified YgjD at different concentrations (20 µg/mL and 10 µg/mL), the strain growth rate significantly increased with the concentration increase. And, the cell growth rate increased to 177.01% and 120.52%, respectively, when compared with the control group (20 µg/mL of the boiled YgjD) and normal group (without additive). It’s worth noting that the growth rate of strain SF-1 also increased to 111.56% in the group with the additive of mutant proteins (E113A). However, the growth rate showed no significant difference among mutant proteins (H115A) group, mutant proteins (H111A+ H115A) group, normal group and control group, which implied the YgjD mutant group with His111+His115 lost its promoting effect on the cell growth. These results further demonstrated that the amino acid (His111 and His115) played an key function in maintaining the structure and activity of YgjD, and the proteinase activity of YgjD involve in the cell division and cell autolysis (Zheng et al., 2007; Bergmiller et al., 2011).
Recovery effect of the recombinant YgjD on the VBNC cells of V. harveyi
The effects of recombinant YgjD on recovery of V. harveyi cells were also analyzed. No visible colonies were observed on the plates added with the purified YgjD protein, which suggested that the YgjD did not have obvious promoting effect on VBNC cells (Table 5).
Discussion
The YgjD protein was regards as one of GCP homologues, that described as a kind of conserved proteases and their homologues widely exist in bacteria. The GCP belong to MK-M22 O-sialoglycoprotein endopeptidase family. Their amino acid sequences are highly conserved. The two histidines are found at conserved sequence of “HXEXH” and consist of the active centre of the proteases. The two histidines of the “HXEXH” are suggested to coordinate Zn2+ and form a catalytic domain (Aravind & Koonin, 1999; Hecker et al., 2007). The GCP isolated from P. haemolytica could hydrolyze glycophorin A (Abdullah et al., 1992). Zn2+, Cu2+, Hg2+ and Ni2+ inhibited protease activity of the GCP of P. haemolytica (Cladman et al., 1996). But the GCP of S. aureus did not show any proteolytic activity against glycophorin A. The author explained that the GCP might have some specific proteolytic activities which are critical for the cell viability (Zheng et al., 2005). It has been reported that deletion of gcp 1 gene (ygjD) in E. coli leads to cessation of the cell growth. The GCP1 is related with regulating the cell division. The GCP1 did not show proteolytic activities against β-casein, hemoglobin, azocoll and albumin. The author found that GCP1 mutant with the two conserved histidines was not able to rescue the lethal conditional gcp 1 mutant phenotype. This result suggested that the two histidines were essential for cell viability of E. coli (Lei et al., 2011). The author also found that the N-terminus of the GCP1 is essential for protein to maintain the bacteria viability. Some proteases were reported to hydrolyze low molecular weight peptides and were unable to work on proteins in bacteria (Wiame et al., 2002). Katz reported that the GCP of E. coli could metabolize the glycated proteins (Katz et al., 2010). GCP depletion in E. coli reduced the growth rate and accumulated active AMPs. The GCP could bind the glycated proteins, elongation factor Tu and pyruvate dehydrogenase (PDH) (Weiß, 2009). Meanwhile, the protease activity of GCP was related to the virulent of microbiome, proteases-mutant strains of GCP was used to live vaccine for vibriosis as described in literatures (Ma, Zhang & Zhao, 2010; Wagner et al., 2013; Mohd-Aris et al., 2019), indicating that GCP proteins have potential applications in microbial growth, resuscitation and vaccine development.
Figure 4: SDS-PAGE analysis of the YgjD mutants and wild-type YgjD.
M denotes the middle-ranged protein marker. Bands 1–4 denote the purified YgjD mutants with His111, His115, His111+His115 and Glu113 by Ni-ID Sepharose CL-6B column, and the wild-type YgjD was used as control (band 5).| Proteins | Substrates | Specific activity (u/mg) | Percent activity retained (%) |
|---|---|---|---|
| YgjD | BAPNA | 8100 ± 400 | 100.00 |
| ATEE | 59000 ± 5000 | 100.00 | |
| BTEE | 53700 ± 1700 | 100.00 | |
| H111A | BAPNA | 0 | 0 |
| ATEE | 64000 ± 6000 | 108.39 ± 0.98 | |
| BTEE | 28700 ± 3300 | 53.30 ± 4.46 | |
| E113A | BAPNA | 6400 ± 600 | 78.83 ± 3.51 |
| ATEE | 41000 ± 6000 | 69.12 ± 4.32 | |
| BTEE | 0 | 0 | |
| H115A | BAPNA | 9000 ± 500 | 111.07 ± 0.69 |
| ATEE | 0 | 0 | |
| BTEE | 0 | 0 | |
| H111A+H115A | BAPNA | 0 | 0 |
| ATEE | 0 | 0 | |
| BTEE | 0 | 0 |
Figure 5: Growth curves of the V. harveyi cells in 2216 E broth with addition of the different amount of wild-type YgjD and YgjD mutants.
As the abovementioned description, the conserved sequence “HXEXH” of the GCP homologues and the HSP70-actin fold play an important role in the growth and rescues of microorganism. In fact, the same conserved structure also were all found in the YgjD of V. harveyi, which showed that the sequence of the ygjD gene was highly conserved among bacteria. The ygjD gene from V. harveyi strain SF-1 consists of 338 amino acids. That showed 100% similarity with other V. harveyi strains. It also showed high similarities of 89%–100% with other Vibrio species. The similarity with Gram-negative E. coli and Gram-positive B. cereus was 76% and 43% respectively. Further it showed 31% of the similarity with eukaryotic organism S. cerevisiae. The purified protein YgjD did not showed proteolytic activity against azocasein and albumin, but showed the enzymic activity when determined with ATEE, BTEE and BAPNA as substrates. 1.0 mM of Zn2+ increased the protease activity of the purified recombinant YgjD, but Cu2+, Mn2+, Ca2+ and Co2+ inhibited the enzyme activities to some extent. The amino acids of the conserved sequence “HXEXH” of the YgjD in V. harveyi were substituted with alamine. The enzyme activities of the YgjD mutants with His111, Glu113 and His115were greatly decreased, and the YgjD mutant with His111+His115 lost its proteolytic activity completely. These results showed that the conserved sequence “HXEXH” of YgjD in V. harveyi was very important for keeping its protease activity. Some studies have found that the study of Vibrio protease is important for understanding its pathogenic mechanism (Liu et al., 2019).
| 12 h | 24 h | 48 h | 72 h | 96 h | 120 h | |
|---|---|---|---|---|---|---|
| YgjD | 0 | 0 | 0 | 0 | 0 | 0 |
| Inactivated protein | 0 | 0 | 0 | 0 | 0 | 0 |
| Sterile water | 0 | 0 | 0 | 0 | 0 | 0 |
| Sodium pyruvate | 0 | 0.4 ± 0.012 | 1.7 ± 0.048 | 2.9 ± 0.036 | 4.4 ± 0.016 | 5.1 ± 0.062 |
Notes:
Culturable counts are CFU/mL (×103).
In this work, the resuscitation promoting effect of the recombinant YgjD on the VBNC cells of V. harveyi was detected, but no visible colonies were observed on the plates added with the purified recombinant protein. Interestingly, with the addition of the purified YgjD to the normal bacteria culture of V. harveyi strain SF-1, the protein could increase the cell growth, and the growth promoting effect was related to the concentrations of the YgjD. But the YgjD mutant with His111+His115 lost its effects on cell growth, which suggested that the biological function of YgjD might be partly related to the protease activity. The mechanism for its promoting effect remained to be further studied.
Conclusion
The conserved ygjD gene from V. harveyi strain SF-1 was cloned and the protein was expressed in E. coli BL21. The conserved sequence “HXEXH” of the protease and HSP70-actin fold were found in the YgjD. The purified recombinant YgjD showed protease activities when determined with ATEE, BTEE and BAPNA. Zn2+ could increase protease activity, Cu2+, Ca2+, Mn2+, Co2+, EDTA, EGTA and PMSF could partly inhibit protease activity. The two conserved His111 and His115of YgjD played very important roles in maintaining its protease activity. The purified recombinant YgjD protein promoted the normal cell growth, but did not show obvious promoting effect on VBNC cells of V. harveyi strain SF-1.