LitR directly upregulates autoinducer synthesis and luminescence in Aliivibrio logei
- Published
- Accepted
- Received
- Academic Editor
- Vladimir Uversky
- Subject Areas
- Biophysics, Genetics, Microbiology, Molecular Biology
- Keywords
- Quorum sensing, litR, Aliivibrio, luxR, Transcriptional regulation, Luminescence
- Copyright
- © 2021 Bazhenov et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2021. LitR directly upregulates autoinducer synthesis and luminescence in Aliivibrio logei. PeerJ 9:e12030 https://doi.org/10.7717/peerj.12030
Abstract
LitR is a master-regulator of transcription in the ainS/R and luxS/PQ quorum sensing (QS) systems of bacteria from Vibrio and Aliivibrio genera. Here, we for the first time directly investigated the influence of LitR on gene expression in the luxI/R QS system of psychrophilic bacteria Aliivibrio logei. Investigated promoters were fused with Photorhabdus luminescens luxCDABE reporter genes cassette in a heterological system of Escherichia coli cells, litR A. logei was introduced into the cells under control of Plac promoter. LitR has been shown to upregulate genes of autoinducer synthase (luxI), luciferase and reductase (luxCDABE), and this effect doesn’t depend on presence of luxR gene. To a much lesser degree, LitR induces luxR1, but not the luxR2 — the main luxI/R regulator. Enhanced litR expression leads to an increase in a LuxI-autoinducer synthesis and a subsequent LuxR-mediated activation of the luxI/R QS system. Effect of LitR on luxI transcription depends on lux-box sequence in luxI promoter even in absence of luxR (lux-box is binding site of LuxR). The last finding indicates a direct interaction of LitR with the promoter in the lux-box region. Investigation of the effect of LitR A. logei on luxI/R QS systems of mesophilic Aliivibrio fischeri and psychrophilic Aliivibrio salmonicida showed direct luxR-independent upregulation of luxI and luxCDABE genes. To a lesser degree, it induces luxR A. fischeri and luxR1 A. salmonicida. Therefore, we assume that the main role of LitR in cross-interaction of these three QS systems is stimulating the expression of luxI.
Introduction
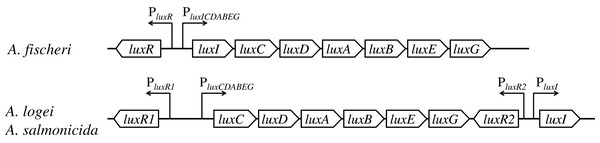
In lux regulons of some Aliivibrio bacteria, such as Aliivibrio fischeri, the regulatory gene luxR and structural genes luxICDABEG are divergently transcribed from a pair of promoters situated between them; the main regulatory operator is lux-box (the site of LuxR binding) (Devine, Countryman & Baldwin, 1988; Devine et al., 1989; Meighen, 1991; Stevens, Dolan & Greenberg, 1994). Structures of lux regulons of mesophilic and psychrophilic bacteria of the Aliivibrio genus significantly differ: lux regulons of psychrophilic ones, Aliivibrio logei and Aliivibrio salmonicida, are divided into two groups of genes: luxR1-luxCDABEG and luxR2-luxI (Fig. 1) (Fidopiastis, Sørum & Ruby, 1999; Khrulnova et al., 2010; Manukhov et al., 2011).
Figure 1: Structural organization of lux regulons of bacteria from Aliivibrio genus (Manukhov et al., 2011).
The luxA and luxB genes encode α- and β-subunits of the luciferase, the luxC, luxD, and luxE genes encode subunits of the reductase, the luxR, luxR1 and luxR2 genes encode regulatory proteins, activators of lux operon transcription, and luxI encodes LuxI synthase, which produces autoinducer (AI). Promoters with their directions of transcription are indicated by angular arrows with the letter P.luxR and luxI genes constitute a quorum sensing (QS) system. luxI-luxR system provides regulation of the expression of lux regulon genes (cell luminescence control) in dependence on cell culture density (Nealson & Hastings, 1979; Fuqua, Winans & Greenberg, 1994; Zavilgelsky & Manukhov, 2001). There are several proteins, which can modulate this QS system’s work. H-NS and CRP regulate the transcription of luxR genes (Dunlap & Greenberg, 1988; Ulitzur et al., 1997; Melkina, Goryanin & Zavilgelsky, 2017; Melkina et al., 2019). The content of the native protein LuxR is post translationally regulated by GroEL/ES chaperons and the Lon protease (Dolan & Greenberg, 1992b; Zavil’gel’skiĭ & Manukhov, 1994; Manukhov, Kotova & Zavil’gel’skiĭ, 2006; Manukhov et al., 2010), in psychrophilic bacteria it is so for the LuxR2 protein, but not the LuxR1(Khrulnova et al., 2016; Konopleva et al., 2016).
LitR in A. fischeri, being the master regulator of the ainS/R and luxS/PQ QS systems, can positively regulate the luxI/R system by activation of a luxR transcription (Fidopiastis et al., 2002; Miyashiro & Ruby, 2012). The deletion of the litR gene in A. salmonicida leads to an autoinducer (AI, 3-oxo-C6-homoserine lactone) concentration decrease detected at late growth phase (Hansen et al., 2015). It is known that Vibrio harveyi LuxR, which is a close homologue to LitR from A. fischeri, A. logei and A. salmonicida, does not require binding to signaling molecules (Fidopiastis et al., 2002; Rutherford et al., 2011). In dependence on its own concentration in the cell V. harveyi LuxR binds to the DNA in promoter regions and activates transcription by a direct interaction with RNA-polymerase (Rutherford et al., 2011; Van Kessel et al., 2013). The concentration of LitR is an object of LuxO-mediated regulation by the luxS/PQ and ainS/R QS systems and it increases when these QS pathways are activated (Miyashiro et al., 2010; Rutherford et al., 2011; Van Kessel et al., 2013).
Thus, the effect of LitR enhanced expression on the lux promoters after activation of ainS/R and luxS/PQ QS systems was investigated in mesophilic bacteria A. fischeri, but not the psychrophilic ones (A. logei, A. salmonicida), which have other lux regulon structure. Previously it was supposed that A. salmonicida LitR could activate the luxR1, luxR2 and luxI genes promoters (Khider et al., 2019), but it was not tested experimentally. The aim of this study was to determine whether there is a direct effect of litR on the expression of lux regulon of psychrophilic bacteria A. logei and A. salmonicida, and, if there is such an effect, to determine the mechanism of the effect of litR on proteins or promoters of lux regulon. The effect of A. logei LitR on the expression of regulatory luxR1 and luxR2 and structural luxCDABEG and luxI genes from A. logei and A. salmonicida was investigated in a heterologous model on Escherichia coli cells. Promoters of the A. fischeri lux genes were used as a control.
Materials and Methods
Bacterial strains and plasmids
All experiments were conducted with the E. coli MG1655 F-, rph−1 strain (Guyer et al., 1981), which were obtained from VKPM collection (Russia). Plasmids used in this study are listed in Table 1 and illustrated in Fig. S1. Primers and a detailed description of plasmids constructed in this study are presented in a Supplemental File (see Plasmid constructions). Genes and promoters of Aliivibrio bacteria were from A. fischeri MGU-6, A. logei KCh1 (Khrulnova et al., 2010) and A. salmonicida NCIMB 2262T (provided by Jesus L. Romalde (Egidius, Wiik & Andersen, 1986)).
| Plasmid | Relevant characteristics | Source or References |
|---|---|---|
| p15Tc-lac | Vector for cloning of regulatory gene under control of Plac; p15 origin, lacI, Tcr | Bazhenov et al. (2021) |
| p15Tc-litR | litR gene of A. logei under control of Plac; p15 origin, lacI, Tcr | This study |
| pDEW201 | Promoter probe vector with promoterless operon Photorhabdus luminescens luxCDABE genes and the replication origin of pBR322; Apr | Van Dyk & Rosson (1998) |
| pDEW201-derivative biosensor plasmids with luxCDABE-reporter for promoters investigation: | ||
| pALR1 | lux-reporter for A. logei PluxR1 + luxR1 | Melkina et al. (2019) |
| pDewP2rev | lux-reporter for A. logei PluxR2 | This study |
| pIVA | lux-reporter for A. logei PluxCDABEG + luxR1 | Khrulnova et al. (2016) |
| pSV16 | lux-reporter for A. logei PluxI + luxR2 | Khrul’nova, Manukhov & Zavil’gel’skiĭ (2011) |
| pAS1 | lux-reporter for A. salmonicida PluxCDABEG + luxR1 | This study |
| pAS2 | lux-reporter for A. salmonicida PluxI + luxR2 | This study |
| pR2 | lux-reporter for A. logei PluxI without luxR2 | Bazhenov et al. (2021) |
| pAFR | lux-reporter for A. fischeri PluxR + luxR | Melkina et al. (2019) |
| pVFR1 | lux-reporter for A. fischeri PluxICDABEG + luxR | Manukhov, Kotova & Zavil’gel’skiĭ (2006) |
| pD-lb1 | lux-reporter for A. logei PluxCDABEG | This study |
| pD-lb2 | lux-reporter for A. logei PluxCDABEG with changed lux-box (matches lux-box of luxI promoter) | This study |
Note:
Apr, ampicillin resistant; Tcr, tetracycline resistant.
Culture media and growth conditions
Bacteria were grown in Luria-Bertani (LB) broth. The liquid LB medium was composed of 1% tryptone, 0.5% yeast extract and 0.5% NaCl, for the preparation of solid medium, agar was added to a final concentration of 1.5% m/v. The medium was supplemented with appropriate antibiotics: 100 μg/ml ampicillin, 10 μg/ml tetracycline, or their composition. Overnight cultures were grown at 37 °C with continuous agitation and then were used to inoculate liquid LB. The resulting cultures were grown at 22–25 °C with continuous agitation (200 rpm). The optical density (OD) of cell suspensions was measured with a KFK-3 photometer (Zagorsk Optical-Mechanical Plant, Russia). An induction of the litR gene expression (plasmid p15Tc-litR) was made at the early exponential growth phase (OD about 0.1) by addition of IPTG (isopropyl β-D-1-thiogalactopyranoside) to a final concentration of one mM.
DNA isolation, restriction, ligation and transformation
Plasmid DNA was isolated by alkaline lysis. Cell transformation with hybrid plasmids followed standard protocols. Endonuclease restriction, DNA fragment ligation, agarose gel electrophoresis and isolation of DNA fragments from agarose gel were performed according to Green & Sambrook (2012). Restriction and ligation reactions were carried out using enzymes from Promega (USA): EcoRI, BamHI and KpnI restriction enzymes and T4 DNA ligase were used for constructing new plasmids (see Supplemental File for details).
Chemical substances
Isopropyl β-D-1-thiogalactopyranoside (IPTG) was from Anatrace (USA).
Measurement of bioluminescence
Bioluminescence intensities were measured in volume of 200 μl in plastic microtubes placed in front of a photomultiplier photocathode at room temperature using Biotox-7MB (BioPhisTech, Russia) or in 96-well plates using SynergyHT (Biotek Instruments, Winooski, VT, USA). Luminescence values were expressed in relative light units (RLU).
Determination of LitR-dependent promoter regulation
The effect of LitR on the expression of genes from lux regulons of the Aliivibrio genus bacteria was investigated for three species: A. fischeri, A. logei and A. salmonicida. For this purpose we used the heterological system of E. coli cells transformed with different biosensor plasmids from a set (Table 1, Fig. S1) that carry luxCDABE genes of P. luminescens transcriptionally fused with promoters of interest. To introduce the litR gene from A. logei into this system, the p15Tc-litR plasmid was constructed. It comprises a p15 origin, the litR A. logei gene under a lac promoter, and a lacI gene, which lowers the base transcription from Plac and allows regulating litR with IPTG.
Luminescence induction factor calculation
The culture of E. coli cells carrying the A. logei litR gene under the IPTG-inducible promoter on the p15Tc-litR plasmid and the P. luminescens luxCDABE genes under control of the promoter of interest on a biosensor plasmid was grown to OD~0.1 in liquid LB medium, then divided into two equal parts: “control” and “induced”. The “induced” portion was supplemented with one mM IPTG. Further, the cultures were grown under equal conditions with periodic measurements of optical density and luminescence. The induction factor is the induced/control ratio of cell cultures’ luminescence. All figures show representative kinetic curves of biological independent triplicates.
Statistics
Error bars at the graphs are the SD of three independent experiment replications.
Computer analysis
Amino acid sequences alignment was performed using the Vector NTI software (Thermo Fisher Scientific, Waltham, MA, USA). The intrinsic disorder of proteins was predicted by their aa sequences with “Predictor of Naturally Disordered Regions” (pondr.com), one can find description of VL-XT, VSL2, VL3 and Charge-Hydropathy algorithms in Obradovic et al. (2003).
Results
Effect of LitR A. logei on promoters of regulatory genes luxR, luxR1 and luxR2
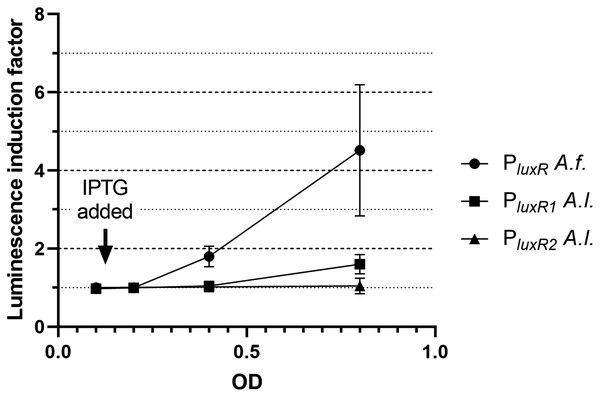
Firstly, the effect of LitR on the expression of luxR A. fischeri, luxR1 A. logei and luxR2 A. logei was investigated. Figure 2 shows the coefficient of increase in luminescence of biosensors after the activation of the litR gene expression.
Figure 2: The induction of promoters PluxR A. fischeri, PluxR1 A. logei and PluxR2 A. logei by enhancement of the litR A. logei expression in E. coli cells.
The curves represent the increase in luminescence of E. coli MG1655 cells carrying the luxCDABE genes of P. luminescens under control of promoter of luxR A. fischeri (pAFR plasmid, PluxR A.f. curve), promoter of luxR1 A. logei (pALR1 plasmid, PluxR1 A.l. curve), or promoter of luxR2 A. logei (pDewP2rev plasmid, PluxR2 A.l. curve) in combination with the litR A. logei gene under control of Plac (p15Tc-litR) in response to the addition of one mM IPTG. IPTG was added at OD~0.1.The data presented in Fig. 2 shows that LitR from A. logei is capable of a significant activation of the luxR A. fischeri gene transcription. This result is consistent with previous work (Fidopiastis et al., 2002) and means that the LitR homologues from A. fischeri and A. logei are interchangeable and have a common function. The amino acid sequence of LitR is highly conservative among the Aliivibrio species. In particular, LitR proteins from psychrophilic bacteria have almost the same sequences, comparing of them with mesophilic LitR A. fischeri gives that DNA-binding regions three compared proteins are identical and whole “helix-turn-helix” domain differs only by several aa substitutions (Fig. S2). Interesting that psychrophilic LitR variants are more ordered than mesophilic one according to analysis with VL-XT, VSL2, VL3 and Charge-Hydropathy instruments at pondr.com (Fig. S3). The increase in LitR content in the cell activates the expression of luxR1 A. logei, although to a lesser extent than that of A. fischeri luxR, and does not affect the expression of the luxR2 A. logei gene. The obtained result was in conflict with the data of Norwegian colleagues on a decrease in the production of an autoinducer in A. salmonicida litR mutant cells (Hansen et al., 2015); therefore, it was decided to test the effect of litR on the transcription of the “rightward” promoters of lux regulon.
Effect of LitR A. logei on promoters of structural genes luxICDABEG
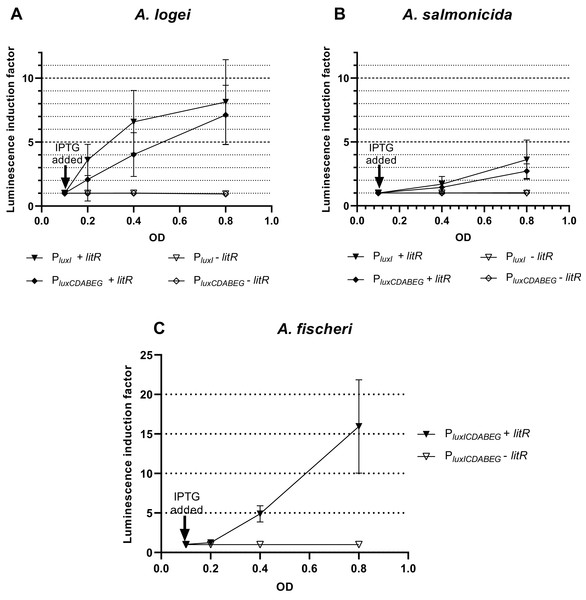
Results of investigation of LitR A. logei effect on expression of autoinducer synthase (luxI), luciferase and reductase (luxCDABE) genes are given in Fig. 3. Promoters of different Aliivibrio species were tested with LitR A. logei in the heterological system of E. coli cells.
Figure 3: The induction of promoters PluxI and PluxCDABEG of A. logei (A), A. salmonicida (B) and A. fischeri (C) by enhancement of the litR A. logei expression in E. coli cells.
Curves represent the increase in luminescence of E. coli MG1655 cells carrying the luxCDABE genes of P. luminescens under control of (A) PluxI A. logei (pSV16), PluxCDABEG A. logei (pIVA), (B) PluxI A. salmonicida (pAS2), PluxCDABEG A. salmonicida (pAS1), or (C) PluxICDABEG A. fischeri (pVFR1) promoters in combination with the litR A. logei gene under control of Plac (p15Tc-litR) in response to the addition of one mM IPTG at OD~0.1. The “−litR” curve corresponds to a negative control-analogous cell lines, which lacks the litR gene (p15Tc-lac vector used).In response to the addition of one mM IPTG at OD~0.1 there was an increase in luminescence of the E. coli MG1655 cells carrying combinations of the plasmid p15Tc-litR (litR under Plac) with different biosensor plasmids containing P. luminescens luxCDABE genes under control of PluxI and PluxCDABEG promoters of A. logei (Fig. 3A) and A. salmonicida (Fig. 3B). The following plasmids with the investigated promoters were used: pSV16 with PluxI A. logei lux-reporter, pIVA with A. logei PluxCDABEG lux-reporter, pAS2 with A. salmonicida PluxI lux-reporter, and pAS1 with A. salmonicida PluxCDABEG lux-reporter (Table 1, Fig. S1).
As can be seen from the graphs, LitR activates the transcription from the promoters of the luxI and luxCDABEG genes of both psychrophilic luminescent bacteria species A. logei and A. salmonicida. It should be noted that in both cases the activation of the luxI gene promoter in response to an increased expression of the litR gene was somewhat stronger than the activation of the luxCDABEG gene promoter. It can be seen that the amplitude of activation of A. salmonicida promoters is almost twice lower than that of A. logei promoters. This can be explained by the influence of other regulatory elements. These promoters are still the subject of research, but it is already known that their regulation is influenced by many factors, such as Lon, GroEL/ES, H-NS, CRP. We assume that the observed twofold difference may be due to differences in the sequences of promoters and surrounding regions, although the lux-box elements for these species are identical.
The data from Fig. 3C indicates that the induction of litR A. logei in the heterological system of E. coli leads to a notable (more than one order of magnitude) activation of expression from the A. fischeri PluxICDABEG promoter. This effect has not been previously described and may indicate that litR is able to induce LuxI/R QS system through stimulation of both luxI and luxR gene expressions in A. fischeri cells. Moreover, the effect on luxI expression was higher in magnitude. The data obtained contradicts the results published by (Fidopiastis et al., 2002), which investigated the effect of litR on the expression of lacZ of the gene inserted into luxC ORF. Perhaps it could be connected with the differences in the sensitivity of the reporter genes used. However, LitR proteins from mesophilic and psychrophilic bacteria have some differences and it needs more detailed investigation.
LuxR-independent activation of promoters by LitR depends on the lux-box sequence
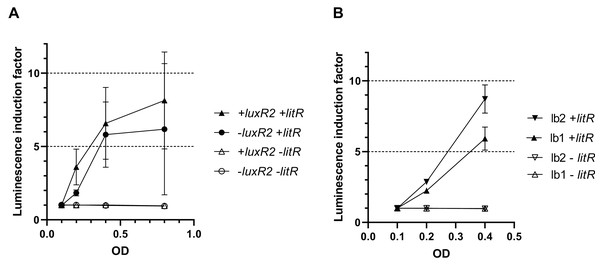
To make sure that the observed activation of “rightward” promoters determined by LitR occurs independently of the presence of regulatory genes from the luxI/R QS system in the cell, measurements of LitR-dependent activation of A. logei luxI gene transcription were carried out in the presence of the luxR2 regulatory gene and without it. Plasmids pSV16 and pR2 were used (Table 1, Fig. S1), in which the luxCDABE P. luminescens genes are under control of the A. logei PluxI promoter with and without the luxR2 gene, respectively. The litR gene was introduced as before on a separate plasmid p15Tc-litR. Measurement results are shown in Fig. 4A, the increase in luminescence was calculated as the ratio of luminescence of IPTG-induced cells to the luminescence of the same cells without IPTG. As a negative control, cells carrying the p15Tc-lac vector without the litR gene were used.
Figure 4: Effect of the luxR2 gene (A) and the lux-box sequence (B) on LitR-mediated activation of the A. logei lux regulon promoters in E. coli cells.
E. coli MG1655 cell were used with different combinations of the following plasmids: p15Tc-lac vector (marked as −litR), p15Tc-litR with the litR gene under control of Plac (+litR), pSV16 − PluxI promoter and luxR2 gene (+luxR2), pR2 − pSV16 with the truncated luxR2 gene (marked as –luxR2), pD-lb1 − PluxCDABEG promoter (lb1), pD-lb2 is pD-lb1 with the lux-box sequence from PluxI (lb2).We assumed that LitR could bind DNA in the promoter upstream region, the same site LuxR binds to (lux-box). To assess the effect of the lux-box sequence on the transcription activation of A. logei PluxCDABEG by the LitR protein, we compared the LitR-dependent activation of two variants of the A. logei PluxCDABEG promoter, which differ only in the lux-box sequences (Fig. 4B). The native PluxCDABEG promoter was truncated exactly upstream of the lux-box and introduced into pDEW201 upstream of P. luminescens luxCDABE gene cassette (line lb1 on Fig. 4B, the pD-lb1 plasmid was used); to obtain another promoter variant, the lux-box was changed to match PluxI (lb2 lines on Fig. 4B, the pD-lb2 plasmid was used). The difference between the obtained chimeric promoters is limited to only five bp (Fig. S4), in both cases, the cells don’t contain any of the luxR, luxR1 or luxR2 genes. All genetic elements, with the exception of lux-boxes, were identical in the compared cultures. The litR gene, as in previous experiments, was introduced into the cell on the p15Tc-litR plasmid, where it was under control of Plac.
As can be seen from the data presented in Fig. 4A, LitR activates PluxI and PluxCDABEG A. logei promoters regardless of the presence of LuxR2 in the cells: luminescence induction in response to litR expression stimulation is almost equal for cells with luxR2 gene and without it (p15Tc-litR in combination with pSV16 and pR2, correspondently). The observed induction of PluxI and PluxCDABEG A. logei and consequently luminescence of cells is completely determined by the litR gene: in its absence (vector p15Tc-lac in combination with pSV16, pR2, pDlb1, or pD-lb2 biosensor plasmids), the luminescence of cells does not change in response to the addition of IPTG. The data in Fig. 4B shows that the change in the lux-box sequence affects the activation of the promoter by LitR. This indicates that the LitR binding site in the promoter region of A. logei luxCDABEG genes coincides or at least intersects with the LuxR binding site.
Discussion
The luxR-luxI regulon could be used for development of expression systems with target protein expression regulation in dependence on bacterial population density (Swennen & Nocadello, 2011; Nocadello & Swennen, 2012). luxI-luxR regulation is modulated by a large number of intracellular factors that ensure its sensitivity to external conditions (Manukhov, Kotova & Zavil’gel’skiĭ, 2006; Konopleva et al., 2016), LitR becomes on a par with them, giving a connection with other QS systems (Fidopiastis et al., 2002).
Our experiments showed that promoters of luxI and luxCDABEG genes of all three investigated species of the Aliivibrio genus are activated by LitR from A. logei. Apparently, the interaction between LuxR and LitR proteins does not occur, since Fig. 4 shows the absence of the effect of the luxR2 gene on litR-mediated activation of the luxI gene promoter. That is, the interaction between litR and the lux operon is realized by the direct influence of LitR on promoters. In the psychrophilic species A. logei and A. salmonicida, the induction of promoters from which the luxI gene is transcribed is notably higher than induction of the other promoters of their lux regulons. As LuxI is an autoinducer synthase, its expression induction could stimulate whole QS system. The expression of luxCDABEG genes increases in the presence of LitR too, but the amplitude of this induction is too low to make Aliivibrio cell luminescence visible. Thus, we assume that the main effect of LitR on lux regulons of psychrophilic bacteria of the Aliivibrio genus is the independent of LuxR stimulation of LuxI synthesis (Figs. 3 and 4A). When ainS/R and luxS/PQ QS systems triggered before the luxI/R one due to stress conditions, stimulation of LuxI synthesis by LitR brings forth the increase in AI production and gives an initial impulse to activate luxI/R system. Higher concentrations of AI stabilize LuxR2 from A. logei/A. salmonicida, and the luxI/R QS system is triggered (Khrulnova et al., 2016). This LitR-dependent luxI induction could appear necessary in stress conditions, when the chaperonin GroEL/ES lacks or the protease Lon content is increased and consequently the QS is not activated even at a high culture density due to the lack of functional LuxR2 (Dolan & Greenberg, 1992a; Zavil’gel’skiĭ & Manukhov, 1994; Khrulnova et al., 2010, 2016).
The LitR binding site is not defined well and is still an object of discussion (Van Kessel et al., 2013; Chaparian et al., 2016). Here we showed that the sequence of lux-boxes has significance for the transcription activation of luxICDABEG by LitR regardless of the presence of luxR1 or luxR2 genes in the cell (Fig. 4). It is an obvious sign that LitR binds to the lux-box or directly next to it.
Conclusion
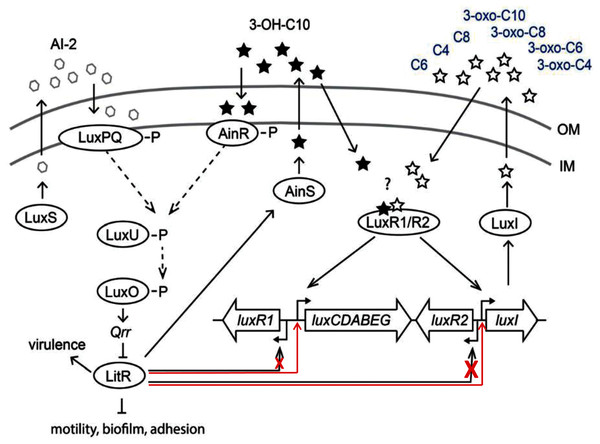
The results of this study demonstrated that LitR of psychrophilic bacteria A. logei is able to directly stimulate (independently of LuxR presence) transcription from promoters of luxI and luxCDABEG genes and to a lesser degree from promoter of luxR1 gene. Thus, 3-OH-C10 and AI-2 in the medium induce expression of litR and could enhance luxI expression and 3-O-C6 synthesis (Fig. 5). But for the effect of 3-OH-C10 and AI-2 on luxI/R system to be significant, the conditions are needed preventing this system from normal function. Our finding could be a clue to understanding the cross-interaction of the luxS/PQ and ainS/R QS systems with the luxI/R one in psychrophilic bacteria of A. logei and A. salmonicida species. Such effect was not previously described for the luxI/R QS system of well-known mesophilic bacteria A. fischeri. Main effect of this cross-interaction is LitR-dependent stimulation of autoinducer synthesis through PluxI induction and this pathway could be activated in conditions, when luxI/R QS system was not activated by its own AI until high culture density.
Figure 5: Scheme of A. logei and A. salmonicida QS regulation.
The autoinducer synthases LuxS, LuxI and AinS, produce the different acyl homoserine lactones and autoinducer-2 (AI-2) which are transported across the inner membrane (IM) and the outer membrane (OM). In presence of AI-2 or 3-hydroxo-decanoyl-homoserine lactone (3-OH-C10) LitR synthesis is activated through the AinS/R and LuxS/PQ pathways. LitR is expressed and regulates the production of the AinS AHL, as well as activities such as motility, biofilm, adhesion, virulence and bioluminescence, in accordance with (Bjelland et al., 2012). A. logei has the same gene combination in the QS systems as A. salmonicida. Red arrows indicate that LitR, contrary to previous assumption, is able to activate luxI and luxCDABEG genes, but not the luxR2 (very low effect on the luxR1 gene). Redrawn from (Hansen et al., 2015).Supplemental Information
Supplementary materials, Supplemental Figures, and plasmids constructing description.
Raw data of measurements.
Measurements of luminescence of E. coli cells transformed promoter-reporter plasmids in combination with plasmid carrying litR A. logei gene under control of Plac promoter. Luminescence was used for calculation of induction coeffitients.