The relative importance of DNA methylation and Dnmt2-mediated epigenetic regulation on Wolbachia densities and cytoplasmic incompatibility
- Published
- Accepted
- Received
- Academic Editor
- Irene Newton
- Subject Areas
- Genetics, Microbiology, Parasitology
- Keywords
- Wolbachia pipientis , DNA methylation, Cytoplasmic incompatibility, Dnmt2 , Drosophila melanogaster , DNA methyltransferase, Immunity
- Copyright
- © 2014 LePage et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2014. The relative importance of DNA methylation and Dnmt2-mediated epigenetic regulation on Wolbachia densities and cytoplasmic incompatibility. PeerJ 2:e678 https://doi.org/10.7717/peerj.678
Abstract
Wolbachia pipientis is a worldwide bacterial parasite of arthropods that infects germline cells and manipulates host reproduction to increase the ratio of infected females, the transmitting sex of the bacteria. The most common reproductive manipulation, cytoplasmic incompatibility (CI), is expressed as embryonic death in crosses between infected males and uninfected females. Specifically, Wolbachia modify developing sperm in the testes by unknown means to cause a post-fertilization disruption of the sperm chromatin that incapacitates the first mitosis of the embryo. As these Wolbachia-induced changes are stable, reversible, and affect the host cell cycle machinery including DNA replication and chromosome segregation, we hypothesized that the host methylation pathway is targeted for modulation during cytoplasmic incompatibility because it accounts for all of these traits. Here we show that infection of the testes is associated with a 55% increase of host DNA methylation in Drosophila melanogaster, but methylation of the paternal genome does not correlate with penetrance of CI. Overexpression and knock out of the Drosophila DNA methyltransferase Dnmt2 neither induces nor increases CI. Instead, overexpression decreases Wolbachia titers in host testes by approximately 17%, leading to a similar reduction in CI levels. Finally, strength of CI induced by several different strains of Wolbachia does not correlate with levels of DNA methylation in the host testes. We conclude that DNA methylation mediated by Drosophila’s only known methyltransferase is not required for the transgenerational sperm modification that causes CI.
Introduction
Wolbachia pipientis, an obligate intracellular bacteria, is estimated to infect approximately 40% of all arthropod species (Zug & Hammerstein, 2012). This widespread prevalence can be attributed to efficient maternal transmission of the infection, intermediate rates of horizontal transmission to new hosts, and strong manipulations of the host reproductive system to enhance its maternal transmission (Stouthamer, Breeuwer & Hurst, 1999; Serbus et al., 2008). These sexual alterations all act to increase the number of infected females within a population and include male-killing, feminization, parthenogenesis, and cytoplasmic incompatibility (CI). CI is the most common defect observed in Wolbachia-infected hosts and has been documented in numerous species (Serbus et al., 2008).
CI acts as a post-fertilization mating barrier by preventing the development of embryos from uninfected females that are mated with Wolbachia-infected males. This zygotic defect can be rescued, however, by females infected with the same strain of Wolbachia present in the male. This rescue capability gives a strong fitness advantage to Wolbachia-infected females and can lead to rapid sweeps of the infection through host populations. For instance, CI-inducing Wolbachia have been able to spread across most of the Drosophila simulans population in eastern Australia in less than a decade (Kriesner et al., 2013). CI is also a major isolation barrier between young sibling species (Bordenstein, O’Hara & Werren, 2001; Jaenike et al., 2006; Miller, Ehrman & Schneider, 2010) and is currently being used as a genetic drive mechanism to eliminate dengue virus in Aedes aegypti populations (Moreira et al., 2009; Bian et al., 2010; Walker et al., 2011) and to generally reduce mosquito population sizes (Laven, 1967; O’Connor et al., 2012).
The evolutionary, ecological, and medical importance of CI has fueled decades of research seeking to understand its underlying mechanisms. However, apart from studies that suggest the host genes JhI-26 and HIRA are involved (Zheng et al., 2011; Liu et al., 2014), it remains unknown how Wolbachia in the testes encode a sperm modification that renders embryos inviable. Previous work elucidated a few post-fertilization hallmarks of CI, most of which are associated with defects in the paternal genome during embryogenesis. These changes include a failure of maternal histones to deposit correctly, prolonged or incomplete replication of the paternal DNA, and failed condensation of the paternal chromosomes (Breeuwer & Werren, 1990; Callaini, Dallai & Riparbelli, 1997; Landmann et al., 2009). The alterations of the paternal chromatin and host cell cycle lead to a failure of the first mitosis followed by embryonic death. Interestingly, Wolbachia are not actually present within the sperm of their hosts, indicating a semi-permanent modification of the paternal genome that is transgenerationally transmitted to the egg (Clark et al., 2008).
Several assumptions can be made about the paternal genome modification underlying cytoplasmic incompatibility including:
-
It targets host pathways that are highly conserved across numerous host species.
-
It involves a semi-permanent but reversible alteration to the paternal genome.
-
It must be able to affect histone recruitment, DNA replication, and chromosome condensation.
Working under these assumptions, we selected the host DNA methylation pathway as a probable target for Wolbachia. Methylation is a stable, yet reversible, modification to DNA that could be sex-specific and easily rescued by infected females. It also has the capability to modulate many cell cycle functions including chromosome condensation and histone recruitment (Bird, 2001; Harris & Braig, 2003; Weber & Schübeler, 2007) and has previously been hypothesized to play a role in CI (Negri, 2011; Saridaki et al., 2011; Ye et al., 2013b; Liu et al., 2014). While the role of DNA methylation in insects is not fully understood, it is a highly conserved pathway that shows strong upregulation during embryogenesis (Field et al., 2004). Finally, the ability of bacteria to alter host methylation and chromatin structure is increasingly recognized (Gómez-Díaz et al., 2012; Bierne, Hamon & Cossart, 2012) and previous work shows that Wolbachia infection in particular alters the host methylation profile in both leafhoppers and mosquitoes (Negri et al., 2009; Ye et al., 2013a).
Here we use the model organism Drosophila melanogaster infected with the wMel strain of Wolbachia to determine the role of host DNA methylation in CI. D. melanogaster flies utilize just one canonical DNA methyltransferase, Dnmt2 (Lyko, Ramsahoye & Jaenisch, 2000), which enables easy genetic manipulation of the host methylation pathway without the confounding influence of other DNA methyltransferases (Dnmt1 and Dnmt3) present in most other insect species (Werren et al., 2010). While the role of Dnmt2-dependent methylation is debated and multifaceted (Schaefer & Lyko, 2010; Raddatz et al., 2013; Takayama et al., 2014), evidence demonstrates that the methylation machinery in D. melanogaster is not only present but also functional (Lyko, Ramsahoye & Jaenisch, 2000; Kunert et al., 2003; Schaefer, Steringer & Lyko, 2008; Gou et al., 2010). Moreover, overexpression of the mouse Dnmt3a in D. melanogaster induces CI-like defects such as reduced rates of cell cycle progression and altered chromosome condensation (Weissmann et al., 2003).
Materials and Methods
Fly rearing and dissections
All flies were reared on a cornmeal and molasses-based media at 25 °C. The Dnmt2 loss-of-function mutant has been previously described (Goll et al., 2006). Briefly, the mutant contains a 28bp insertion with multiple stop codons as well as a frameshift within the coding region of Dnmt2. Overexpressing flies were created through the Gal4-UAS system. Crosses were performed between virgin nos-Gal4 driver females (y1w*; P{w[ + mC] = GAL4-nos.NGT}40 (either Wolbachia-infected or uninfected)) and 5–6 uninfected UAS-Dnmt2 (Kunert et al., 2003), UAS-GFP or W1118 males. Crosses for Fig. S1 were conducted between virgin Act5c-Gal4 driver females (y1w*; P{w[ + mC] = Act5C-GAL4}25FO1/CyO, y+, Wolbachia infected or uninfected depending on desired progeny) and UAS-Dnmt2 males. For Act5c-Gal4 crosses, straight-winged progeny were assumed to be overexpressing Dnmt2 while CyO expressing lines were used as the wild-type expressing lines. Wolbachia-uninfected lines were created through tetracycline treatment (20 ug/mL for 3 generations) and infection status was confirmed through PCR using the following primers: WolbF (GAAGATAATGACGGTACTCAC) and WolbR3 (GTCACTGATCCCACTTTAAATAAC) which target the 16S rRNA gene of Wolbachia. These lines were further reared for at least three generations on undrugged media before experimentation to avoid detrimental paternal effects seen in other systems (Zeh et al., 2012).
The wAu, wNo, and wRi (also known as wRi Agadir) strains of Drosophila simulans were kindly provided by Charlat Sylvain (University of Lyon, France). All testes and ovary dissections were performed in cold phosphate buffered saline (PBS). Males were dissected within 24 h of emergence while females were aged 3–4 days before dissections. Testes samples consisted of tissue obtained from a minimum of 20 males while ovary samples were pooled from 10 females each. Tissues were frozen and stored at −80 °C before analysis.
Hatch rate assays
Assays were performed using a grape juice/agar media in 30 mm plates for egg laying. For each cross 32–48 individual crosses of one male and one female were set up in separate mating chambers with individual grape juice plates. A minimal amount of a 1:2 dry yeast and water mix was added to each plate and the parents were allowed to mate for 16 h before the grape juice plates were discarded. Fresh plates were then used for 24 h, removed, and the number of eggs laid was counted for each cross. The number of unhatched eggs was counted again at 36 h after the plates had been removed to determine hatch rates.
MethylFlash quantification of DNA methylation
Genomic levels of cytosine methylation (5-mC) were measured using the MethylFlash kit (Epigentek, Farmingdale, NY, USA). 8–10 replicate sets of testes (20–40 testes pairs each replicate) were dissected and DNA was isolated using the Puregene Tissue kit (Qiagen, Venlo, Netherlands). 100 ng of genomic DNA from each sample was used and each sample was analyzed in duplicate on a BMG LabTech FLOUstar OPTIMA plate reader (Ortenberg, Germany) according to manufacturer instructions.
Wolbachia density
Eight replicates each of whole animals (pools of 3), testes (pools of 20 pairs), and ovaries (pools of 10 pairs) were collected and DNA was isolated. All males were less than 24 h old while females had been aged 3–4 days. Quantitative PCR was performed on a Bio-Rad CFX96 Real-Time System using iTaq Universal SYBR Green Supermix (Bio-Rad, Hercules, CA, USA). groEL copy number, determined against a standard curve, was compared to counts for the host gene Actin, also determined against a standard curve. It was assumed that one copy of groEL was present in each Wolbachia genome and 1 or 2 copies of Act5c (for males and females, respectively, as the gene is on the X chromosome) in each Drosophila genome. Primers: Act5c (231bp product, Forward: ATGTGTGACGAAGAAGTTGCT Reverse: GTCCCGTTGGTCACGATACC), groEL (97bp product, Forward: CTAAAGTGCTTAATGCTTCACCTTC Reverse: CAACCTTTACTTCCTATTCTTG). qPCR conditions: 50°10 min, 95°5 min, 40×(95°10 s, 55°30 s), 95°30 s. Followed by melt curve analysis (0.5°steps from 65–95°for 5 s each).
Gene expression
Quantitative PCR was performed on a Bio-Rad CFX96 Real-Time System using iTaq Universal SYBR Green Supermix (Bio-Rad, Hercules, CA, USA). RNA was isolated from 8 sets of testes (20 pairs each) using the RNeasy Mini kit (Qiagen, Venlo, Netherlands) and DNA was removed with the TURBO DNA-free DNase kit (Ambion, Grand Island, NY, USA). cDNA was synthesized using a SuperScript III First-Strand kit (Invitrogen, Grand Island, NY, USA) and diluted 1:20. All calculations were done using delta delta Ct with Rp49 expression used for normalization of results. Primers: Dnmt2 (150bp product, Forward: CCGTGGCGTGAAATAGCG Reverse: ACACCGCTTTCGGAGGACG), Rp49 (154bp product, Forward: CGGTTACGGATCGAACAAGC Reverse: CTTGCGCTTCTTGGAGGAGA). qRT-PCR conditions are the same as used in qPCR for Wolbachia densities.
Bisulfite sequencing
One hundred testes were dissected in PBS from Wolbachia infected (y1w*) and uninfected males and flash frozen. gDNA was then isolated using the Puregene kit (Qiagen, Venlo, Netherlands) and fragmented by Covaris shearing. gDNA was submitted to Vanderbilt Technologies for Advanced genomics (VANTAGE) where the PE-75 bp library was generated using the TruSeq sample preparation kit (with methylated adapters), bisulfite treated, PCR amplified (EpiMark and ZymoTaq) and sequenced (Illumina HiSeq 2000, 86bp PE read). Sequences with ≥10× coverage were analyzed using Bismark (Krueger & Andrews, 2011) and cytosines which were methylated in at least one read were counted.
Results and Discussion
Wolbachia wMel increases levels of testes DNA methylation
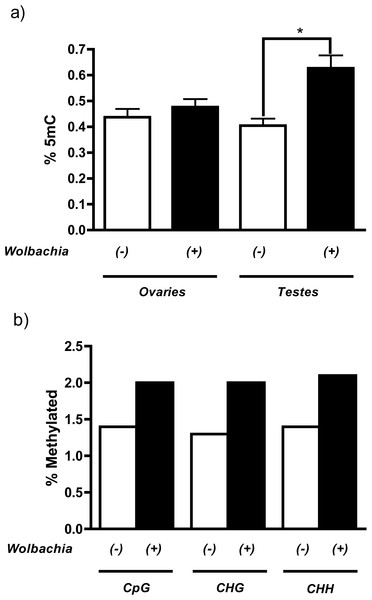
MethylFlash analysis of host DNA from testes revealed that infection with the Wolbachia strain wMel in Drosophila melanogaster increases levels of genome-wide cytosine methylation (Fig. 1A). More importantly, this methylation is specific to the host testes (55% increase, P = 0.0015, Mann Whitney U test) and is not observed in the ovaries, consistent with the prediction that only the paternal genome is modified during cytoplasmic incompatibility. The overall levels of methylation are extremely low, which is consistent with previously reported levels of methylation in Drosophila melanogaster (Lyko, Ramsahoye & Jaenisch, 2000; Kunert et al., 2003). Conflicting reports over the strength and prevalence of DNA methylation in D. melanogaster (Lyko, Ramsahoye & Jaenisch, 2000; Raddatz et al., 2013; Schaefer & Lyko, 2010) led us to test the validity of our initial results with genome-wide bisulfite sequencing. Results indicate that, contrary to most other species, DNA methylation in Drosophila melanogaster is not CpG specific and is evenly distributed over cytosine residues (Fig. 1B and Table S1). Sequencing results also mirror those of MethylFlash and show that infection with wMel increases testes DNA methylation 46% across all cytosine residues with a range of 43–54% depending upon the type of cytosine residue (CpG, CHG, or CHH) (Fig. 1B and Table S1). The minor discrepancies between MethylFlash and bisulfite sequencing (55% and 46% increase in methylation, respectively) are likely due to the sensitivity of the MethylFlash system on such low quantities of methylation. A more thorough investigation of the bisulfite sequencing, including changes in promoter and gene body methylation, is ongoing.
Figure 1: Wolbachia increase host levels of DNA methylation.
(a) Wolbachia infection (wMel) of Drosophila melanogaster increases DNA methylation in host testes by 55% (P = 0.0015, Mann–Whitney U (MWU) test, two-tailed), as measured by the ELISA-based MethylFlash kit. This increase is not observed in host ovaries (P = 0.25). Bars denote standard error of the mean (SEM) (b) Bisulfite sequencing of Drosophila melanogaster testes DNA shows that infection by wMel increases methylation of all cytosine residues including CpG (43%), CHG (54%), and CHH (50%).Overexpression of Dnmt2 neither induces nor strengthens CI
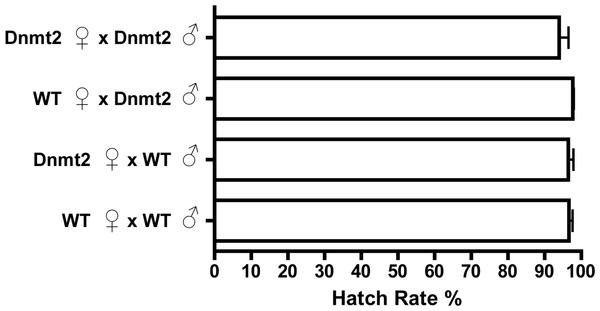
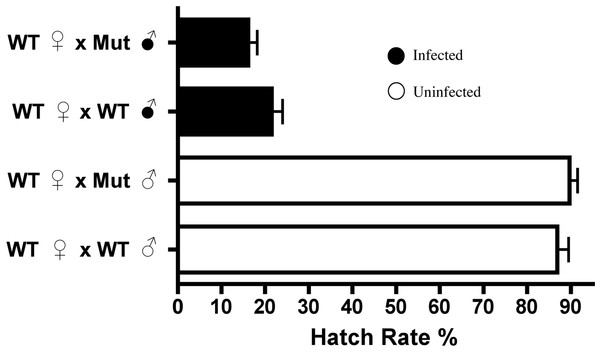
Drosophila melanogaster possess just one canonical DNA methyltransferase, Dnmt2, and overexpression of this enzyme in fruit flies has previously been shown to increase levels of DNA methylation (Kunert et al., 2003; Schaefer, Steringer & Lyko, 2008). Utilizing the Gal4-UAS expression system, we overexpressed Dnmt2 in uninfected males to test if an increase in host methylation alone could induce the CI defect of reduced embryo hatching rates. Figure 2 shows that there was no discernable difference in hatching rates with uninfected males expressing increased or wild type levels of Dnmt2 (P = 0.91, MWU). The result was confirmed using an Actin-based driver that again yielded no discernable differences in hatch rates compared to wild type flies (Fig. S1, P = 0.83, MWU). These findings specify that amplified levels of Dnmt2-mediated epigenetic regulation are not sufficient to recapitulate cytoplasmic incompatibility.
Figure 2: Expression of DNA methyltransferase 2 does not induce CI.
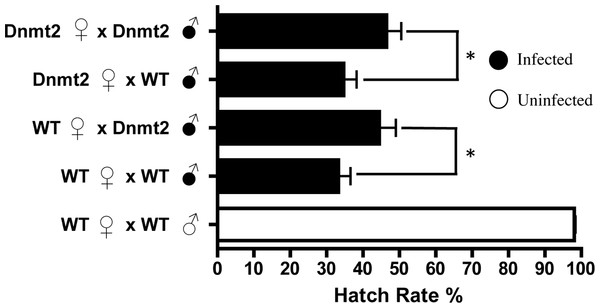
Overexpression of the DNA methyltransferase Dnmt2 in uninfected males, utilizing the Gal4-UAS system with a nos driver, does not reduce hatching rates. Dnmt2, overexpressing flies; WT, wild type flies. Bars denote standard error of the mean (SEM).If multiple factors are responsible for CI, it is possible that overexpression of Dnmt2, while unable to induce CI in uninfected flies, may be able to strengthen the modification in the presence of Wolbachia. To test this hypothesis, we overexpressed Dnmt2 in Wolbachia-infected males that were then mated to uninfected virgin females. Surprisingly, Dnmt2 overexpression in males decreased the level of cytoplasmic incompatibility by an average of 17.4% (Fig. 3). This effect is not dependent on the Dnmt2 expression status of the female and suggests that increased methylation of host DNA can diminish the penetrance of cytoplasmic incompatibility.
Figure 3: Overexpression of Dnmt2 reduces levels of CI.
The overexpression of Dnmt2 in Wolbachia-infected males decreases rates of CI (P < 0.05, Mann–Whitney U test). Dnmt2 expression in the mother has no effect. Bars denote standard error of the mean (SEM). Dnmt2, overexpressing flies; WT, wild type flies.Overexpression of Dnmt2 reduces Wolbachia titers in host testes
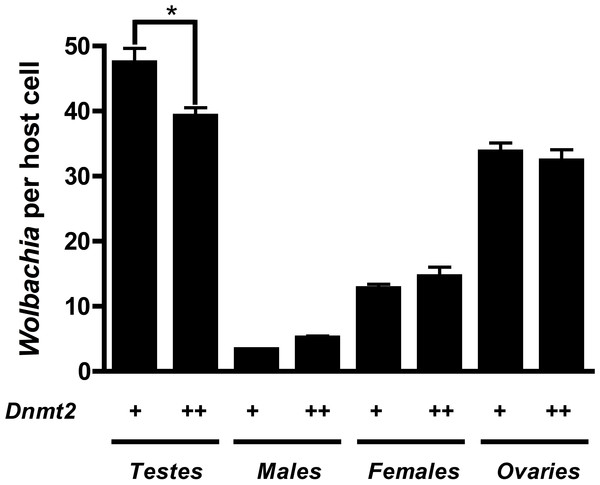
Previous work suggested that Dnmt2 is detrimental to Wolbachia proliferation in mosquitoes. In fact, Wolbachia strain wMel Pop-CLA utilizes a host miRNA to downregulate Dnmt2 expression when infecting Aedes aegypti (Zhang et al., 2013). We observed no differences in Dnmt2 expression between wMel infected and uninfected D. melanogaster testes (data not shown) but hypothesized that overexpression of Dnmt2 in the host may adversely affect Wolbachia titers. In support of this prediction, we found that Wolbachia density (as measured by the ratio of Wolbachia groEL gene copy number/Drosophila Actin gene copy number) decreased by 17.3% in adult testes overexpressing Dnmt2 transcripts by 9.6% (Fig. 4 and Fig. S2, respectively). The low level of transcript overexpression could be specific to the developmental stage of the experimental sample or due to usage of a pUAST vector for germline expression instead of the more efficient pUASP (Kunert et al., 2003). While the upregulation of Dnmt2 in infected males is not statistically significant, it remains possible that actual protein levels are much higher than those represented by RNA transcripts. Strong protein expression, as measured by Western blot, was seen in uninfected ovaries (data not shown). Nevertheless, the 17.3% decrease in Wolbachia titers compares well with the 17.4% reduction in CI penetrance reported above. Expression of the negative control green fluorescent protein (GFP) did not reduce Wolbachia titers, as expected (Fig. S3). As the bacterial and phage density models of CI specify that Wolbachia titers in the testes are linked to the strength of CI (Breeuwer & Werren, 1993; Bordenstein et al., 2006), we conclude that the reduction of CI observed in Dnmt2-overexpressing males is likely due to reduced Wolbachia density.
Figure 4: Dnmt2 overexpression alters Wolbachia titers.
Overexpression of Dnmt2 reduces Wolbachia titers within the testes (P < 0.01, MWU test) but has no affect on titers within ovaries or whole flies. Wolbachia infection is derived from the y1w∗ Drosophila background. Bars denote standard error of the mean (SEM). Dnmt2 + +, overexpressing flies; Dnmt2 +, wild type flies. (P = 0.007, Mann–Whitney U test, two-tailed).Even though we do not observe any change in Dnmt2 mRNA levels after Wolbachia infection, we cannot rule out that Wolbachia may be affecting intracellular Dnmt2 localization rather than levels of gene expression. An increase in localization of Dnmt2 to the nucleus would not only protect the cytosolic Wolbachia but also explain the additional genomic methylation associated with infection. In this scenario, the testes-specific increase in host methylation initially observed would simply be a by-product of high Wolbachia activity. Additionally, an immunomodulatory role for Dnmt2 in Drosophila has already been documented in protection against RNA viruses (Durdevic et al., 2013) though we believe the findings in this report are the first evidence for a putative antibacterial role for Dnmt2 in fruit flies.
Hosts defective in DNA methylation still exhibit CI
As Dnmt2 overexpression did not induce nor increase cytoplasmic incompatibility, we next tested the strength of CI in hosts defective in the methyltransferase pathway. Knockout mutants for Dnmt2 characterized by Goll et al. (2006) were acquired and found by PCR and amplicon sequencing to be infected by the wMel strain of Wolbachia. The strain is hereafter referred to as Mut and was tetracycline treated for three generations to create the uninfected line MutT. We show by MethylFlash that the increase in host DNA methylation induced by Wolbachia infection is abolished in the knockout Mut background (Fig. S4) and is thus Dnmt2-dependent. However, loss of this crucial enzyme in the DNA methylation pathway has no effect on the penetrance of CI (Fig. 5), as shown in comparisons between mutant and wild type males mated to uninfected females (P = 0.13, MWU). The low level of DNA methylation still present in mutants has recently been observed by others (Boffelli, Takayama & Martin, 2014) and suggests a possible mechanism of DNA methylation in Drosophila that is independent of canonical DNA methyltransferases. Thus, it is possible that CI could be induced by alterations in genomic methylation but in a Dnmt2-independent manner.
Figure 5: Dnmt2 mutants express wild-type levels of CI.
Crosses with Dnmt2-mutant males (“Mut”) show that Dnmt2 expression within the father is not necessary for expression of CI. Mut, Dnmt2 mutant flies; WT, wild type flies. Bars denote standard error of the mean (SEM).Curiously, despite the previously observed role for Dnmt2 in host immunity (Zhang et al., 2013; Durdevic et al., 2013), the mutants observed here exhibit no increase in Wolbachia titers within any of the tissues tested (Fig. S5). It is interesting to note that Dnmt2 mutant Drosophila, derived from the W1118 background line, have titers that are, on average, half of those seen in y1w* background lines (see Fig. 4). This difference has been observed several times in our experiments and suggests either a differing ability of the host lines to control Wolbachia titers or an as yet unclassified difference in the wMel strains infecting these flies.
Host levels of DNA methylation do not correlate with strength of CI
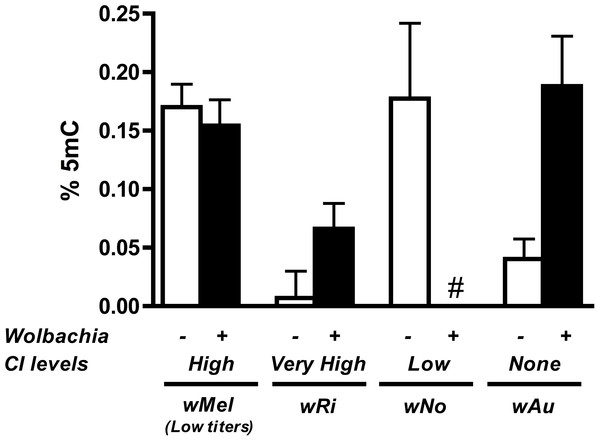
To substantiate the claim that DNA methylation is not involved in the induction of CI for other Wolbachia strains and/or host species, we tested the DNA methylation status of testes DNA from Drosophila species infected with various strains of Wolbachia. These taxa include D. simulans infected with strains wRi, wNo, and wAu, which express strong, moderate, and no CI, respectively. We also tested a different D. melanogaster-infecting strain wMel derived from the W1118 background strain instead of y1w*. As previously mentioned, while the W1118 line induces strong CI, Wolbachia titers are much lower in these animals compared to the infection found in y1w*.
Results show that methylation status of the infected host testes is random with regards to the strength of CI (Fig. 6). While infection with the high CI-inducer wRi exhibits higher methylation in infected testes as compared to uninfecteds, this effect is marginally insignificant (P = 0.072, MWU) and is countered by data from the wAu strain, which causes no CI but still significantly increases host DNA methylation in testes (P = 0.0047, MWU). Furthermore, infection with the wNo strain of Wolbachia, which causes moderate CI, actually has less methylation in host testes. Finally, a low-titer infection of wMel (W1118), while still inducing CI, does not induce the same level of DNA methylation associated with a high-density infection (y1w*).
Figure 6: Levels of host DNA methylation do not correlate with strength of CI.
Testing of several different Wolbachia infections, capable of inducing various levels of CI in their respective hosts, shows that levels of host DNA methylation and strength of CI are not correlated. Bars denote standard error of the mean (SEM) of testes DNA methylation, as measured by MethylFlash. White bars (−) denote uninfected flies and black bars (+) denote infected flies. # indicates levels of methylation too low for detection.Conclusions
The underlying mechanism of Wolbachia-induced CI largely remains elusive after several decades of research. Here we show that host DNA methylation, a promising candidate pathway hypothesized to play a role (Negri, 2011; Saridaki et al., 2011; Ye et al., 2013b; Liu et al., 2014), does not seem to be involved in the induction of CI. While Wolbachia infection preferentially increases host DNA methylation in Drosophila melanogaster testes (Fig. 1), this modification is not conserved across other CI-causing strains of Wolbachia (Fig. 6) and overexpression of a host methyltransferase neither induces nor increases rates of CI. We have also found that Wolbachia-induced changes in host methylation are dependent on the DNA methyltransferase Dnmt2 (Fig. S4) but that Drosophila melanogaster lacking Dnmt2 still suffer from CI (Fig. 5). Finally, we found Dnmt2 has anti-Wolbachia properties, as previously reported in Aedes aegypti (Zhang et al., 2013), and overexpression of Dnmt2 reduces the strength of CI.
Taken together, we show that one of the canonical chromatin modification pathways, Dnmt2-dependent DNA methylation, likely has no role in Wolbachia-induced CI. Wolbachia infection can be associated with changes in host methylation levels, but it is most likely a consequence of the bacteria modulating host immune response or the host defending itself against the infection. The possibility also remains that infection alters gene-specific, and Dnmt2-independent, levels of methylation that our current study of genomic methylation levels has not detected. While further investigation of the Dnmt2 epigenetic pathway will not elucidate a CI mechanism, it may be useful in studying the complex nature of pathogen-host interactions between Wolbachia and the many species it infects. It remains possible that a novel methyltransferase, recently suggested to exist in Drosophila (Takayama et al., 2014; Boffelli, Takayama & Martin, 2014), could affect CI.
Supplemental Information
Dnmt2 overexpression does not induce CI
Overexpression of Dnmt2 by an Actin-Gal4 driver does not induce CI in uninfected males. Bars denote SEM. Dnmt2, overexpressing flies; WT, wild type flies.
Overexpression of Dnmt2 in host testes
Dnmt2 is overexpressed 9.6% compared to wild type in testes using ananos-Gal4 driver. Bars denote SEM. WT, wild type; Dnmt2, Dnmt2 overexpressing. Rp49 is used as a control for gene expression.
Wolbachia titers are not decreased by GFP expression
Expression of green fluorescent protein (GFP) does not reduce Wolbachia titers, as measured in whole males, females, testes, and ovaries. Wolbachia infection arises from the y1w∗ background. Bars denote SEM. ± indicates whether sample express GFP.
Wolbachia-induced change in host methylation is Dnmt2 dependent
Testes from Drosophila melanogaster Dnmt2 mutants do not exhibit Wolbachia-induced increase in DNA methylation as measured by MethylFlash. Wolbachia infection arises from the W1118 background. Bars denote SEM. MutT, uninfected, Dnmt2 mutant; Mut, Wolbachia infected, Dnmt2 mutant.
Dnmt2 mutants harbor normal Wolbachia titers
Loss of Dnmt2 does not affect Wolbachia titers in Drosophila melanogaster. Bars denote SEM. Dnmt2 +, wild type flies; Dnmt2 −, Dnmt2 mutant flies.