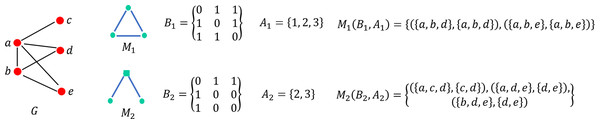COMICS: a community property-based triangle motif clustering scheme
- Published
- Accepted
- Received
- Academic Editor
- Yilun Shang
- Subject Areas
- Algorithms and Analysis of Algorithms, Graphics, Network Science and Online Social Networks
- Keywords
- Community property, Triangle motif, Large network, Clustering
- Copyright
- © 2019 Feng et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ Computer Science) and either DOI or URL of the article must be cited.
- Cite this article
- 2019. COMICS: a community property-based triangle motif clustering scheme. PeerJ Computer Science 5:e180 https://doi.org/10.7717/peerj-cs.180
Abstract
With the development of science and technology, network scales of various fields have experienced an amazing growth. Networks in the fields of biology, economics and society contain rich hidden information of human beings in the form of connectivity structures. Network analysis is generally modeled as network partition and community detection problems. In this paper, we construct a community property-based triangle motif clustering scheme (COMICS) containing a series of high efficient graph partition procedures and triangle motif-based clustering techniques. In COMICS, four network cutting conditions are considered based on the network connectivity. We first divide the large-scale networks into many dense subgraphs under the cutting conditions before leveraging triangle motifs to refine and specify the partition results. To demonstrate the superiority of our method, we implement the experiments on three large-scale networks, including two co-authorship networks (the American Physical Society (APS) and the Microsoft Academic Graph (MAG)), and two social networks (Facebook and gemsec-Deezer networks). We then use two clustering metrics, compactness and separation, to illustrate the accuracy and runtime of clustering results. A case study is further carried out on APS and MAG data sets, in which we construct a connection between network structures and statistical data with triangle motifs. Results show that our method outperforms others in both runtime and accuracy, and the triangle motif structures can bridge network structures and statistical data in the academic collaboration area.
Introduction
In all aspects of human endeavor, we are in the world of large-scale data, embracing the aspects of biology, medicine, social, traffic, and science (Ning et al., 2017). These data sets describe the complicated real-world systems from various and complementary viewpoints. Generally, the entities in real-world systems are modeled as nodes, whose connections and relationships are modeled as edges. Those networks become new carriers of rich information from domain-specific areas, such as the reciprocity among people in online social networks (Koll, Li & Fu, 2013). More than that, human beings are inclined to cooperate or participate in group activities, which can be reflected in social and academic collaboration networks. To be more specific, in academic area, big scholarly data grows rapidly, containing millions of authors, papers, citations, figures, tables, and other massive scale related data, such as digital libraries and scholarly networks (Xia et al., 2017). As collaboration behaviors among scholars are becoming frequent, collaboration networks are generally in large-scale and contain rich collaboration information, reflecting the cooperation patterns among scholars in different research areas. Bordons et al. (1996) regard the academic teams as scientists communities, in which scholars can share research methods, materials, and financial resources rather than institutions organized by fixed structures (Barjak & Robinson, 2008). Furthermore, the ternary closures in social networks constitute a minimal stable structure; that is, a loop with three nodes. The number of ternary closures in social networks changes over time, which reveals the evolvement of human social behaviors. Besides, the definition of a clustering coefficient is based on the distributions of ternary closures. Milo et al. (2002) defined small network structures as motifs to present interconnections in complex networks by numbers that are significantly higher than those in randomized networks. Motifs can define universal classes of networks, and researchers are carrying on the motif detection experiments on networks from different areas, such as biochemistry, neurobiology, and engineering, to uncover the existence of motifs and the corresponding structure information in networks (Ribeiro, Silva & Kaiser, 2009; Bian & Zhang, 2016). Hence, triangle motifs can be used to uncover relationships in networks.
Connectivity is a fundamental character in both graph theory and network science. When networks are in small-scale, the dense areas can be easily identified. However, with the rapid growth of network scale and diversity, many graph partition methods, community detection, and clustering algorithms fail to uncover the information of graph structure. Graph partition and mining algorithms consume a large amount of time when dealing with large-scale networks, for example, the gSpan algorithm (Yan & Han, 2002) and the Min–Cut algorithm (Stoer & Wagner, 1997), which overlook the elementary network structures. The clusters and subgraphs of a large network are generally have small internal distances and large external distances among nodes. Considering the ternary closures, triangle network motifs have been regarded as elementary units in networks. However, a general method to cluster the communities and analyze the relationships with community properties and triangle motifs effectively is still lacking.
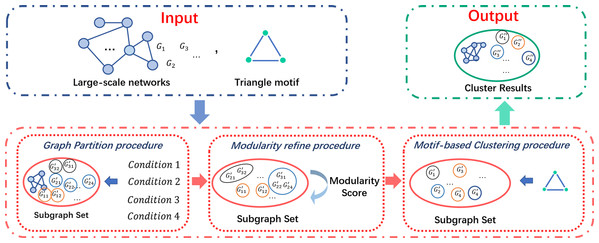
In this paper, we propose a community property-based triangle motif clustering scheme (COMICS) to cluster network communities, and analyze the relationships with triangle motifs. In this method, we partition networks with the edge connection properties and regard the undirected and unweighted complete triangle motifs as the element clustering units. The partition operations are based on four network cutting conditions, whose definitions are based on the network connectivity to maintain the massive links in networks. More than that, by considering the American Physical Society (APS) and Microsoft Academic Graph (MAG) data sets in the academic analysis area, we regard each cluster generated from the input network as an academic team, and define three metrics: teamwork of collaborator variance (TCV), teamwork of paper variance (TPV), and motif variances of scholars (MSV) to evaluate the behaviors of the detected academic teams. Our contributions can be summarized as follows:
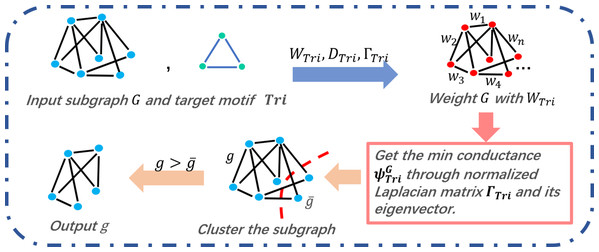
By jointly considering time complexity and clustering accuracy, we construct the COMICS, which mines the structure information with complete triangle motifs. A series of speed-up and refining methods, graph partition and refining techniques, are integrated to improve the performance of the basic clustering process.
We prove the time complexity of the presented algorithm is O(rn3), where r is the number of the clustered subgraphs from the original large network, and n is the number of nodes.
We regard the undirected and unweight complete triangle motif as the elementary unit instead of nodes in the clustering procedure. Our work verifies that the complete triangle motif is available in network analysis.
We define three metrics to analyze the hidden information in academic collaboration networks. Performance evaluations show that the academic teams with high quantity of scholar motif variances also have high values of TCVs.
The roadmap of this paper is illustrated as follows. We briefly illustrate the related works in the following section. After that, a series of fundamental definitions, problem statement, and some necessary notations are described. Then, we describe the architecture of COMICS in details. We evaluate the performance of our method with three large-scale networks as case studies in the experiment section. Finally, we conclude this paper.
Experiments
In this section, we compare COMICS with K-means and co-authorship team detection algorithm from the perspectives of network clustering accuracy and time complexity, respectively. We choose four large-scale networks, including two social network, that is, Facebook and gemsec-Deezer data sets (Leskovec & Krevl, 2014; Rozemberczki et al., 2018) and two academic collaboration networks, that is, APS and MAG data sets.
We analyze the accuracy of the clustering results by calculating compactness and separation. We demonstrate the efficiency of our solution in both academic collaboration and social networks. We also consider other statistical data information of academic networks, TCVs, TPVs, and MSV. All those corresponding metrics are illustrated in this section. All experiments are conducted on a desktop with Intel(R) Xeon(R) CPU E5-2690 v3 @ 2.60 GHz (two processors) and 128 GB memory. The operating system is Window 10, and the codes are written in python.
The American Physical Society data set (2009–2013) consists of 96,908 papers associated with 159,724 scholars in the physical field. Meanwhile, the MAG data set (1980–2015) on computer science includes 207,432 scholars with 84,147 papers in the computer science area. Edges in the academic networks represent two authors have coauthored at least one paper. The Facebook social network data set in our experiments contains eight networks, 134,833 nodes and 1,380,293 edges. We list the eight social networks in Table 1. In that case, we cluster the social networks by the different categories listed in the data set. Gemsec-Deezer data set collected by Deezer (November 2017) is also experimentalized in this paper. This data set contains 143,884 nodes and 846,915 edges from three countries, Romania (41,773 nodes and 125,826 edges), Croatia (54,573 nodes and 498,202 edges) and Hungray (47,538 nodes and 222,887 edges).
| TV shows | Politician | Government | Public figures | |
| Node | 3,892 | 5,908 | 7,057 | 11,565 |
| Edge | 17,262 | 41,729 | 89,455 | 67,114 |
| Athletes | Company | New Sites | Artist | |
| Node | 13,866 | 14,113 | 27,917 | 50,515 |
| Edge | 86,858 | 52,310 | 206,259 | 819,306 |
Experiment settings
In this subsection, we describe the settings of our experiments from three aspects, that is, time cost, clustering accuracy and academic teamwork behavior analysis with complete triangle motif in academic areas. In academic collaboration networks, we consider two algorithms. The Facebook social networks do not contain any statistical information. Therefore, we merely compare our method with K-means algorithm in the social network:
K-means clustering algorithm (Ding & He, 2004): This method proves that principal components are the continuous solutions to cluster membership indicators for K-means clustering. It takes principal component analysis into the clustering process, which is suitable for the scholar science and social data sets.
Co-authorship algorithm (Reyes-Gonzalez, Gonzalez-Brambila & Veloso, 2016): This algorithm considers all the principal investigators and collaborators, and defines knowledge footprints1 of the groups to calculate the distances between scholars and the group. Based on the distance, the academic groups can be detected in an accurate way. This method iterates all the researchers with their collaborator and institution similarities until they are assigned to a academic team can be applied to understand the self-organizing of research teams and obtain the better assessment of their performances.
To demonstrate the runtime efficiency and the accuracy of our clustering results in large-scale networks, we divide the APS and MAG data sets into different parts with various sizes by years, respectively, so that we can get the collaboration networks with distinct number of nodes (from 1,000 to 200,000). Considering the integrality and veracity of the academic research teams in data sets, we take the whole APS and MAG data sets as the collaboration networks to detect the collaborative relationships.
Evaluation metrics
To evaluate and analyze the accuracy of network clustering results of our proposed COMICS, we use two metrics, that is, compactness and separation, to evaluate node closeness in clustering results and the distances among clusters. In academic collaboration networks, we combine the statistical paper publishing data with network structures together, and calculate three metrics to find the characteristics discovered through the target triangle motif to uncover the hidden collaboration patterns and teamwork of scholars in academic networks.
Compactness and separation (Halkidi, Batistakis & Vazirgiannis, 2002) are used to evaluate the accuracy of clustering results by different methods. Compactness is a widely used metric to quantify the tightness of clustering subgraphs, representing the distances between nodes in a subgraph. Separation calculates the distances among the cores of different subgraphs. That is, if a clustering subgraph is with lower compactness value and higher separation value, the subgraph can be detected effectively. Compactness is expressed by Eq. (11), (11)
Here, R is the clustering result set, vi is one of the nodes in the subgraph, and w is defined as the core of the subgraph cluster, because w is the node with the maximum degree in a cluster. The value of |vi − w| means the shortest distance between node vi and the cluster core node w. SP is defined as in Eq. (12). (12) wherein, k is the number of subgraphs in the result set and wi is the core of the given subgraph i, which is the same as wj. The value of |wi − wj| equals to the shortest distance between wi and wj.
In collaboration networks, we assume the clusters as academic teams, in which scholars work together. Therefore, three metrics are defined to analyze the collaboration behaviors through triangle motif: TCV, TPV, and MSV.
TCV: This metric reflects the tightness and volatility among members in a team. For one scholar i in a team, we define the TCV as follows, (13)
Herein, n is the number of team members, coi is the number of scholars that scholar i has collaborated within the same team, and coave is the average number of collaborators collaborated with scholars in a team.
TPV: An academic team with high performance refers that the members in team have published a large number of paper. Similarly, in a stable team, the gaps of published paper numbers among team members are small. To evaluate the academic levels and stability of a team, we define TPV as follows: (14) where σqtt means scholar i’s variance of publishing papers in the detected team, qi is the number of papers that scholar i has published, and qave is the average number of papers in the team.
MSV: This metric calculates the difference of motif number that the scholar nodes are included in the collaboration networks. We define the MSV as follows, (15)
Herein, ti is the number of target motif that scholar i owns, and tave is the average motifs of a team.
To uncover the collaboration patterns mined by triangle motifs among scholars in academic teams, we use the above three arguments to analyze relationships between productions and motifs of the clustered academic teams.
Results and Discussion
In this section, we evaluate the experimental results by comparing with K-means and co-authorship algorithm in both runtime and the effectiveness. In the view of internal and external connections, we calculate compactness and separation values for each algorithm results.
The time cost results of three networks are shown in Tables 2–4, respectively. “K” in the tables represents thousand, for example, “1K” means a network with one thousand nodes. N/A means that the clustering procedure takes more than 5 days.
| COMICS | Co-authorship | K-means | |
|---|---|---|---|
| 1 K | 36.32 s | 2.12 s | 1.73 s |
| 3 K | 435.67 s | 17.45 s | 207.06 s |
| 10 K | 3,058.21 s | 1,084.83 s | 3.47 h |
| 30 K | 1.03 h | 2,856.47 s | 5.73 h |
| 50 K | 1.83 h | 4.82 h | 13.36 h |
| 80 K | 2.29 h | 9.87 h | >24 h |
| 120 K | 5.46 h | 16.36 h | >24 h |
| 150 K | 9.97 h | >24 h | N/A |
| COMICS | Co-authorship | K-means | |
|---|---|---|---|
| 1 K | 24.74 s | 3.79 s | 2.04 s |
| 3 K | 343.17 s | 21.05 s | 237.93 s |
| 10 K | 2,956.64 s | 345.29 s | 3.53 h |
| 30 K | 1.08 h | 2,636.95 s | 6.62 h |
| 50 K | 2.47 h | 2.93 h | 12.48 h |
| 80 K | 3.35 h | 4.07 h | >24 h |
| 120 K | 5.09 h | 8.27 h | >24 h |
| 150 K | 8.91 h | 21.83 h | N/A |
| 200 K | 14.68 h | >24 h | N/A |
| COMICS | K-means | |
|---|---|---|
| TV shows | 573.62 s | 322.86 s |
| Politician | 1,394.05 s | 786.42 s |
| Government | 1.03 h | 2.71 h |
| Public figures | 1.26 h | 1.60 h |
| Athletes | 1.58 h | 2.04 h |
| Company | 0.98 h | 3.07 h |
| New sites | 4.78 h | 9.32 h |
| Artist | 6.89 h | 23.42 h |
| Romania | 3.46 h | 17.68 h |
| Hungray | 3.96 h | 18.07 h |
| Croatia | 6.04 h | 38.42 h |
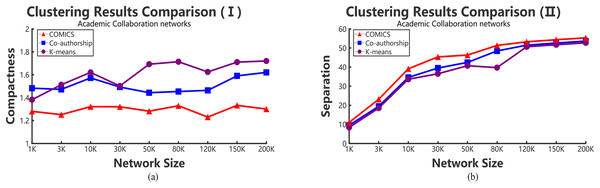
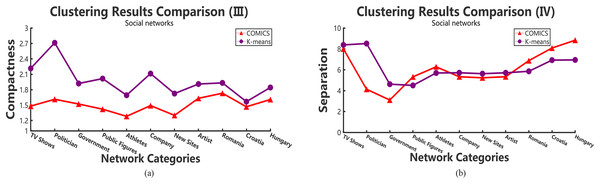
According to Tables 2–4, it can be concluded that, in small networks (less than 30,000 nodes), the three methods make little differences in running time. However, as the size of network increases, our clustering algorithm costs the least time. The time costs in different data sets make little differences. However, the results show the same trend and the proposed method takes more time in small networks and outperforms other large networks. As shown in Tables 2 and 3, when academic collaboration networks contain more than 30,000 nodes, COMICS takes the least time than the other two algorithms. More than that, in social networks, the time cost of our method is also satisfied in large size networks. Therefore, it can be concluded that though the partition operations cost a lot of time, it is necessary to apply the speeding up techniques in clustering. Moreover, for different types of networks, topological structures, density are also vital factors that can effect the clustering procedures and results. Figures 4A and 5A show the compactness values generated by our algorithm and the comparing algorithms on different sizes of networks, respectively. As the figures show, in collaboration networks, compactness values corresponding to different networks are lower than those in co-authorship algorithm and K-means algorithm, which are similar with that in social networks. Our algorithm performs better than the two comparing algorithms. Figures 4B and 5B plot the separation values of the three algorithms with the network growth in both academic, Facebook social and gemsec-Deezer networks. It can be seen that with the growing network size, COMICS achieves the highest separation values. This means subgraphs clustered by our method have greater separation values all the time. According to Figs. 4B and 5B, we can conclude that the distances among core nodes in each cluster are close no matter what algorithms are used. The reason is that no matter what algorithms are used in the target network, the core nodes of clusters are almost the same. All the core nodes are with the maximum degrees. In all, our clustering algorithm achieves the best subgraph clustering results obviously.
Figure 4: The variation tendency of compactness and separation values of collaboration network clustering results with COMICS, co-authorship and K-means algorithms.
(A) Compactness in academic collaboration networks and (B) separation in academic collaboration networks.Figure 5: The variation tendency of compactness and separation values of the clustering results in social networks with COMICS and K-means algorithms.
(A) Compactness in social networks and (B) separation in social networks.Analysis in academic collaboration networks
After analyzing the time complexity and effectiveness of our system above, in this subsection, we analyze the clustering results with the triangle motifs in academic collaboration networks. The results prove the triangle motif structures can reflect the hidden statistical information and connections with network structures. For example, as the analysis results show, collaboration patterns as well as the correlations of network structure and team productions can be summarized in the academic collaboration networks.
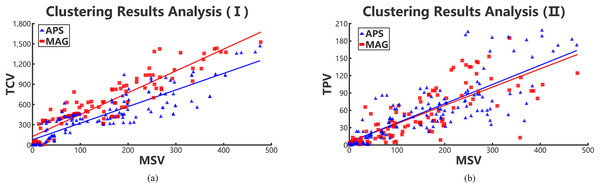
We regard the cluster results of each academic collaboration network as an academic team. Then the values of three variances, that is, TPV, TCV, and MSV are calculated, and the results are shown in Figs. 6A and 6B. Hence, we can see that the number of high-order triangle motif can reflect the performance of an academic team to some extent.
Figure 6: Positive relations in collaboration networks through collaboration variances, paper variances and motif variances of each clustering.
Red rectangles and blue triangles represent the collaboration academic teams clustered from MAG and APS data sets, respectively. (A) Relationships between TCV and MSV and (B) relationships between TPV and MSV.According to Figs. 6A and 6B, we conclude that the TPV and TCV are both proportional to the MSV. Meanwhile, the TPV is also approximately positive linear with the MSV. That means, the lower the MSV is in a cluster team, the performance of team members are in smaller gaps. Therefore, it can be concluded that the value of MSV can reflect the gap of collaboration relationships in teams and performance of team members. However, we can infer that the scholars with few number of complete triangle motifs, have collaborated with only few scholars in the team. Those scholars are probably students or new team members, resulting in the high collaboration and paper variances. Hence, in collaboration networks, we can use MSV to evaluate the gaps of team collaboration relationships and the performance of team members. The two teamwork gaps in different periods represent the stability and volatility of academic teams.
Conclusion
In this paper, we put forth the high-order motif-based clustering system to get a subgraph set from the large-scale networks. In the constructed system, we take graph partition and refining techniques to speed up algorithm runtime. Through network cutting, we check the four cutting conditions from the aspect of network connectivity, which can prevent damaging the global structures of large-scale networks. Experiments are carried on four large networks, that is, APS and MAG from the academic area, Facebook and gecsec-Deezer networks from the social area, respectively. The results demonstrate the effectiveness of our method in time cost and accuracy in large-scale network clustering.
Furthermore, the collaboration teamwork analysis verifies the availability of complete triangle motif, which represents the smallest collaboration unit in the collaboration networks. We analyze the collaboration clustering results with three metrics, that is, TCV, TPV, and MSV. The results show that both TCV and TPV are proportional to MSV. Therefore, it can be concluded that the value of MSV can reflect the two gaps, that is, collaborative relationships and performance of different team members. Besides, the two gaps in different periods can also reflect the dynamic change of team members. In the future, we will focus on dynamic motif clustering for real-time network management (Ning et al., 2018; Ning, Huang & Wang, 2019; Wang et al., 2018a). In addition, network security (Wang et al., 2018b, 2019) and crowdsourcing based methods (Ning et al., 2019a, 2019b) also deserve to be investigated.