Suppression of hesA mutation on nitrogenase activity in Paenibacillus polymyxa WLY78 with the addition of high levels of molybdate or cystine
- Published
- Accepted
- Received
- Academic Editor
- Pedro Silva
- Subject Areas
- Microbiology, Molecular Biology
- Keywords
- Paenibacillus, Cystine, Nitrogenase, hesA, Molybdenum
- Copyright
- © 2019 Liu et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2019. Suppression of hesA mutation on nitrogenase activity in Paenibacillus polymyxa WLY78 with the addition of high levels of molybdate or cystine. PeerJ 7:e6294 https://doi.org/10.7717/peerj.6294
Abstract
The diazotrophic Paenibacillus polymyxa WLY78 possesses a minimal nitrogen fixation gene cluster consisting of nine genes (nifB nifH nifD nifK nifE nifN nifX hesA and nifV). Notably, the hesA gene contained within the nif gene cluster is also found within nif gene clusters among diazotrophic cyanobacteria and Frankia. The predicted product HesA is a member of the ThiF-MoeB-HesA family containing an N-terminal nucleotide binding domain and a C-terminal MoeZ/MoeB-like domain. However, the function of hesA gene in nitrogen fixation is unknown. In this study, we demonstrate that the hesA mutation of P. polymyxa WLY78 leads to nearly complete loss of nitrogenase activity. The effect of the mutation can be partially suppressed by the addition of high levels of molybdate or cystine. However, the nitrogenase activity of the hesA mutant could not be restored by Klebsiella oxytoca nifQ or Escherichia coli moeB completely. In addition, the hesA mutation does not affect nitrate reductase activity of P. polymyxa WLY78. Our results demonstrate hesA is a novel gene specially required for nitrogen fixation and its role is related to introduction of S and Mo into the FeMo-co of nitrogenase.
Introduction
Nitrogenase, nitrate reductase, and a number of other enzymes require molybdenum and sulfur for their activity (Johnson, 1980; Lester & Demoss, 1971; Pienkos, Shah & Brill, 1977; Shah et al., 1984). Molybdenum and sulfur are found in these molybdoenzymes as parts of low-molecular-weight cofactors (Johnson, 1980; Shah & Brill, 1977). Two different molybdenum cofactors have been described: iron-molybdenum cofactor (FeMo-co), which is found only in nitrogenase (Pienkos, Shah & Brill, 1977; Shah & Brill, 1977), and molybdenum cofactor (Mo-co), which is found in other molybdoenzymes.
Most biological nitrogen fixation is catalyzed by molybdenum-dependent nitrogenase, which is distributed within bacteria and archaea. This nitrogenase is a two-component enzyme consisting of the Fe and MoFe proteins (Hu & Ribbe, 2011; Rubio & Ludden, 2008). The MoFe protein is an α2β2 heterotetramer (encoded by nifD and nifK) that contains two metalloclusters; FeMo-co, a [Mo-7Fe-9S-C-homocitrate] cluster which serves as the active site of substrate binding and reduction and the P-cluster, a [8Fe-7S] cluster which shuttles electrons to FeMo-co. The Fe protein (encoded by nifH) is a γ2 homodimer bridged by an intersubunit [4Fe-4S] cluster that serves as the obligate electron donor to the MoFe protein (Oldroyd & Dixon, 2014). It is now well established from genetic and biochemical analysis that nifE, nifN, nifX, nifB, nifQ, nifV, nifY and nifU, nifS and nifH contribute to the synthesis and insertion of FeMo-co into nitrogenase (Hu & Ribbe, 2011; Rubio & Ludden, 2008). Of these genes, nifQ serves as a molybdenum donor for FeMo-co synthesis, nifU and nifS are involved in [Fe-S] cluster assembly. NifS is a cysteine desulfurase that provides sulfur for the assembly of transient [Fe-S] clusters onto the molecular scaffold NifU.
Mo-co consists of the molybdopterin cofactor with molybdenum bound to its dithiolene moiety. Mo-co is found in nitrate reductase of some fungi and bacteria, such as nitrate reductase of Escherichia coli (Leimkühler, Wuebbens & Rajagopalan, 2011; Mendel, 2013). In E. coli, several loci (moa, mob, moe and mog) are involved in the biosynthesis of the Mo-co (Leimkühler, Wuebbens & Rajagopalan, 2001). In Mo-co biosynthesis, MoeB functions to sulfurylate MoaD, and IscS functions as a primary sulfur-donating enzyme by interacting specifically with MoeB and MoaD in the biosynthesis of molybdopterin in E. coli (Sambasivarao et al., 2002; Zhang et al., 2010).
Diazotrophic (nitrogen-fixing) Gram-positive and endospore-formed Paenibacillus spp. have potential uses as a bacterial fertilizer in agriculture. Comparative genomic analysis shows that a nif gene cluster composed of nine genes nifBHDKENXhesAnifV is conserved in the genomes of Paenibacillus polymyxa WLY78 and other the diazotrophic Paenibacillus species and strains. Whereas, the Gram-negative diazotrophic model Klebsiella oxytoca has 20 nif genes (nifJHDKTYENXUSVWZMFLABQ) organized in seven transcriptional units and co-located within a 24 kb cluster (Arnold et al., 1988). Compared to K. oxytoca, P. polymyxa WLY78 has no nifQ, nifS and nifU, but contains a hesA gene. The hesA gene is also highly conserved within the nif gene clusters of Cyanobacteria and Actinobacteria (Frankia). The predicted product HesA is a member of the ThiF-MoeB-HesA family containing an N-terminal nucleotide binding domain and a C-terminal MoeZ/MoeB-like domain. However, the function of hesA gene in nitrogen fixation is unknown.
In this study, the function of the P. polymyxa hesA gene in nitrogen fixation is investigated. Mutation of P. polymyxa hesA leads to no N2-fixation in the media containing low levels of molybdate. The effect of the mutation can be suppressed by the addition of high levels of molybdate or cystine. The hesA mutation does not affect nitrate reductase activity of P. polymyxa WLY78. Our results demonstrate hesA is specially required for nitrogen fixation.
Materials and Methods
Bacterial strains and media
Bacterial strains used in this study are listed in Table S1. P. polymyxa WLY78, mutants and the recombinant E. coli strains were grown in LB medium at 37 °C. When appropriate, antibiotics for E. coli strains and P. polymyxa WLY78 mutants were required. For P. polymyxa WLY78 mutant strains, antibiotics were added at five mg/L chloramphenicol (Cm), 12.5 mg/L tetracycline. While for E. coli, antibiotics were added at 100 mg/L ampicillin, 25 mg/L tetracycline and 25 mg/L Cm for maintenance of plasmids.
Nitrogen-limited medium (per liter contains: 10.4 g Na2HPO4, 3.4 g KH2PO4, 26 mg CaCl2·2H2O, 30 mg MgSO4, 0.3 mg MnSO4, 36 mg Ferric citrate, 7.6 mg Na2MoO4·2H2O, 10 μg p-aminobenzoic acid, 10 μg biotin, four g glucose as carbon source and 0.3 g glutamic acid as nitrogen source) was used for assay of nitrogenase activity (Wang et al., 2013). When necessary, different concentrations of Na2MoO4 or cystine or sulfate were supplemented in the nitrogen-limited medium.
To determine nitrate reductase activity, P. polymyxa WLY78 and mutant strains were cultured in mineral salts medium (per liter contains: 9.16 g K2HPO4∙3H2O, 1.0 g (NH4)2SO4, 0.22 g MgSO4∙7H2O, 2.0 g KH2PO4, 8.0 g nutrient broth, 10 g KNO3, 10 g glucose) (Sperl & Demoss, 1975).
Enzyme assays
For nitrogenase activity assays, P. polymyxa WLY78, mutants, the complemented strains and the recombinant E. coli strains were grown in 20 mL of LB media (Supplemented with antibiotics) in 50 mL flasks shaken at 200 rpm overnight at 37 °C. The cultures were collected by centrifugation, precipitations were washed three times with sterilized water and then resuspended in nitrogen-limited medium containing different concentrations of Na2MoO4 (supplemented with antibiotics for mutants and the engineered E. coli strains when necessary) to a final OD600 nm of 0.2–0.5. The nitrogenase activity was determined by the procedure described by Xie et al. (2012).
For the assay of nitrate reductase, P. polymyxa WLY78 and mutant strains were grown in 50 mL of LB media in 100 mL flasks shaken at 200 rpm for 16 h at 37 °C. The cultures were collected by centrifugation, washed three times with sterilized water and then resuspended in mineral salts medium. The cultures were grown at 200 rpm at 37 °C anaerobically. The nitrate reductase was determined by the method described by Garzón (Garzón et al., 1992). Nitrate reductase activity was expressed as nanomoles of nitrite∙minute−1∙milligram−1 of cell protein.
Growth of the wild type and mutant strains
To measure growth of strains cultured in different concentrations of Na2MoO4, cells were grown in 20 mL of LB media in 50 mL flasks shaken at 200 rpm at 37 °C overnight. The cultures were collected by centrifugation, washed three times with sterilized water and then resuspended in sufficient nitrogen medium (nitrogen-limited medium supplemented with 100 mM NH4Cl) containing different concentrations of Na2MoO4 to a start OD600 nm of 0.1. After incubating the cultures for 48 h at 37 °C with shaking at 250 rpm, the maximum growth of P. polymyxa WLY78 and mutant strains were determined by absorbancy at 600 nm.
Construction of Paenibacillus hesA mutant strain
The Paenibacillus hesA mutant was constructed through homologous recombination with pRN5101 (Hou et al., 2016), which carries a temperature-sensitive region from plasmid pE194ts (Villafane et al., 1987) containing two replication origins: one is pMB1 which can be reproduced in E. coli and the other replicon is from a plasmid pE194 which can be reproduced in Gram-positive Bacillus (Gryczan, Contente & Dubnau, 1978). A 3432 bp DNA fragment, including a 1,119 bp upstream sequence and a 1,116 bp downstream fragment of the hesA coding region, was cloned to pRN5101. Then, a 1,197 bp DNA fragment, including the promoter and coding region of Cm resistant gene, was amplified from plasmid pPR9TT and inserted between upstream and downstream homologous arms in the opposite direction from the nif genes, generating plasmid pRN5101-Cm. The plasmid pRN5101-Cm was introduced into P. polymyxa WLY78 by electroporation (BTX, ECM 399) for homologous recombination. Transformants resistant to Cm were grown at 30 °C and transferred to five mL of LB media (supplemented with five mg/L Cm) in 20 mL test tubes shaken at 200 rpm for 36 h at 39 °C for about five generations. The strains were then plated onto LB agar plates supplemented with Cm and incubated for 24 h at 39 °C. Integration of the recombinant plasmid was confirmed by PCR and sequencing. The deletion mutant strain was designated as ΔhesA.
Construction of recombinant plasmids for genetic complementation assays
The genes of hesA, nifQ and moeB were PCR amplified from genomic DNA of P. polymyxa WLY78, K. oxytoca M5a1 and E. coli K12, respectively. These PCR products, being under the control of nifB promoter of P. polymyxa WLY78, were cloned to the plasmid pHY300PLK, respectively. Then the recombinant plasmids were introduced into P. polymyxa hesA mutant, respectively.
For complementation of the hesA deletion of the recombinant E. coli 78-7 which carries a nif gene cluster composed of nine genes (nifBHDKENXhesAnifV) from P. polymyxa WLY78 (Wang et al., 2013), the P. polymyxa hesA coding region, being under the control of nifB promoter of P. polymyxa WLY78 was cloned to vector pBluescript II SK (+) that contained a Cm expression cassette, producing the recombinant plasmid pBluescript II SK (+)-hesA. Primers for PCR are listed in Table S2. Recombinant plasmids and strains are listed in Table S3.
Quantitative real-time PCR analysis of nifV expression
For quantitative real-time-PCR (qRT-PCR), cultures of P. polymyxa WLY78 and ΔhesA were grown in N2-fixing conditions (two mM glutamate and without O2) and harvested after 20 h of incubation. Total RNA was isolated using TRIzol (cat. no. 9108; Takara Bio, Dalian, China) (Rio et al., 2010). The possibility of contamination of genomic DNA was eliminated by digestion with RNase-free DNase I (cat. no. RR047A; Takara Bio, Dalian, China). The integrity and size distribution of the RNA were verified by agarose gel electrophoresis, and the concentrations were determined spectrophotometrically. Synthesis of cDNA was carried out using RT Prime Mix according to the manufacturer’s specifications (cat. no. RR047A; Takara Bio, Dalian, China). 0.4 μg of cDNA was used for qRT-PCR. The relative transcript levels of nifV were determined with 16S rDNA as a control by SYBR Premix Ex Taq (Tli RNaseH Plus) kit (cat. no. RR820A; Takara Bio). Primers for nifV and 16S rDNA used for qRT-PCR are listed in Table S2.
Statistical analysis
Statistical analyses were performed using SPSS 20.0 (SPSS, Chicago, IL, USA). The least significant test was calculated at the 0.05 or 0.01 probability level for different treatment mean comparisons of nitrogenase activity.
Results
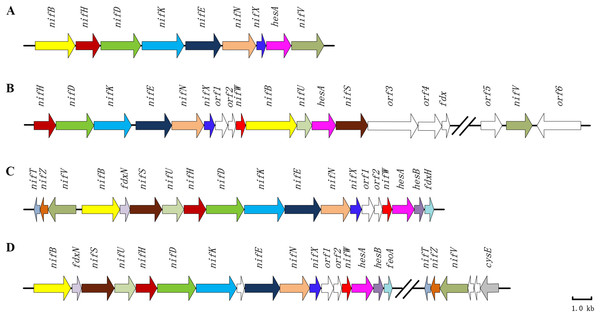
A hesA gene is highly conserved in nif gene clusters among diazotrophic Paenibacillus, cyanobacteria and Frankia
Paenibacillus polymyxa WLY78 possesses a minimal nif gene cluster consisting of nine genes (nifB nifH nifD nifK nifE nifN nifX hesA and nifV). Our previous results demonstrated that the recombinant E. coli 78-7 which was generated by introducing the P. polymyxa nif gene cluster into E. coli JM109 could produce active nitrogenase (Wang et al., 2013). Notably, a hesA gene is contained within the nif gene cluster, and its predicted product HesA is a member of the ThiF-MoeB-HesA family containing an N-terminal nucleotide binding domain and a C-terminal MoeZ/MoeB-like domain. NCBI blast analyses reveal that hesA gene is significantly conserved within nif gene clusters among diazotrophic Paenibacillus, cyanobacteria and Frankia (Fig. 1). HesA from P. polymyxa WLY78 at amino acid sequence level shares more than 90% identity with those of other diazotrophic Paenibacillus spp. (Xie et al., 2014) and 46.8%, 43.1% and 49.2% identity with HesA of Frankia alni ACN14a (Chang et al., 2012), Cyanothece sp. ATCC 51142 (Welsh et al., 2008) and Anabaena sp. PCC 7120 (Mitschke et al., 2011), respectively.
Figure 1: Clusters of nitrogen-fixation related genes between P. polymyxa WLY78 and other nitrogen-fixing bacteria.
(A) P. polymyxa WLY78, (B) Frankia sp. EAN1pec, (C) Cyanothece sp. ATCC 51142, and (D) Anabaena sp. PCC 7120.Furthermore, we find that P. polymyxa HesA has 32%, 30%, 26% and 24% identity with MoeB of Pseudomonas fluorescens, K. oxytoca, E. coli and Bacillus subtilis, respectively, whereas it shows only 9% identities with NifQ of K. oxytoca and A. vinelandii and 8–18% with these NifQs from other N2-fixing bacteria. The data indicate that HesA exhibits higher similarity to MoeB than to NifQ. Also, there are no moeB-like genes other than hesA in Paenibacillus.
The hesA mutation of P. polymyxa WLY78 leads to nearly complete loss of nitrogenase activity
As described in Materials and Methods, the ΔhesA mutant was constructed by inserting a 1,197 bp Cm resistance gene cassette into the nif gene cluster (nifBHDKENXhesAnifV) and substituting for hesA. qRT-PCR further revealed that the mRNA level of nifV gene was not affected by the hesA mutation (Fig. S1), suggesting that the insertion of Cm-cassette into nif gene cluster did not generate polarity effect.
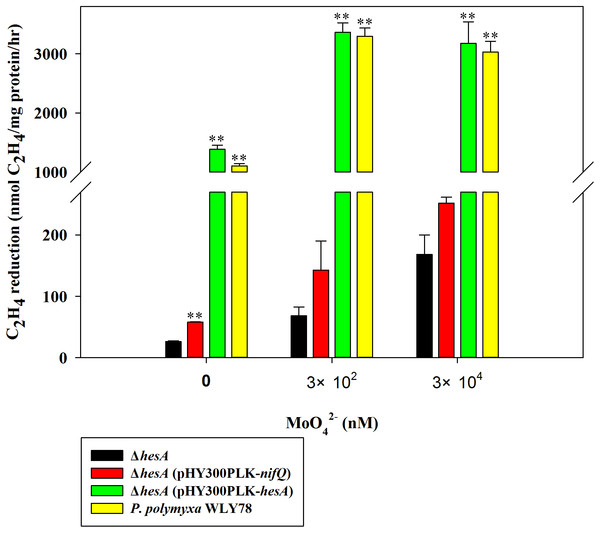
Here, the nitrogenase activities of the ΔhesA mutant and wild-type P. polymyxa WLY78 grown in medium containing 0.25 mM sulfate as sulfur source and supplemented with 0.3 and 30 μM molybdate, respectively, were comparely analyzed (Fig. 2). The ΔhesA mutant showed a significant decrease in nitrogenase activity compared to wild type, suggesting that hesA is involved in nitrogen fixation. We noticed that the hesA mutant and P. polymyxa WLY78 also exibited some activity in medium without molybdate, which might be due to trace molybdenum in the medium.
Figure 2: Relative nitrogenase activity of P. polymyxa WLY78, the ΔhesA mutant, the complemented strains ΔhesA (pHY300PLK-hesA) and ΔhesA (pHY300PLK-nifQ).
x-Axis indicates the concentration of molybdate added to the culture medium with 0.25 mM sulfate as sulfur source. The nitrogenase activity of the ΔhesA mutant was used as a control. Date represent the mean ± SD (n = 3). ** Indicates significant differences between control and other treatments determined by LSD at P < 0.01.Furthermore, we demonstrated that P. polymyxa hesA could completely restore the activity of hesA mutant to the level of wild-type Paenibacillus, confirming that hesA is required for nitrogen fixation. We also tried to complement with K. oxytoca nifQ and found that the nifQ could not suppress the effect of hesA mutation completely (Fig. 2). The data suggest that K. oxytoca nifQ could not act as a molybdenum donor for FeMo-co synthesis of nitrogenase in P. polymyxa.
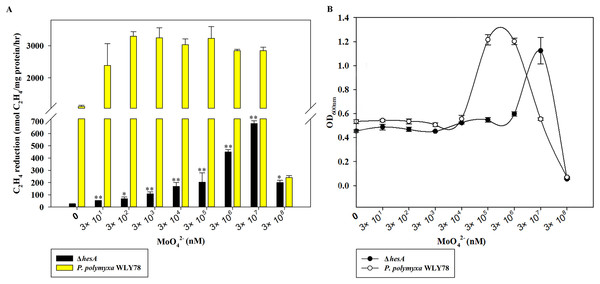
The hesA mutant requires high levels of molybdate for nitrogenase activity
The molybdate requirement for nitrogenase activity in the ΔhesA mutant was studied, with P. polymyxa WLY78 as a control. As shown in Fig. 3A, P. polymyxa WLY78 had nitrogenase activity even in the absence of added sodium molybdate, because there were always traces of Mo in other chemicals, the glassware and water. The maximum activity of wild-type P. polymyxa WLY78 was obtained at 0.3 μM molybdate, and the nitrogenase activity did not increase as the concentration of molybdate increased. In contrast, the nitrogenase activity of the ΔhesA mutant improved as the molybdate concentration increased, and the maximum activity was obtained at 30 mM molybdate. The maximum activity of the ΔhesA mutant at 30 mM molybdate was 25% of that of the wild type obtained at 0.3 μM molybdate, indicating that the addition of very high levels of molybdate could partially suppress the effect of the mutation.
Figure 3: Effect of the molybdate concentration on nitrogenase activity (A) and maximum growth (B) of P. polymyxa WLY78 and the ΔhesA mutant.
x-Axis indicates the concentration of molybdate added to the culture medium with 0.25 mM sulfate as sulfur source. The maximum growth means culture absorbance after 48 h under diazotrophic growth conditions. The nitrogenase activity of the ΔhesA mutant in the absence of added sodium molybdate was used as a control. Date represent the mean ± SD (n = 3). * or ** indicate significant differences between control and other treatments determined by LSD at P < 0.05 or P < 0.01.Diazotrophic growth of the wild-type P. polymyxa WLY78 and ΔhesA mutant showed the same dependence on the molybdate added (Fig. 3B). Compared with the wild type, the ΔhesA mutant needed higher concentration of molybdate for the maximum growth. Millimolar concentrations of molybdate obviously inhibited both activity and diazotrophic growth. Meanwhile, the ΔhesA mutant showed a higher tolerance.
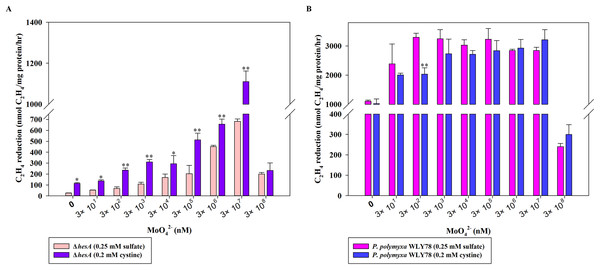
Effect of two sulfur sources on nitrogenase activity of the ΔhesA mutant
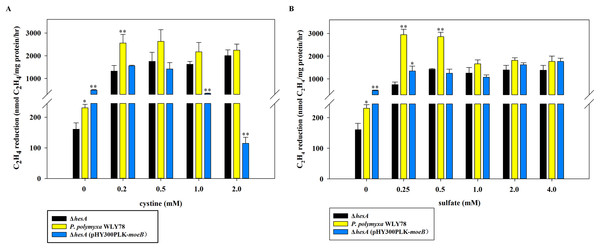
Here, the effects of two sulfur sources on nitrogenase activity of the ΔhesA mutant were determined. As shown in Fig. 4A, the nitrogenase activity of the ΔhesA mutant in medium containing cystine as the only sulfur source was two to five times higher than that in medium containing sulfate at all of different concentrations of molybdenum except for a high inhibitory level of 0.3M molybdenum. However, no difference in nitrogenase activity of wild-type Paenibacillus was observed when using either sulfate or cystine as sulfur source (Fig. 4B). These results demonstrated that the use of cystine as the only sulfur source could suppress the effect of hesA mutation on nitrogenase activity, but the addition of cystine could not affect the requirement of molybdenum for maximum nitrogenase activity. The results indicated that hesA might be involved in sulfur transport.
Figure 4: Effect of the sulfur sources on the nitrogenase activity of the ΔhesA mutant and P. polymyxa WLY78.
(A) Nitrogenase activity of ΔhesA with two sulfur sources. (B) Nitrogenase activity of P. polymyxa WLY78 with two sulfur sources. x-Axis indicates the concentration of molybdate added to the culture medium with 0.25 mM sulfate or 0.2 mM cystine or the combination of sulfate and cystine as sulfur source. Date represent the mean ± SD (n = 3). * or ** indicate significant differences among different treatments determined by LSD at P < 0.05 or P < 0.01.High levels of cystine restore the nitrogenase activity of the ΔhesA mutant, but E. coli moeB cannot
As shown in Fig. 5A, the nitrogenase activity of the ΔhesA mutant was improved with the addition of cystine, and higher concentration of cystine was required for the ΔhesA mutant to reach the maximum activity of the wild type. Similarly, the activity of the ΔhesA mutant increased with the addition of sulfate, but a higher concentration (above 1.0 mM) of sulfate was required for the ΔhesA mutant to reach the maximum activity of the wild type (Fig. 5B).
Figure 5: Effect of the different concentrations of cystine (A) or sulfate (B) on nitrogenase activity of ΔhesA, P. polymyxa WLY78 and the complemented strain ΔhesA (pHY300PLK-moeB).
(A) Nitrogenase activity of the strains with different cystine concentrations. (B) Nitrogenase activity of the strains with different sulfate concentrations. x-Axis indicates the concentration of cystine or sulfate added to the culture medium with 30 mM molybdate. The nitrogenase activity of the ΔhesA mutant was used as a control. Date represent the mean ± SD (n = 3). * or ** indicate significant differences between control and other treatments determined by LSD at P < 0.05 or P < 0.01.In addition, we tried to complement the ΔhesA mutant with E. coli moeB and found that the expression of moeB in the mutant did not improve nitrogenase activity (Fig. 5). In fact, the nitrogenase activity in the ΔhesA mutant expressing moeB declined when the concentration of cystine exceeded 1.0 mM. The data suggested that E. coli moeB could not completely suppress the effect of the hesA mutation when sulfate or cystine was used as the only sulfur source. It has been reported that MoeB, MPT synthase sulfurase, is involved in sulfuration of MPT synthase (Leimkühler, Wuebbens & Rajagopalan, 2001; Pitterle & Rajagopalan, 1993; Rajagopalan, 1997).
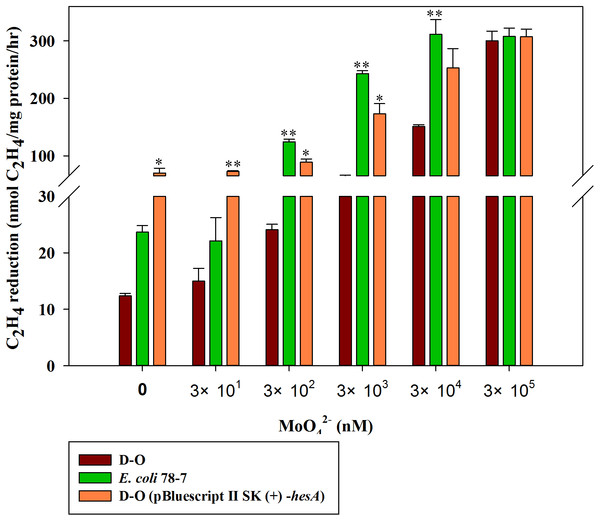
A high level of molybdate is required for nitrogenase activity of the recombinant E. coli 78-7 with a hesA deletion
In order to confirm the role of the hesA in nitrogen fixation, we compared the activities of the recombinant E. coli 78-7 strain carrying a nif gene cluster (nifBHDKENXhesAnifV) of P. polymyxa WLY78 (Wang et al., 2013) and the hesA deletion strain (D-O), which was a derivative of E. coli 78-7 strain without the hesA gene. As shown in Fig. 6, the nitrogenase activity of the D-O strain was improved as the concentration of molybdate increased. The D-O strain required 10 times as much molybdate to achieve the level of activity of the E. coli 78-7 strain. The P. polymyxa hesA gene could complement the D-O strain and restore the activity of the hesA deletion strain to that of the E. coli 78-7 strain, suggesting that the addition of very high levels of molybdate can suppress the effect of the hesA mutation. The current result was consistent with the results obtained in the P. polymyxa hesA mutant.
Figure 6: Effect of the molybdate concentration on nitrogenase activity of the recombinant E. coli 78-7, the hesA deletion strain (D-O) and the complemented strain D-O (pBluescript II SK (+)- hesA).
x-Axis indicates the concentration of molybdate added to the culture medium with 0.25 mM sulfate as sulfur source. The nitrogenase activity of the D-O strain was used as a control. Date represent the mean ± SD (n = 3). * or ** indicate significant differences between control and other treatments determined by LSD at P < 0.05 or P < 0.01.Cystine can enhance activity of the hesA deletion in E. coli 78-7
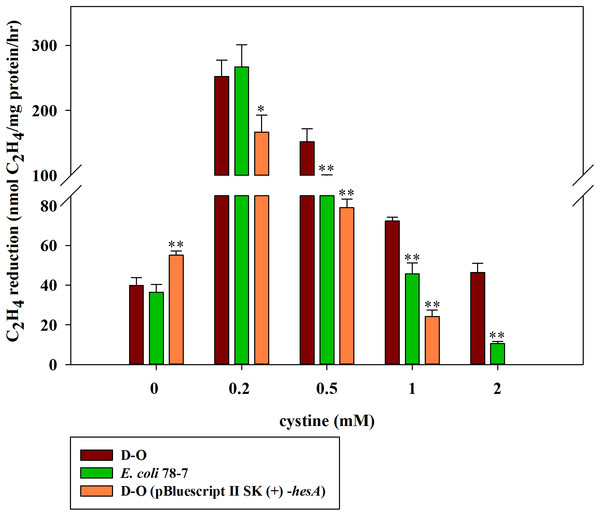
Here, we further investigated the effect of cystine on the activities of the hesA deletion strain (D-O) which carried other nif genes (nifBHDKENXV) without hesA when it was grown in medium supplemented with 30 μM molybdate and with different concentrations of cystine. As shown in Fig. 7, cystine could enhance activity of the hesA deletion strain (D-O), but it did not have the similar effects on the complemented strain D-O (pBluescript II SK (+)-hesA).
Figure 7: Effect of the cystine concentration on nitrogenase activity of the recombinant E. coli 78-7, the hesA deletion strain (D-O) and the complemented strain D-O (pBluescript II SK (+)- hesA).
x-Axis indicates the concentration of cystine added to the culture medium with 30 μM molybdate. The nitrogenase activity of the D-O strain was used as a control. Date represent the mean ± SD (n = 3). * or ** indicate significant differences between control and other treatments determined by LSD at P < 0.05 or P < 0.01.The P. polymyxa hesA mutation does not affect nitrate reductase activity
Nitrate reductase, like nitrogenase, has been characterized as a molybdoenzyme (Taniguchi & Itagaki, 1960) and molybdenum is necessary for nitrate reductase (Lester & Demoss, 1971). Here, the nitrate reductase activities of the ΔhesA mutant and wild-type P. polymyxa WLY78 were comparatively analyzed. The ΔhesA mutant (1,048.12 ± 28.80 nmol nitrite·min−1·mg−1protein) and wild-type P. polymyxa WLY78 (1,057.37 ± 5.95 nmol nitrite·min−1·mg−1protein) showed the similar nitrate reductase activity, indicating that the hesA gene product is required for synthesis of FeMo-co, but not implicated in the biosynthesis of Mo-co.
Discussion
A nif gene cluster composed of nine genes nifBHDKENXhesAnifV is conserved in the genomes of P. polymyxa WLY78 and other the diazotrophic Paenibacillus spp. We find that the nif gene cluster has a hesA, but no nifQ and nifSU. The genome of P. polymyxa WLY78 has molybdate transport genes, including modA, modC and modF. It also contains moaA, moaB, moaC, moaD, moaE, mobA, mobB and moeA except for moeB, and contains a complete suf (sufCBSUD) operon, a partial suf (sufABC) operon, a partial isc system (iscSR and fdx) and two nifS-like genes (Shi et al., 2016).
The function of hesA gene of diazotrophic Paenibacillus is here for the first time to be investigated. Mutation of P. polymyxa hesA leads to nearly complete loss of nitrogenase activity. Similar result was reported that loss of hesA resulted in about a 50% loss of nitrogenase in Anabaena (Borthakur et al., 1990). Furthermore, we demonstrated that the effect of the mutation could be suppressed by the addition of high levels of molybdate to the medium. We also showed that deletion of hesA from the nif gene cluster made the recombinant E. coli 78-7 require high concentration of molybdate for nitrogenase activity. Our complementation studies demonstrated that P. polymyxa hesA could restore the nitrogenase activity of hesA mutant to that of wild-type Paenibacillus or that of the recombinant E. coli 78-7, but K. oxytoca nifQ could not, indicating that K. oxytoca NifQ cannot replace HesA in the original host. The results indicate that P. polymyxa hesA gene plays a role in molybdenum transport to nitrogenase. Similar results that NifQ− and Mol− mutants of K. oxytoca required high concentration of molybdate for nitrogenase activity were reported (Imperial et al., 1984, 1985). There were no functional differences among the NifQ− and Mol− mutants of K. oxytoca with respect to FeMo protein when molybdate was in excess (Imperial et al., 1985). Our results and their results suggest that NifQ is not necessary when molybdate is in excess. It was reported that the FeMo-co of nitrogenase could be completely synthesized in vitro by Fe2+, S2−, MoO42− and R-homocitrate, using purified NifB, NifEN and NifH proteins (Curatti et al., 2007). Curatti et al.’s reports suggested that NifQ is not necessary when molybdate is in excess in vitro synthesis of FeMo-co. One possible explanation is that NifQ cannot act as Mo donor in Paenibacillus.
Sulfur is an essential element for FeMo-co of nitrogenase and Mo-co of other enzymes and it can be obtained from various compounds. Generally, sulfate (SO42−) and thiosulfate (S2O32−) are the preferred sulfur sources for the majority of N2-fixing organisms (Aguilar-Barajas et al., 2011). It was shown that the physiological sulfur donor for the formation of the dithiolene group of molybdopterin in Mo-co biosynthesis was likely to be L-cysteine and IscS functioned as a primary sulfur-donating enzyme by interacting specifically with MoeB and MoaD in the biosynthesis of molybdopterin in E. coli (Sambasivarao et al., 2002; Zhang et al., 2010). MoeB contains several conserved cysteine residues, one of which has been postulated to be involved in the formation of a thioester linkage between MoaD and MoeB (Rogers, Crescenzo & Söll, 1995).
In this study, we demonstrated that cystine did not modify the requirement of molybdate for nitrogenase activity in the wild-type P. polymyxa WLY78. Whereas, the nitrogenase activity of the ΔhesA mutant was higher when it was grown in medium containing cystine as the only sulfur source than sulfate as the only sulfur source. Moreover, cystine could enhance activity of hesA deletion of E. coli 78-7. However, P. polymyxa hesA gene could not complement the hesA deletion of E. coli 78-7 when cystine was high in the medium. We do not know whether E. coli moeB plays a role in nitrogen fixation of the hesA mutant from the recombinant E. coli 78-7. It was reported that the effects of nifQ and mol mutations of K. oxytoca on nitrogenase could be suppressed by the addition of cystine, suggesting that a sulfur donor and molybdenum interact at an early step in the biosynthesis of the iron-molybdenum cofactor (Dos Santos & Dean, 2008). Sulfate interferes with the utilization of molybdate, which becomes apparently only when the nifQ or mol products are not active in K. oxytoca (Dos Santos & Dean, 2008). A competition between sulfate and molybdate was found in Clostridium pasteurianum (Elliott & Mortenson, 1975) and Aspergillus niger (Tweedie & Segel, 1970) at the level of transport into the cell. The transport of sulfate and molybdate in K. oxytoca are independent and specific, and that the sulfur source effect on molybdate requirement is intracellular.
Furthermore, we comparatively analyzed the nitrate reductase activity of the ΔhesA mutant and wild-type P. polymyxa WLY78, and our results showed that the hesA mutation did not affect nitrate reductase activity. The results confirm that the P. polymyxa hesA is involved in FeMo-co synthesis, but not in Mo-co synthesis.
Supplemental Information
The relative transcript levels of nifV were determined by qRT-PCR.
The transcript level of nifV of the wild-type P. polymyxa WLY78 was used as a control. Data represent the mean ± SD of three independent experiments performed in triplicate.