Diversity and evolution of the endosymbionts of Bemisia tabaci in China
- Published
- Accepted
- Received
- Academic Editor
- Laura Boykin
- Subject Areas
- Agricultural Science, Entomology
- Keywords
- Arsenophonus, Portiera aleyrodidarum, Bemisia tabaci, Phylogeny, Cardinium, Prevalence
- Copyright
- © 2018 Tang et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2018. Diversity and evolution of the endosymbionts of Bemisia tabaci in China. PeerJ 6:e5516 https://doi.org/10.7717/peerj.5516
Abstract
The whitefly Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) is a cryptic species complex, including members that are pests of global importance. This study presents a screening of B. tabaci species in China for infection by the primary endosymbiont, Portiera aleyrodidarum, and two secondary endosymbionts, Arsenophonus and Cardinium. The results showed that P. aleyrodidarum was detected in all B. tabaci individuals, while Arsenophonus was abundant in indigenous species of B. tabaci Asia II 1, Asia II 3, and China 1 but absent in the invasive species, Middle East-Asia Minor 1 (MEAM1); Cardinium presented in the Mediterranean (MED), Asia II 1 and Asia II 3 species but was rarely detected in the MEAM1 and China 1 species. Moreover, phylogenetic analyses revealed that the P. aleyrodidarum and mitochondrial cytochrome oxidase 1 (mtCO1) phylograms were similar and corresponding with the five distinct cryptic species clades to some extent, probably indicating an ancient infection followed by vertical transmission and subsequent co-evolutionary diversification. In contrast, the phylogenetic trees of Arsenophonus and Cardinium were incongruent with the mtCO1 phylogram, potentially indicating horizontal transmission in B. tabaci cryptic species complex. Taken together, our study showed the distinct infection status of endosymbionts in invasive and indigenous whiteflies; we also most likely indicated the co-evolution of primary endosymbiont and its host as well as the potential horizontal transfer of secondary endosymbionts.
Introduction
Bemisia tabaci is a cryptic species complex comprising a minimum of 40 morphologically similar species (De Barro et al., 2011; Dinsdale et al., 2010; Hu et al., 2018; Wang, Li & Liu, 2017). Among members of the complex, the Middle East-Asia Minor 1 (MEAM1) and Mediterranean (MED) groups (commonly known as the B and Q biotypes, respectively) have drawn much attention due to their global invasion and vectoring important plant pathogens (e.g., tomato yellow leaf curl virus) (Brown, 1994; Cohen & Nitzany, 1966). In China, B. tabaci was not considered as a serious pest until the arrival of the MEAM1 group in the mid-1990s (Qiu et al., 2007). In 2004, the MED group was detected, and rapidly became widely distributed in China, causing considerable damage to a wide range of vegetables, fibers, and ornamental crops (Chu et al., 2005). It is interesting that MED has been replacing the earlier invader MEAM1 as well as several indigenous species of whiteflies (e.g., Asia II and China 1) in many regions (Liu et al., 2007; Sun et al., 2013).
Associations between insects and endosymbionts are quite common in nature. It has been estimated that at least 15−20% of all insect species live in symbiotic relationships with bacteria (Douglas, 1998; Gosalbes et al., 2010). Endosymbionts associated with insects can be classified into primary endosymbionts (P-endosymbionts) and secondary endosymbionts (S-endosymbionts) (Baumann, 2005). The P-endosymbionts are obligate and usually have mutualist relationships with their hosts. Besides, P-endosymbionts are generally localized in bacteriocytes of bacteriome (Caspi-Fluger et al., 2011) and transmitted vertically from mother to progeny (Werren & O’Neill, 1997). In contrast, the S-endosymbionts are usually facultative symbionts, and they could reside in several host tissues such as gut, hemolymph, Malpighian tubules, salivary glands or ovarian cells (Cicero, Fisher & Brown, 2016; Cooper, Sengoda & Munyaneza, 2014; Dobson et al., 1999; Zchori-Fein, Roush & Rosen, 1998). Infection of secondary endosymbionts can be either maternally inherited or horizontally transmitted (Moran & Baumann, 2000). It has been discovered that the P-endosymbionts Portiera aleyrodidarum and S-endosymbionts such as Arsenophonus, Cardinium, Fritschea, Hamiltonella, Rickettsia, and Wolbachia were infected in whiteflies (Bing et al., 2013; Chiel et al., 2007; Chu et al., 2011; Everett et al., 2005; Karut et al., 2017; Thao & Baumann, 2004; Zchori-Fein, Lahav & Freilich, 2014). Previous studies have investigated the prevalence, diversity and evolution of endosymbionts in the B. tabaci species complex from different countries or regions (e.g., Turkey, China, Brazil, and Africa) (Ahmed et al., 2010; Bing et al., 2013, 2014; Ghosh, Bouvaine & Maruthi, 2015; Hashmi et al., 2018; Jahan et al., 2015; Karut et al., 2017; Marubayashi et al., 2014; Santos-Garcia et al., 2015; Sseruwagi et al., 2018; Thierry et al., 2011, 2015). However, most of these reports only focused on the two invasive cryptic species MEAM1 and MED; furthermore, for studies from China, the sample size and distribution range were limited. For example, regarding prevalence of S-endosymbionts in B. tabaci, only the laboratory samples from two provinces were investigated (Bing et al., 2013); only one S-endosymbiont (Wolbachia) was explored from a slightly larger geographical scale (Bing et al., 2014).
Therefore, although substantial datasets regarding endosymbionts prevalence of B. tabaci species complex are present, their current situation in whole China is still not very clear and further investigation is essential. In the present study, our first goal is to investigate the prevalence and diversity of the P-endosymbionts P. aleyrodidarum and two common S-endosymbionts Arsenophonus and Cardinium (as representatives of S-endosymbionts) within B. tabaci in a wider range of China. We also aimed to explore the evolutionary relationships between these three endosymbionts and their host based on phylogenetic analyses of 16S and 23S ribosomal DNA (rDNA) (from endosymbionts) as well as mitochondrial cytochrome oxidase 1 (mtCO1) gene (from B. tabaci). Our study will not only uncover the current status of endosymbionts infection within B. tabaci in China, but also greatly provide a supplement for studies of B. tabaci endosymbionts worldwide.
Materials and Methods
Sample collection
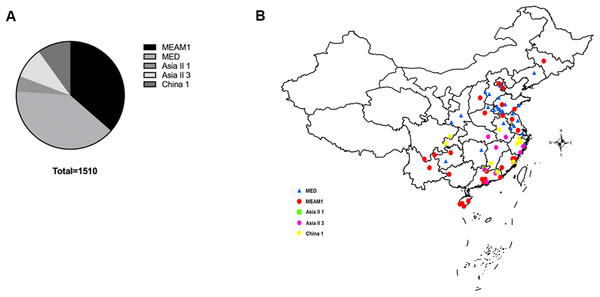
A total of 1,510 B. tabaci individuals were collected from 71 geographical locations, including 19 provinces and four municipalities in China (Fig. 1). At each location, B. tabaci samples were collected from different leaves of separate plants. The collection details, geographical sites and host plants were described in Table S1.
Figure 1: The quantity and distribution of B. tabaci cryptic species in China.
(A) The quantity of each B. tabaci cryptic species based on molecular identification. (B) The locations of the B. tabaci cryptic species populations in China. Names of locations are given in Table S1. Maps were created using Esri’s ArcGIS platform (http://www.esri.com/software/arcgis).DNA extraction and gene amplification
Total DNA was extracted from individual whitefly as described in Luo et al. (2002). The primers of mtCO1 gene was used for whitefly species identification. 16S rDNA primers were used to detect P. aleyrodidarum and Cardinium, and for Arsenophonus, 23S rDNA primers were utilized. The primers, annealing temperature, and predicted PCR products size were shown in Table 1. The Polymerase chain reaction (PCR) reaction mixture contained one U Taq DNA polymerase, five μl (10×) reaction buffer, three μl MgCl2 (final concentration of 25 mmol/l), two μl dNTPs (10 mmol/l), two μl of forward and reverse primers (20 μmol/l each) and two μl of template DNA (Simon et al., 1994). PCR products were visualized by 1.5% agarose gels and sequenced by IGE Biotechnology Co., Ltd (Guangzhou, China). The sequences were deposited in GenBank under accession numbers KP137471–KP137491 for B. tabaci mtCO1 gene, KP201110–KP201126 for P. aleyrodidarum 16S rDNA, KP201103–KP201109 for Arsenophonus 23S rDNA, and KP201127–KP201134 for Cardinium 16S rDNA (Table 2).
| Gene | Primer sequence (5′→3′) | Annealing temperature | Product size (bp) | Reference |
|---|---|---|---|---|
| B. tabaci mtCOI | C1-J-2195: TTGATTTTTTGGTCATCCAGAAGT | 50 °C | 800 | Frohlich et al. (1999) |
| L2-N-3014: TCCAATGCACTAATCTGCCATATTA | ||||
| P. aleyrodidarum 16S rDNA | Pro-F: TGCAAGTCGAGCGGCATCAT | 59 °C | 1,000 | Zchori-Fein & Brown (2002) |
| Pro-R: AAAGTTCCCGCCTTATGCGT | ||||
| Cardinium 16S rDNA | Ch-F: TACTGTAAGAAATAAGCACCGGC | 57 °C | 400 | Zchori-Fein & Perlman (2004) |
| Ch-R: GTGGATCACTTAACGCTTTCG | ||||
| Arsenophonus 23S rDNA | Ars-F: CGTTTGATGAATTCATAGTCAAA | 60.5 °C | 900 | Thao & Baumann (2004) |
| Ars-R: GGTCCTCCAGTTAGTGTTACCCAAC |
| Species | mt COI | P. aleyrodidarum | Arsenophonus | Cardinium | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n. | Re. seq. | Acc. no. | Per. (%) | n. | Re. seq. | Acc. no. | Per. (%) | n. | Re. seq. | Acc. no. | Per. (%) | n. | Re. seq. | Acc. no. | Per. (%) | |
| MEAM1/B | 4 | BHZ-1 | KP137471 | 85.1 | 2 | BST-CA1 | KP201110 | 89.1 | 3 | BGD-CR1 | KP201127 | 92.5 | ||||
| BST-2 | KP137472 | 8.2 | BGW-CA2 | KP201111 | 10.9 | BHC-CR2 | KP201128 | 2.5 | ||||||||
| BSH-3 | KP137473 | 4.9 | BSRF-CR3 | KP201129 | 5 | |||||||||||
| BJC-4 | KP137474 | 1.8 | ||||||||||||||
| MED/Q | 4 | QSQ-1 | KP137475 | 90.3 | 6 | QBJ-CA1 | KP201112 | 88.8 | 2 | QHS-AR1 | KP201103 | 89.7 | 2 | QSH-CR1 | KP201130 | 96.2 |
| QST-2 | KP137476 | 0.5 | QAH-CA2 | KP201113 | 5.8 | QHL-AR2 | KP201104 | 10.3 | QSZ-CR2 | KP201131 | 0.4 | |||||
| QHW-3 | KP137477 | 5.8 | QSL-CA3 | KP201114 | 1.0 | |||||||||||
| QSZ-4 | KP137478 | 3.3 | QSJ-CA4 | KP201115 | 2.0 | |||||||||||
| QSH-CA5 | KP201116 | 1.8 | ||||||||||||||
| QAH-CA6 | KP201117 | 0.5 | ||||||||||||||
| Asia II 3 | 5 | A3AJ-1 | KP137479 | 86.9 | 3 | A3ZW-CA1 | KP201118 | 86.2 | 1 | A3ZL-AR1 | KP201105 | 100.0 | 1 | A3ZL-CR1 | KP201132 | 100.0 |
| A3HW-2 | KP137480 | 1.5 | A3ZL-CA2 | KP201119 | 9.2 | |||||||||||
| A3JN-3 | KP137481 | 0.8 | A3GQ-CA3 | KP201120 | 4.6 | |||||||||||
| A3GQ-4 | KP137482 | 6.2 | ||||||||||||||
| A3ZL-5 | KP137483 | 4.6 | ||||||||||||||
| Asia II 1 | 3 | A1ZQ-1 | KP137484 | 81.4 | 2 | A1GQ-CA1 | KP201121 | 94.3 | 2 | A1ZL-AR1 | KP201106 | 92.3 | 2 | A1GQ-CR1 | KP201133 | 92.0 |
| A1GH-2 | KP137485 | 12.9 | A1JS-CA2 | KP201122 | 5.7 | A1GS-AR2 | KP201107 | 7.7 | A1SL-CR2 | KP201134 | 8.0 | |||||
| A1GH-3 | KP137486 | 5.7 | ||||||||||||||
| China 1 | 5 | CAJ-1 | KP137487 | 94.7 | 4 | CCY-CA1 | KP201123 | 2.7 | 2 | CCY-AR1 | KP201108 | 97.6 | ||||
| CCY-2 | KP137488 | 2.0 | CZS-CA2 | KP201124 | 90.7 | CAJ-AR2 | KP201109 | 2.4 | ||||||||
| CFW-3 | KP137489 | 0.7 | CZH-CA3 | KP201125 | 5.3 | |||||||||||
| CJG-4 | KP137490 | 1.3 | CZJ-CA4 | KP201126 | 1.3 | |||||||||||
| CGM-5 | KP137491 | 1.3 | ||||||||||||||
| Total | 21 | 17 | 7 | 8 | ||||||||||||
Note:
n., haplotype number; Re. Seq., representative sequence; Acc. no., accession number; Per., percentage in each cryptic species.
Sequence alignment and phylogenetic analysis
Sequence fragments were assembled using ContigExpress and aligned using the Clustal X 1.83 program (Chenna et al., 2003). The GenBank database was searched for homologous sequences of mtCO1, 16S rDNA and 23S rDNA using the basic local alignment search tool. Phylogenetic trees were constructed using MrBayes 3.2.1 (Ronquist & Huelsenbeck, 2003). The best-fit substitution model for each of the aligned sequences was selected with the program Modeltest 3.7 (Posada & Crandall, 1998). All the trees were constructed using the GTR+I+G model. The metropolis-coupled Markov chain Monte Carlo algorithm was conducted using four chains. Analyses were initiated with random starting trees, processed for 3 × 106 generations, and sampled every 1,000 generations. For the burn-in period, we discarded 100,000 generations. Posterior clade probabilities obtained from the analysis were used to assess nodal support. Tree information was visualized and edited using FigTree ver. 1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/).
Results
Molecular identification of B. tabaci individuals
Analyses of mtCO1 sequences indicated that our 1,510 B. tabaci samples comprised of two invasive species (MEAM1 and MED) and three indigenous species (Asia II 1, Asia II 3, and China 1). Among the individuals tested, 36.4% (550/1,510) and 39.7% (600/1,510) were identified as MEAM1 and MED, respectively. The remained 4.6%, 9.3%, and 9.9% insects were identified as Asia II 1 (70/1,510), Asia II 3 (140/1,510), and China 1 (150/1,510), respectively (Fig. 1A; Table S1). Moreover, both the MEAM1 and MED whiteflies were widely distributed across China, whereas the three indigenous species (Asia II 1, Asia II 3, and China 1) were relatively less detected and mainly distributed in southeastern part of China (Fig. 1B).
Prevalence of endosymbionts among five species of B. tabaci
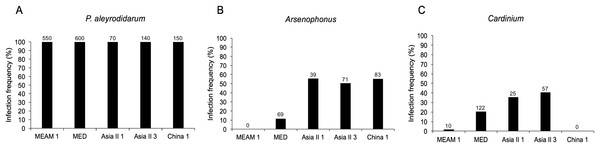
As expected, P. aleyrodidarum was detected in all whitefly individuals and species (Fig. 2A); 21.7% (327/1,510) individuals harbored S-endosymbiont (Arsenophonus or Cardinium); 5.6% (84/1,510) whiteflies were co-infected with both, and the remained majority (72.8%; 1,099/1,510) lacked an infection with either of the two S-endosymbionts. Infection frequencies with S-endosymbionts varied across B. tabaci species. In detail, Arsenophonus was abundant in the indigenous species (Asia II 1, Asia II 3, and China 1), with infection rates ranging from 50.7% to 55.7%; however, it was infrequent in MED (11.5%) and MEAM1 (0.0%) (Fig. 2B). Cardinium was moderately common in MED, Asia II 1 and Asia II 3 populations with frequencies of 20.3–40.7%, but rarely detected in MEAM1 (0.5%) and not found in China 1 (Fig. 2C).
Figure 2: Infection frequency of endosymbionts in five B. tabaci cryptic species.
(A) P. aleyrodidarum; (B) Arsenophonus; (C) Cardinium. Number above bars indicate the number of infection.Genetic diversity of B. tabaci and its endosymbionts
Aligned sequences from B. tabaci (813 bp, mtCO1 gene), P. aleyrodidarum (886 bp, 16s rDNA), Arsenophonus (551 bp, 23S rDNA), and Cardinium (460 bp, 16S rDNA) were used to analyze the genetic variation of whitefly and its endosymbionts. The results showed that 21 haplotypes were identified in whiteflies based on mtCO1 sequences, while 17, seven and eight symbiont haplotypes were defined based on analysis of P. aleyrodidarum, Arsenophonus and Cardinium sequences, respectively (Table 2). These haplotypes sequences were used to construct the phylogenetic trees.
Phylogenetic analysis of B. tabaci and endosymbionts
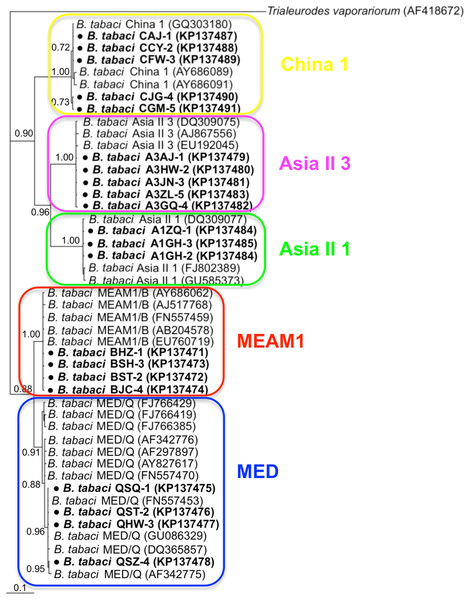
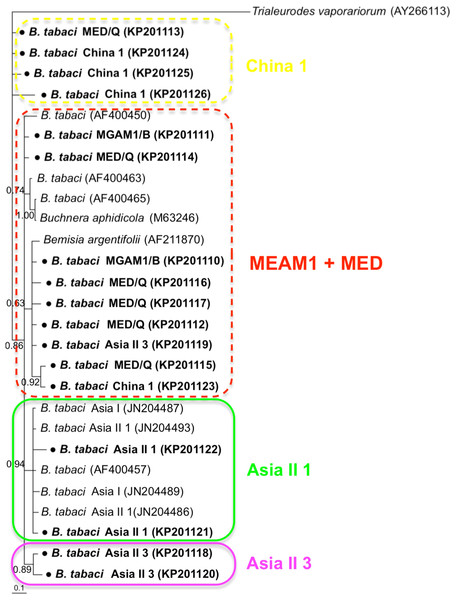
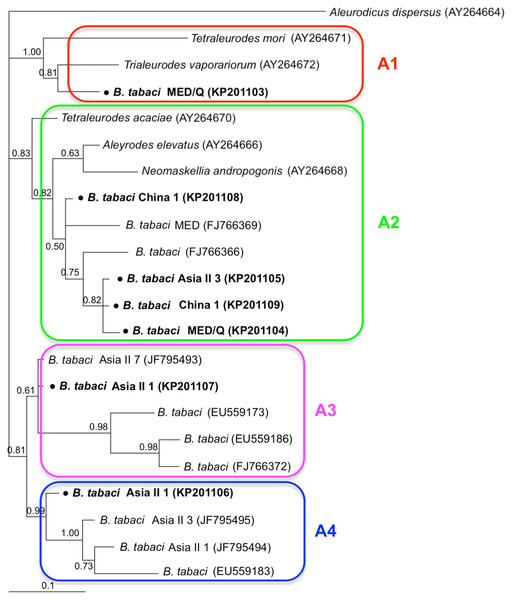
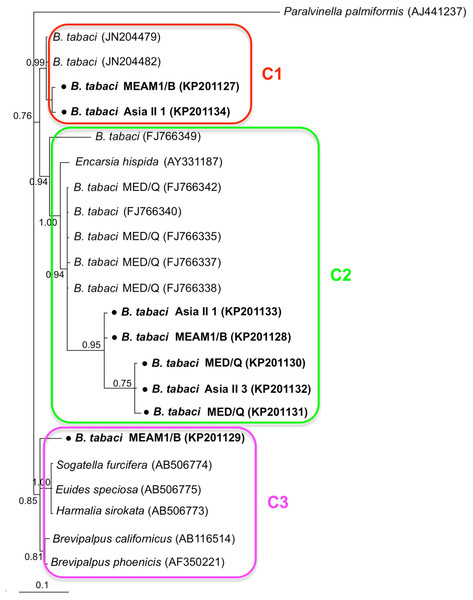
Phylogenetic trees were constructed for B. tabaci, P. aleyrodidarum, Arsenophonus, and Cardinium based on mtCO1 gene, 16s rDNA, 23S rDNA, and 16s rDNA sequences of haplotypes, respectively. We also pulled the related sequences from NCBI to explore the phylogenetic status of our haplotypes. The GenBank number of those related sequences could be found in phylogenetic trees. Based on the mtCO1 gene, we found five distinct genetic groups of B. tabaci, which was corresponding with the five cryptic species including the MEAM1, MED, Asia II 1, Asia II 3, and China 1 (Fig. 3). We can also find that all of our MED individuals belonged to Q1 subclade (Chu et al., 2011). For phylogenetic trees of endosymbionts, several distinct bacterial strains existed within individual bacterium. It is interesting that the phylogenetic tree of P. aleyrodidarum were similar to the mtCO1 tree and exhibited four groups corresponding to MEAM1+MED, Asia II 1, Asia II 3, and China 1 clades to some extent (Fig. 4). While there are still several differences. For example, the sequences of P. aleyrodidarum of China 1 were not well clustered and P. aleyrodidarum from B. tabaci species MED (KP201113) was also present in that China 1 clade. In addition, the MEAM1+MED clade (Fig. 4) contained sequences of P. aleyrodidarum from both MEAM1 and MED; however, it was separated into two distinct clades in the B. tabaci tree (Fig. 3). In contrast, the phylogenetic trees for Arsenophonus and Cardinium were totally incongruent with the mtCO1 tree, exhibiting four (A1–A4) and three groups (C1–C3), respectively (Figs. 5 and 6).
Figure 3: The Bayesian phylogenetic tree of B. tabaci cryptic species based on mtCOI sequences.
The value beside the nodes are posterior probabilities. Trialeurodes vaporariorum (AF418672) is used as outgroup. Accession numbers for mtCOI sequences submitted to GenBank are KP137471–KP137491. All mtCOI sequences of B. tabaci cryptic species used in this study were clustered with other related references sequences from GenBank and their accession numbers are also indicated in the tree. Bold dots indicate the sequences from the present study.Figure 4: The Bayesian phylogenetic tree of P. aleyrodidarum based on 16S rDNA sequences.
The value beside the nodes are posterior probabilities. T. vaporariorum (AY266113) is used as outgroup. Accession numbers for 16S rDNA sequences submitted to GenBank are KP201110–KP201125. All 16S rDNA sequences of P. aleyrodidarum used in this study were clustered with other related references sequences from GenBank and their accession numbers are also indicated in the tree. Bold dots indicate the sequences from the present study. Dotted boxes indicate imperfect cluster of each B. tabaci cryptic species.Figure 5: The Bayesian phylogenetic tree of Arsenophonus based on 23S rDNA sequences.
The value beside the nodes are posterior probabilities. Aleurodicus dispersus (AY264664) is used as outgroup. Accession numbers for 23S rDNA sequences submitted to GenBank are KP201103–KP201109. All 23S rDNA sequences of Arsenophonus used in this study were clustered with other related references sequences from GenBank and their accession numbers are also indicated in the tree. A1–A4 indicate the four clusters.Figure 6: The Bayesian phylogenetic tree of Cardinium based on 16S rDNA sequences.
The value beside the nodes are posterior probabilities. Paralvinella palmiformis (AJ441237) is used as outgroup. Accession numbers for 16S rDNA sequences submitted to GenBank are KP201127–KP201134. All 16S rDNA sequences of Cardinium used in this study were clustered with other related references sequences from GenBank and their accession numbers are also indicated in the tree. C1–C3 indicate the three clusters.Discussion
In this study, we conducted an extensive screening of B. tabaci for the presence of one P-endosymbiont and two common S-endosymbionts, along with phylogenetic analyses of these symbionts to compare with host species from the cryptic B. tabaci complex. The reason we chose Arsenophonus and Cardinium as representatives of S-endosymbionts because they are the very common S-endosymbionts in whiteflies, and Wolbachia has been thoroughly investigated in the study of Bing et al. (2014). The results showed that P-endosymbiont P. aleyrodidarum was detected in all whitefly individuals while S-endosymbionts infection were varied among species. The variation in the prevalence of endosymbionts could be influenced by numerous factors such as host, environmental conditions, geographical location or even climate (Chu et al., 2011; Karut & Tok, 2014; Morag et al., 2012; Skaljac et al., 2010). In our study, Arsenophonus was abundant in Asia II 1, Asia II 3, and China 1 species but absent in the invasive species MEAM1, which is exactly consistent with previous studies (Bing et al., 2013; Karut et al., 2017); Cardinium was present in the MED, Asia II 1 and Asia II 3 species (20.3–40.7%) but was rarely detected in MEAM1 and not detected in China 1. Taken together, it seemed that these two S-endosymbionts had high prevalence in native species rather than invasive species, which is consistent with another S-endosymbiont Wolbachia but in contrast to Hamiltonella; Hamiltonella was found abundant in invasive species rather than native species (Bing et al., 2013).
Previous studies showed that B. tabaci could be co-infected with particular pairs of S-endosymbionts, including Rickettsia and Hamiltonella, Hamiltonella and Cardinium, or Rickettsia and Arsenophonous; others were less common, such as Cardinium and Rickettsia, Hamiltonella and Arsenophonous, Cardinium and Wolbachia, and even three or four endosymbionts together (Gueguen et al., 2010; Karut & Tok, 2014; Marubayashi et al., 2014; Pan et al., 2012; Skaljac et al., 2010). In our study, we found evidence for a low rate (5.6%) of co-infection with Arsenophonous and Cardinium in four B. tabaci species (MEAM1 was the exception since Arsenophonous was not detected in this species), which has also been reported before (Bing et al., 2013; Chu et al., 2011; Parrella et al., 2014; Zchori-Fein, Lahav & Freilich, 2014). However, the reason of so few rate of co-infection by Arsenophonous and Cardinium is that Arsenophonous and Cardinium are potential reproductive manipulators that compete for resources inside the bacteriocytes, thus compromising the fitness of host (Gottlieb et al., 2008). We have one plausible explanation for the co-infection status, that is Cardinium is not restricted to the bacteriocytes (Skaljac et al., 2010), and perhaps the non-overlapping niche makes co-infection of Arsenophonous and Cardinium possible. In addition, the co-infection symbiont system in whiteflies may indicate the roles of dual endosymbionts: work as important mutually dependents to provide full complement of nutrients to their host (Rao et al., 2015) or affect the fitness and biology of the B. tabaci (Ghosh et al., 2018).
Our phylogenetic analyses indicated that B. tabacia mtCO1 sequences could be assigned to five distinct clades, which conformed to existing MEAM1, MED, Asia II 1, Asia II 3, and China 1 clades. Similarly, the sequences of the P-endosymbionts P. aleyrodidarum were assigned to their own clade and the phylogeny was similar with that of B. tabaci genetic groups to some extent. This may potentially indicate an ancient infection followed by vertical transmission and subsequent co-evolutionary diversification (Baumann, 2005). Meanwhile, it is important to note that the correlation was not perfect: sequences of P. aleyrodidarum from MEAM1 and MED were assigned to the same clade instead of the two distinct clades presented in the mtCO1 tree. The reason could be the dissemination of the MEAM1 and MED species; furthermore, the spread of these two invasive species in China has been frequently associated with founder effects that fix specific mtDNA variation(s) along with particular endosymbionts (Chu et al., 2011; Gueguen et al., 2010). Taken together, although there was similarity between the two trees of P. aleyrodidarum and B. tabaci, the genetic variation of primary symbiont might not be an ideal reflecting the genetic variation of the cryptic B. tabaci.
The S-endosymbionts, Arsenophonus and Cardinium, both showed a lack of congruence with the B. tabaci mtCO1 phylogram. This is consistent with the finding from Ahmed et al. (2013), who provided evidence for horizontal transmission of S-endosymbionts in the B. tabaci cryptic species complex based on phylogenies studies. There are substantial phylogenetic evidences showing that S-endosymbionts such as Wolbachia and Arsenophonus, undergoing horizontal transfer among host arthropod species (Ahmed et al., 2013; Chrostek et al., 2017; Kolasa et al., 2017; Li et al., 2017; Russell et al., 2003; Vavre et al., 1999). In some cases, the mechanisms for horizontal transmission of S-endosymbionts are already known, including transferring through parasitoid wasps (Ahmed et al., 2015; Gehrer & Vorburger, 2012), plants (Caspi-Fluger et al., 2012; Li et al., 2017) or even sexual transmission (Moran & Dunbar, 2006). Therefore, the potential horizontal transfer of S-endosymbionts in our samples could be one or combination of the above ways.
In summary, this study reported the varied prevalence of three endosymbionts within five B. tabaci cryptic species. The P-endosymbiont P. aleyrodidarum was detected in all whitefly individuals; the S-endosymbionts Arsenophonus was abundant in native species while Cardinium was common in the invasive species. In addition, the phylogenetic relationships between endosymbionts and their hosts B. tabaci probably indicated the vertical transmission and co-evolution of P. aleyrodidarum and B. tabaci; meanwhile, horizontal transfer of Arsenophonus and Cardinium may happen in our collecting samples. Our study not only reported current infection status of endosymbionts within B. tabaci populations in China, but also demonstrated that S-endosymbionts genetic variation may not reflect host genetic variation and should not be used to infer taxonomic relationships within the host species complex. If funding allows, more endosymbionts should be investigated and future investigations could be the contribution of endosymbionts to invasiveness, population expansion, or even competitiveness of whitefly species.