Embryo polarity in moth flies and mosquitoes relies on distinct old genes with localized transcript isoforms
Figures
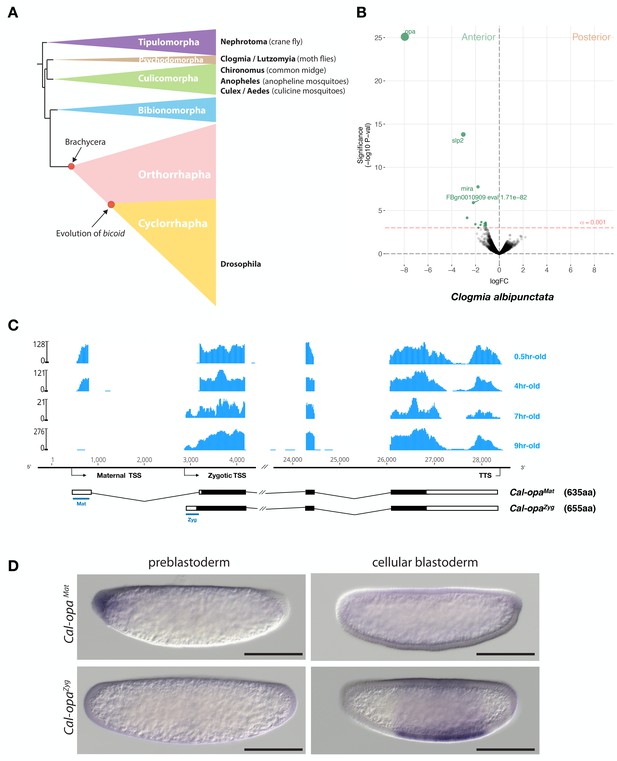
Expression of alternative Cal-opa transcripts in Clogmia embryos.
(A) Phylogenetic relationship of fly species referred to in the text (Wiegmann et al., 2011). (B) Differential expression analysis of maternal transcripts between anterior and posterior halves of 1 hr-old Clogmia embryos. logFC: log fold-change. (C) Stage-specific RNA-seq read coverage of Cal-opa locus. Transcription start sites (TSS) and transcription termination sites (TTS) are indicated on the genomic scaffold (solid line with 1000 bp intervals marked) and were confirmed by RACE. Exon-intron sketches of Cal-opaMat and Cal-opaZyg transcript variants are shown with the open reading frame in black and the position of in situ hybridization probes and dsRNA underlined in blue. (D) RNA in situ hybridization of Cal-opaMat and Cal-opaZyg transcripts in 1 hr-old preblastoderm and 7 hr-old cellular blastoderm embryos. Anterior is left and dorsal up. Scale bar: 100μm.
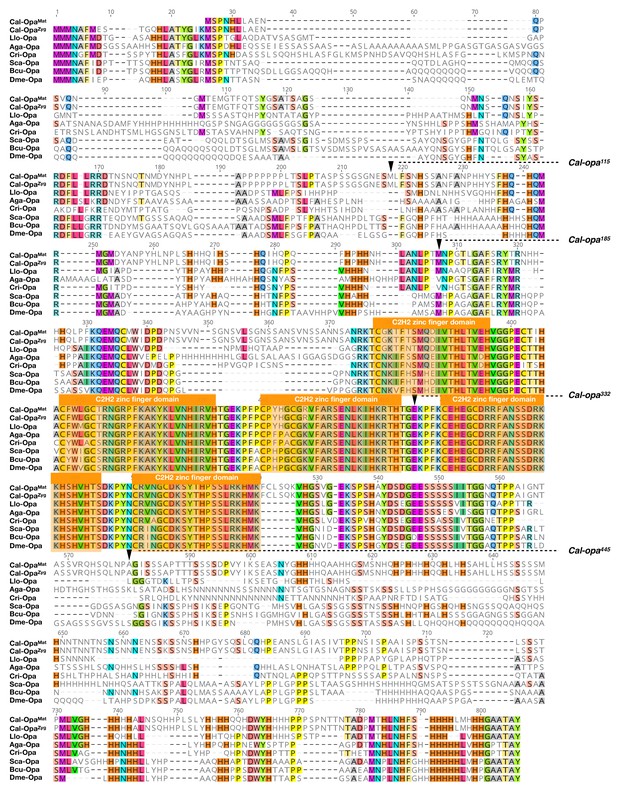
Protein alignment of predicted dipteran Odd-paired orthologs.
Odd-paired orthologs were aligned using ClustalW 2.1 (Larkin et al., 2007). Black triangles mark Cal-Opa truncations referred to in the main text (see Figure 2). Cal: Clogmia albipunctata (Psychodidae); Llo: Lutzomyia longipalpis (Psychodidae); Aga: Anopheles gambiae (Culicidae); Cri: Chironomus riparius (Chironomidae); Sca: Stomoxys calcitrans (Muscidae); Bcu: Bactrocera cucurbitae (Tephritidae); Dme: Drosophila melanogaster (Drosophilidae). Conserved zinc finger domains are indicated with colored boxes. Conserved amino acids are shown color.
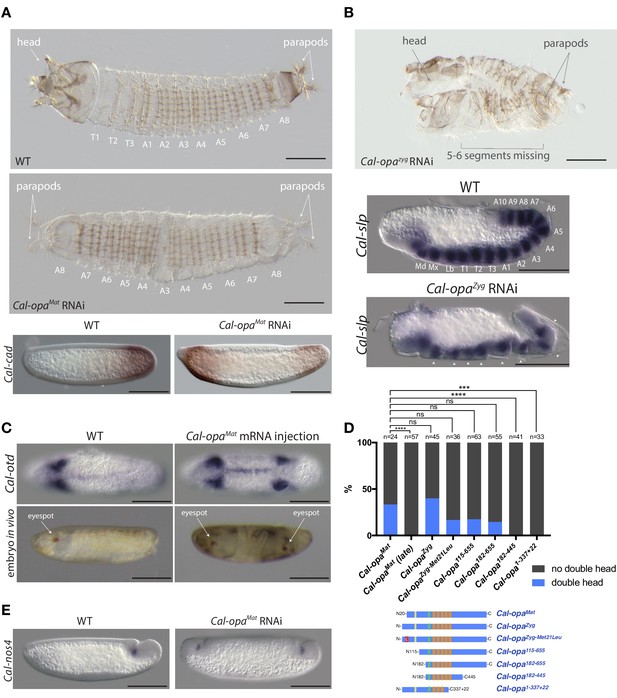
Function of alternative Cal-opa transcripts in early Clogmia embryo.
(A) 1 st instar larval cuticle of wild type (top) and following Cal-opaMat RNAi (middle). RNA in situ hybridization of Cal-cad in a wild-type preblastoderm embryo (bottom left) and stage-matched Cal-opaMat RNAi embryos (bottom right). Anterior is left and dorsal up. T: thoracic segment; A: abdominal segment. Segment numbers in Cal-opaMat RNAi larval cuticle were assigned based on the assumption of polarity reversal. Scale bar: 100μm. (B) 1 st instar larval cuticle phenotype following Cal-opaZyg RNAi (top) and RNA in situ hybridization of Cal-slp in extending wild-type germband (middle) and stage-matched Cal-opaZyg RNAi embryo (bottom). Anterior is left and dorsal up. Md: mandibular segment; Mx: maxillary segment; Lb: labial segment; T: thoracic segment; A: abdominal segment. Scale bar: 100μm. (C) RNA in situ hybridizations of Cal-otd in wild-type gastrula (top left) and stage-matched embryo following posterior Cal-opaMat mRNA injection (top right) are shown in ventral view. A live wild-type embryo (bottom left) and a stage-matched embryo following posterior Cal-opaMat mRNA injection (bottom right) in lateral view. Anterior is left. Scale bar: 100μm. (D) Posterior injection of Cal-opa mRNA and mutated variants. Complete, symmetrical duplication of the bilateral Cal-otd expression domain in gastrulating embryos was counted as double head (blue, see Figure 2C). All other phenotypes, including incomplete duplications and wild type, were conservatively counted as ‘no double head’ (black). Sketches of predicted Cal-Opa proteins are shown with ZIC/Opa conserved motif (ZOC) in yellow, the ZIC family protein N-terminal conserved domain (ZFNC) in green, and zinc finger domains in orange. The Met21Leu mutation in Cal-OpaZyg-Met21Leu is marked in red. Cal-opaMat (late): Cal-opaMat was injected during the syncytial blastoderm stage (4 hr). ns: p>0.05; ***: p<0.001; ****: p<0.0001, Fisher’s exact test. (E) RNA in situ hybridization of Cal-nos4 in a gastrulating embryo (left) and stage-matched Cal-opaMat RNAi embryos (right).
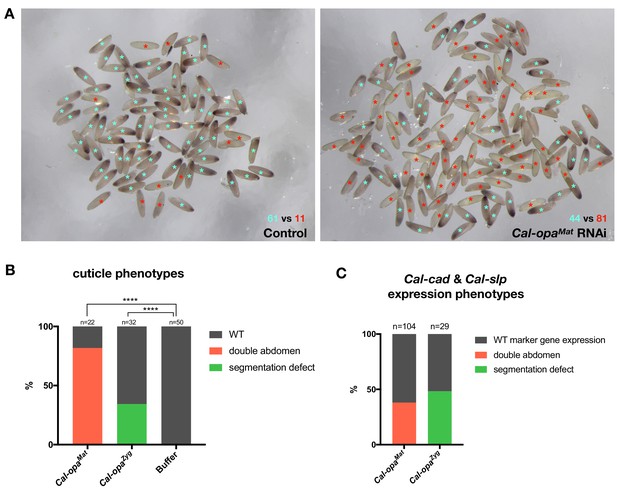
Frequencies of strong Cal-opaMat and Cal-opaZyg RNAi phenotypes.
(A) Cal-opaMat expression in preblastoderm embryos injected with panish dsRNA (negative control, left) or Cal-opaMat dsRNA (right). Asterisk in light blue: WT Cal-opaMat expression; asterisk in red: no Cal-opaMat expression (p<0.0001, Fisher’s exact test). RNAi experiments were performed in parallel using the same dsRNA concentration (1 µg/µl) and embryos were fixed one hour after the injection. RNA in situ hybridizations were performed simultaneously with the same probe concentration and Alkaline Phosphatase incubation time. (B) Cuticle phenotypes of 1 st instar larvae. For representative examples see Figure 2A and Figure 2B. ****: p<0.0001, Fisher’s exact test. (C) Cal-cad and Cal-slp expression phenotypes in embryos. For representative examples see Figure 2A and B. Cal-cad in situ hybridizations were done on Cal-opaMat RNAi embryos that reached cellular blastoderm stage. Cal-slp in situ hybridizations were done on 14- to 18 hr-old Cal-opaZyg RNAi embryos. Embryos that had failed to reach the stage of germband extension were excluded from the analysis.
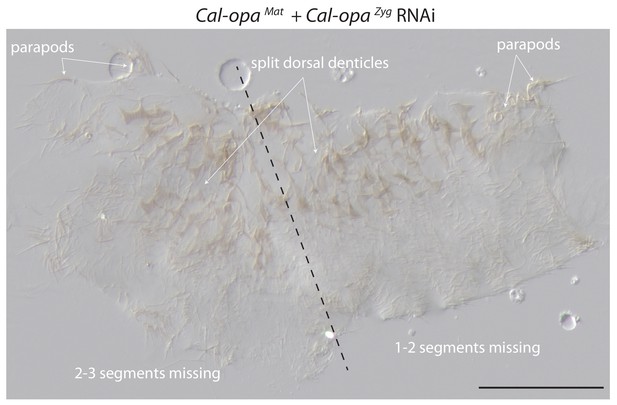
Cuticle phenotype of a 1 st instar larva following Cal-opaMat and Cal-opaZyg double RNAi.
A double abdomen with several missing segments is shown in lateral view. More severe phenotypic defects were not found in 1 st instar larvae, possibly due to embryo death prior to cuticle secretion. Scale bar, 100μm.
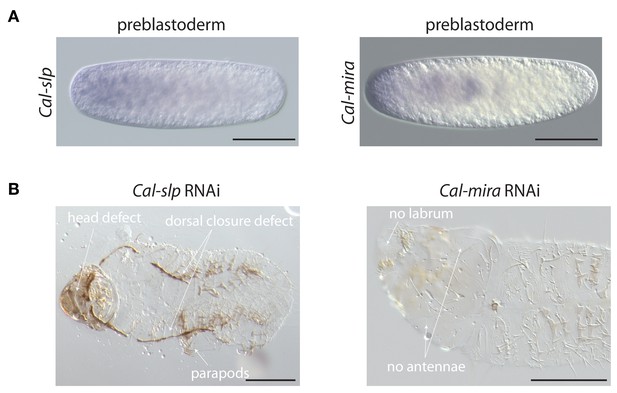
Maternal transcript localization and RNAi phenotypes of Cal-slp and Cal-mira in Clogmia.
(A) Cal-slp and Cal-mira RNA in situ hybridization in embryos at preblastoderm stage. Anterior is left; Scale bar: 100μm. (B) Cuticle phenotypes of 1 st instar larvae following Cal-slp RNAi (left) and Cal-mira RNAi (right) in ventral view. Anterior is left. Scale bar: 100μm.
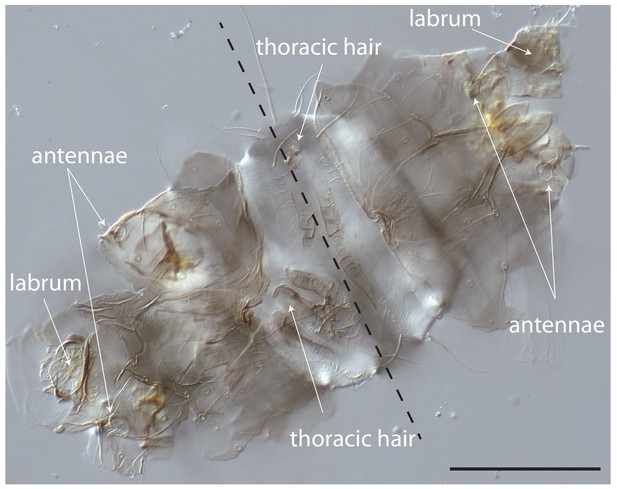
Cuticle phenotype of a 1 st instar Clogmia larva following Cal-opaMat mRNA injection.
Dorsal view. Scale bar: 100μm.
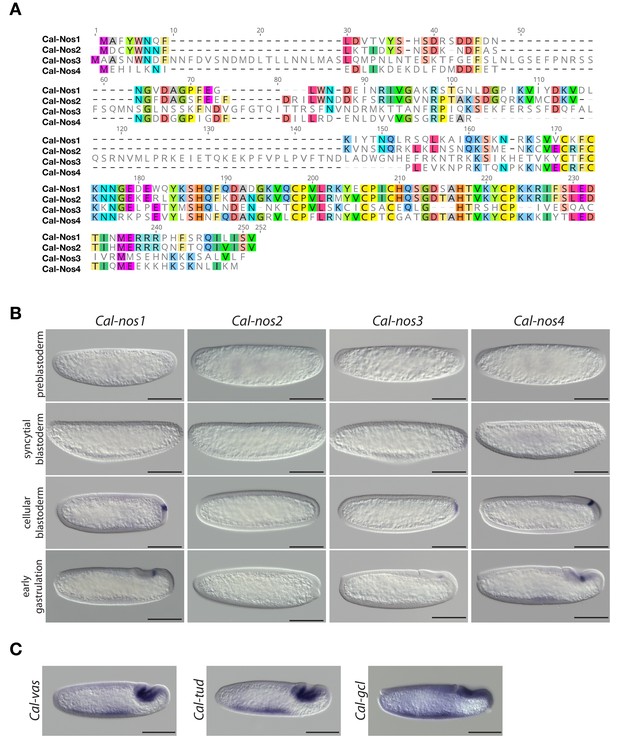
Clogmia homologs of nanos, vasa, tudor, and germ cell-less.
(A) Protein alignment of Cal-Nos paralogs using ClustalW 2.1 (Larkin et al., 2007). Conserved amino acids are highlighted. (B) Cal-nos1, Cal-nos2, Cal-nos3, and Cal-nos4 RNA in situ hybridizations in embryos at preblastoderm, syncytial blastoderm, cellular blastoderm, and gastrulation stages. Anterior is left and dorsal up. Scale bar: 100μm. (C) Cal-vas, Cal-tud, and Cal-gcl RNA in situ hybridizations in embryos at gastrulation stage. Anterior is left and dorsal up. Scale bar: 100μm.
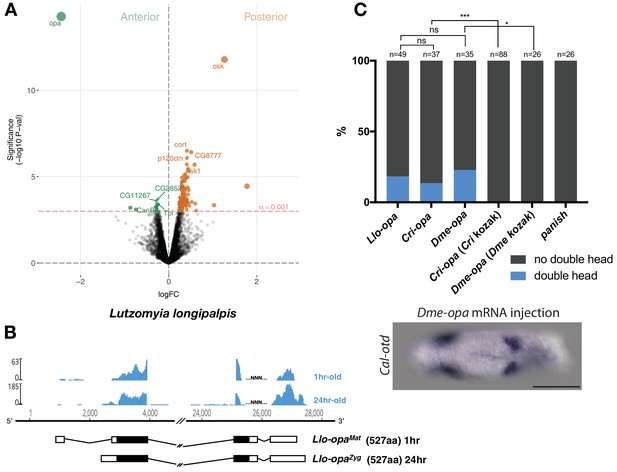
Alternative odd-paired transcript isoforms in Lutzomyia and ectopic head induction by mRNAs derived from Cal-opa orthologs in Clogmia.
(A) Differential expression analysis of maternal transcripts between anterior and posterior halves of 1–2 hr-old Lutzomyia embryos. logFC: log fold-change. (B) Stage-specific RNA-seq read coverage of Llo-opa genomic locus and sketches of Llo-opaMat and Llo-opaZyg transcripts (see also legend to Figure 1A). (C) Posterior injection of odd-paired mRNAs from Lutzomyia, Chironomus, and Drosophila in Clogmia embryo. Phenotypes were counted as in (Figure 2D) and double head from Dme-opa mRNA injection is shown as example. Cri-opa and Dme-opa mRNAs include Kozak sequence of Cal-opaMat (TAAG upstream of the predicted translation start site). Cri-kozak and Dme-kozak refer to odd-paired sequences with donor-specific Kozak-sequences from Chironomus (AAAA) and Drosophila (GACC), respectively. ns.: p>0.05; *: p<0.05. ***: p<0.001, Fisher’s exact test.
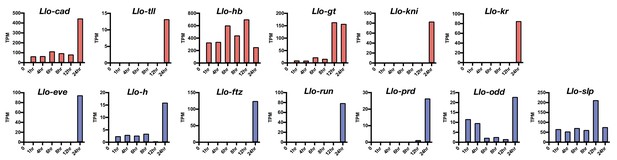
Expression level of select Lutzomyia gap gene (red) and pair-rule gene (blue) homologs.
Based on RNA-seq data from 1, 4, 6, 8, 12, and 24 hr-old embryos. TPM: transcripts per million.
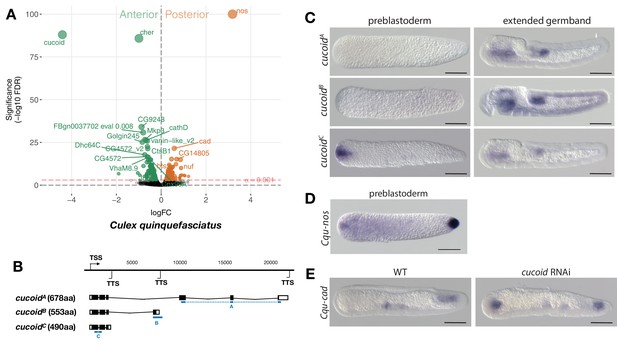
Expression and function of alternative cucoid isoforms in Culex.
(A) Differential expression analysis of maternal transcripts between anterior and posterior halves of 1 hr-old Culex embryos. logFC: log fold-change. (B) Sketches of cucoid transcript isoforms based on RNA-seq and RACE experiments (see also legend to Figure 1A). Blue lines the position of in situ hybridization probes. (C) RNA in situ hybridization of cucoidA, cucoidB, and cucoidC transcripts in 1 hr-old preblastoderm and germband extending embryos. Anterior is left and dorsal up. Scale bar: 100μm. (D) RNA in situ hybridization of Cqu-nos in 1 hr-old preblastoderm embryo. Anterior is left. Scale bar: 100μm (E) RNA in situ hybridization of Cqu-cad in a wild-type gastrulating embryo (left) and in a stage-matched cucoid RNAi embryos (right; 16/48 with cucoid dsRNA versus 0/25 with lacZ control dsRNA; p<0.0005). Anterior is left and dorsal up. Scale bar: 100μm.
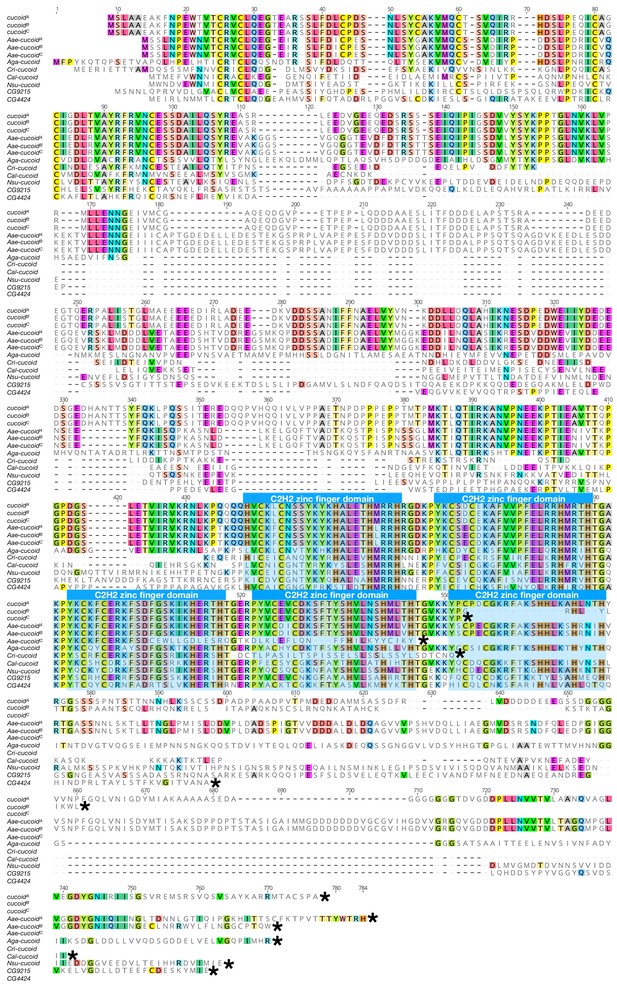
Protein alignment of dipteran cucoid orthologs and a Drosophila paralog CG4424.
Proteins were aligned using ClustalW 2.1 (Larkin et al., 2007). Conserved zinc finger domains are indicated with colored boxes. Black asterisk(*) indicates the last amino acids of proteins. Nsu: Nephrotoma suturalis (Tipulidae); Cal: Clogmia albipunctata (Psychodidae); Aae: Aedes agypti (Culicide); Cqu: Culex quinquefasciatus (Culicidae); Aga: Anopheles gambiae (Culicidae); Cri: Chironomus ripariu (Chironomidae).
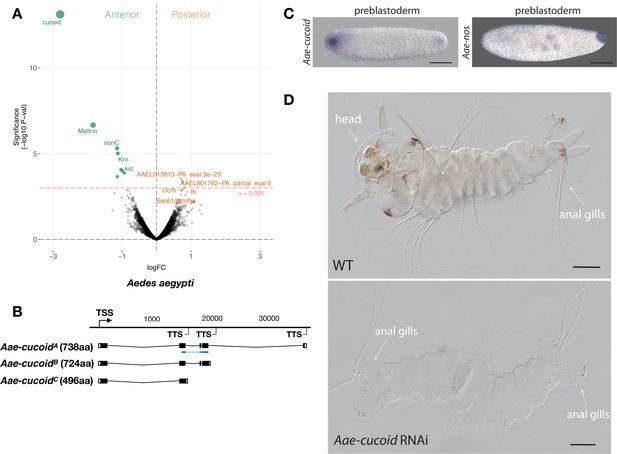
Expression and function cucoid in Aedes.
(A) Differential expression analysis of maternal transcripts between anterior and posterior halves of 1 hr-old Aedes embryos. logFC: log fold-change. (B) Sketches of Aae-cucoid transcript isoforms based on RNA-seq and RACE experiments with the position of the in situ hybridization probe and dsRNA underlined in blue (see also legend to Figure 1A). (C) RNA in situ hybridization Aae-cucoid (left) and Aae-nos (right) transcript in 1 hr-old preblastoderm embryo. Anterior is left and dorsal up. Scale bar: 100μm. (D) 1 st instar Aedes larval cuticle of wild type (top) and following Aae-cucoid RNAi (bottom; 9/26 versus 0/22 with control dsRNA; p<0.005). Scale bar: 100μm.
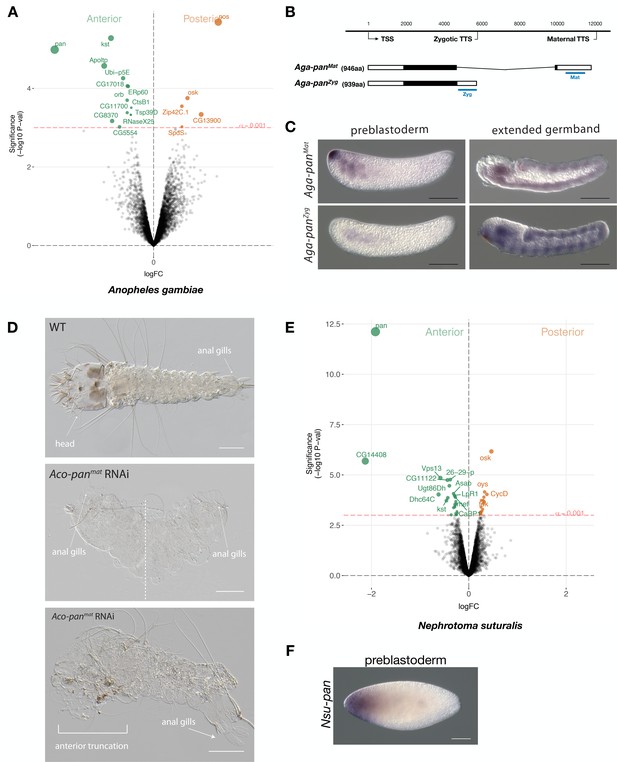
Expression and function of alternative pangolin isoforms in Anopheles.
(A) Differential expression analysis of maternal transcripts between anterior and posterior halves of 1 hr-old Anopheles embryos. logFC: log fold-change. (B) Sketches of Aga-panMat and Aga-panZyg transcripts based on RNA-seq and RACE experiments (see also legend to Figure 1A). Blue lines the position of in situ hybridization probes and dsRNA. (C) RNA in situ hybridization of Aga-panMat and Aga-panZyg transcripts in 1 hr-old preblastoderm and germband extending embryos of A.gambiae. Anterior is left and dorsal up. Scale bar: 100μm. (D) 1 st instar A.coluzzii larval cuticle of wild type (top). Cuticle phenotypes of double abdomen (middle; 3/37) and intermediate, anterior truncation phenotype (bottom; 8/37) following Aco-panMat RNAi. Scale bar: 100μm. (E) Differential expression analysis of maternal transcripts between anterior and posterior halves of 1 hr-old Nephrotoma (Tipulidae) embryos. logFC: log fold-change. (F) RNA in situ hybridization of Nsu-panMat.
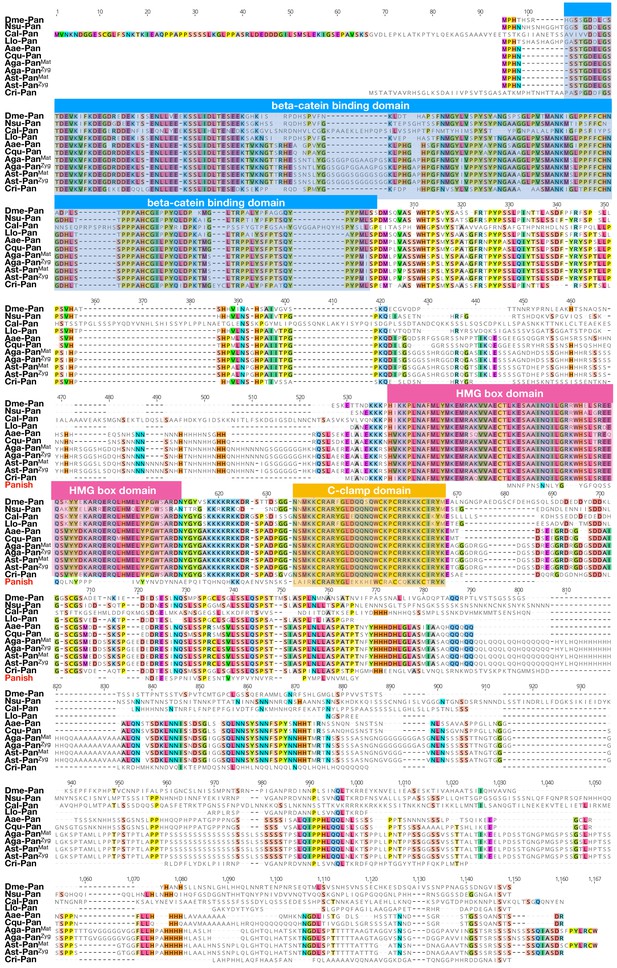
Protein alignment of dipteran Pangolin orthologs and Panish.
Proteins were aligned using ClustalW 2.1 (Larkin et al., 2007). Conserved domains are indicated with colored boxes. Dme: Drosophila melanogaster (Drosophilidae); Nsu: Nephrotoma suturalis (Tipulidae); Cal: Clogmia albipunctata (Psychodidae); Llo: Lutzomyia longipalpis (Psychodidae); Aae: Aedes agypti (Culicide); Cqu: Culex quinquefasciatus (Culicidae); Aga: Anopheles gambiae (Culicidae); Ast: Anopheles stephensi (Culicidae); Cri: Chironomus riparius (Chironomidae). Conserved amino acids are shown color.
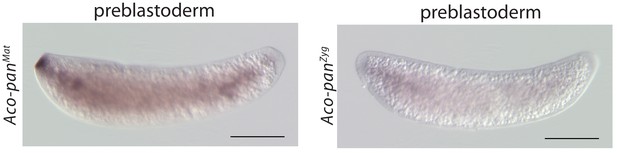
RNA in situ hybridization of Aco-panMat and Aco-panZyg transcripts in 1 hr-old A.coluzzii preblastoderm embryos.
Anterior is left and dorsal up. Scale bar: 100μm.
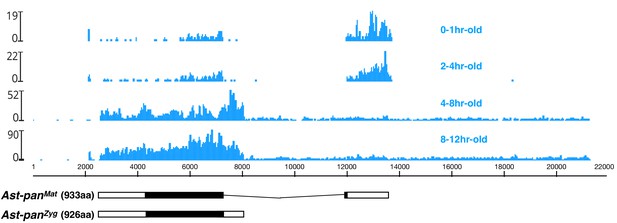
Stage-specific RNA-seq read coverage of Ast-pan genomic locus and sketches of Ast-panMat and Ast-panZyg transcripts.
Black box, open reading frame; white box, untranslated regions. See also legend to Figure 1A.
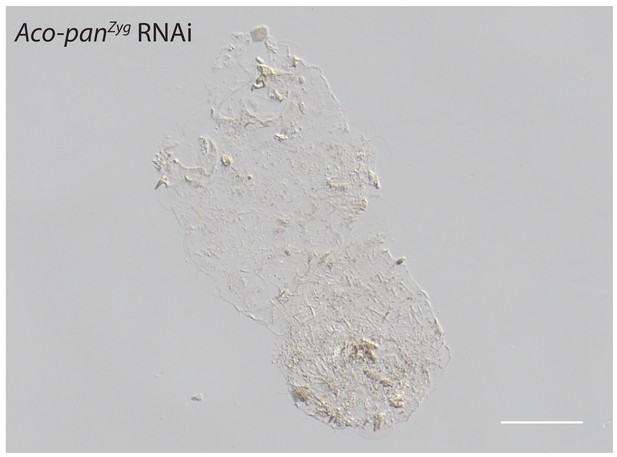
Cuticle phenotype of a A.coluzzii 1 st instar larva following Aco-panZyg RNAi (11/23).
Phenotype difference between Aco-panMat RNAi and Aco-panZyg is significant; p<0.05, Fisher’s exact test. See Figure 5F for a WT cuticle. Scale bar: 100μm.
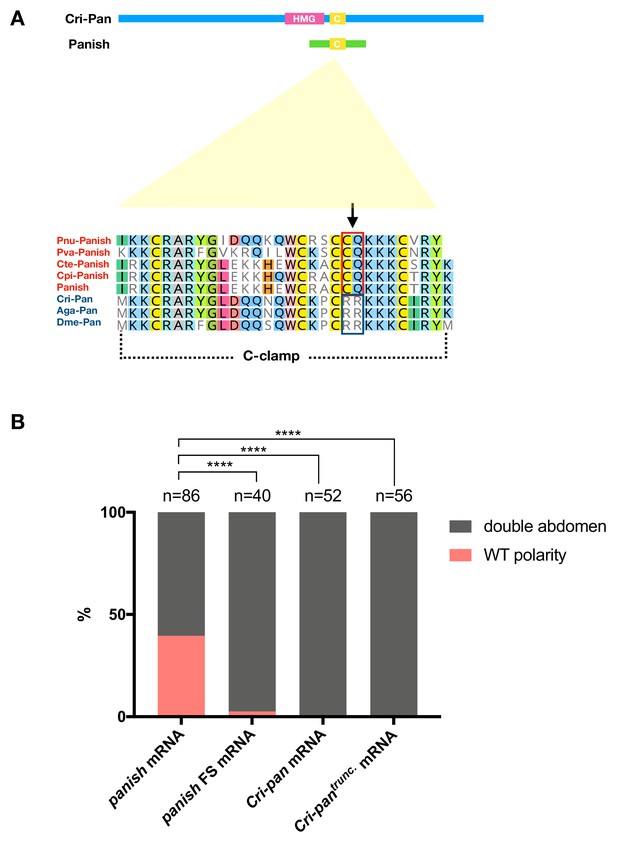
panish RNAi rescue experiments with Cri-pan mRNA.
(A) Sketches of Cri-Pan and Panish (HMG: HMG-box domain; C: Cysteine-clamp domain) and protein alignment of conserved C-clamp domains between Panish and Pan orthologs. Panish-specific sequence change (RR->CQ) is indicated as an arrow. Pnu: Polypedilum nubifer; Pva: Polypedilum vanderplanki; Cte: Chironomus tentans; Cpi: Chironomus piger; Cri: Chironomus riparius; Aga: Anopheles gambiae; Dme: Drosophila melanogaster. (B) panish RNAi rescue experiments using panish mRNA, panish FS (frameshift mutation) mRNA, Cri-pan mRNA, and Cri-pantrunc mRNA. Cri-pantrunc mRNA encodes Cri-Pan protein that lacks N-terminal sequences before C-clamp domain (N-terminal 365 aa deleted, including HMG-box domain) and with Panish-specific sequence change (RR->CQ) in C-clamp domain, as in (A).
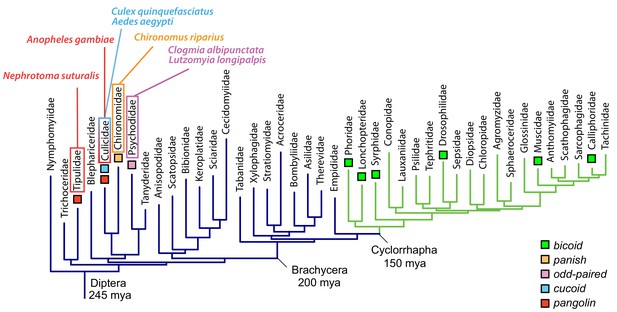
Anterior axis determinants in Diptera.
The phylogenetic tree of dipteran families is based on published data and shows Cylorrhapha in green (Wiegmann et al., 2011). Mya, million years ago.
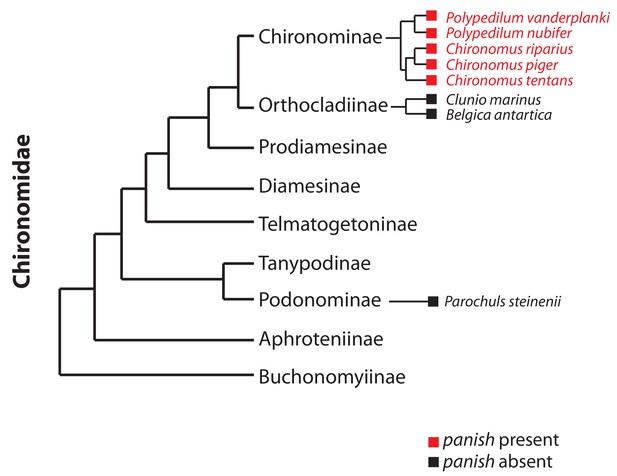
Occurrence of panish in Chironomidae genomes.
Orthologs of panish in Chironomus piger and C. tentans have been reported previously (Klomp et al., 2015). We examined panish in recently published genome sequences of Polypedilum vanderplanki (Gusev et al., 2014), P. nubifer (Gusev et al., 2014), Clunio marinus (Kaiser et al., 2016), Belgica antarctica (Kelley et al., 2014), and Parochuls steinenii (Kim et al., 2017). The phylogeny is based on Cranston et al. (2012).
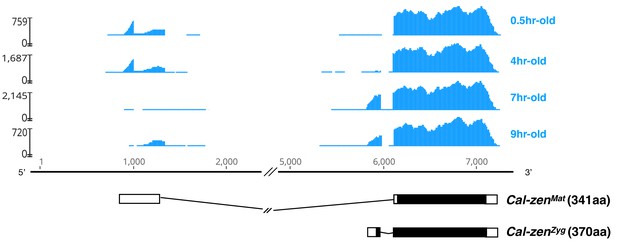
Maternal and zygotic transcript variant of Clogmia zerknüllt (Cal-zen).
Stage-specific RNA-seq read coverage of Cal-zen genomic locus above schematic sketches of Cal-zenMat and Cal-zenZyg transcript variants with open reading frame (black) and recovered UTR sequences. The RNA-seq reads were obtained from preblastoderm (0.5 hr-old), syncytial blastoderm (4 hr-old), cellular blastoderm (7 hr-old), and gastrulation stages (9 hr-old). Maternal zerknüllt expression is common in lower Diptera but was probably lost in the stem lineage of Cyclorrhapha (Stauber et al., 2002). It is possible that zerknüllt evolved a localization signal in this stem lineage, because in Megaselia abdita (Phoridae), a cyclorrhaphan fly with maternal bicoid expression and zygotic zerknüllt expression, the maternal bicoid transcript is localized at the anterior pole of the embryo while the zygotic zerknüllt transcript is localized on the apical side of blastoderm cells (Stauber et al., 1999), presumably through the same microtubule-dependent machinery (Bullock and Ish-Horowicz, 2001).
Videos
Live double head embryo in ventrolateral view.
The larval cuticle of this embryo is shown in Figure 3—figure supplement 1.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Nephrotoma suturalis) | Nsu | https://doi.org/10.1083/jcb.25.1.95 | NA | |
| Strain, strain background (Chironomus riparius) | Cri | https://doi.org/10.1126/science.aaa7105; https://doi.org/10.1111/mec.1411 | CRIP_Laufer | |
| Strain, strain background (Clogmia albipunctata) | Cal | https://doi.org/10.1007/s004270050238 | NA | |
| Strain, strain background (Culex quinquefasciatus) | Cqu | NIAID | NA | |
| Strain, strain background (Anopheles gambiae) | A.gambiae | NIAID | G-3 strain | |
| Strain, strain background (Anopheles coluzzii) | A.coluzzii | Insect transformation facility, University of Maryland | NA | |
| Strain, strain background (Aedes aegypti) | Aae | NIAID | Liverpool ‘Black eye’ | |
| Gene (Clogmia albipunctata) | Cal-opaMat | This paper | MN122104 | See Materials and methods - Cloning procedures and mRNA/dsRNA synthesis; RNA in situ hybridization;Rapid Amplification of cDNA Ends (RACE) |
| Gene (Clogmia albipunctata) | Cal-opaZyg | This paper | MN122105 | See Materials and methods - Cloning procedures and mRNA/dsRNA synthesis; RNA in situ hybridization; Rapid Amplification of cDNA Ends (RACE) |
| Gene (Clogmia albipunctata) | Cal-slp | This paper | MN122106 | See Materials and methods - Cloning procedures and mRNA/dsRNA synthesis; RNA in situ hybridization |
| Gene (Clogmia albipunctata) | Cal-mira | This paper | MN122107 | See Materials and methods - Cloning procedures and mRNA/dsRNA synthesis; RNA in situ hybridization |
| Gene (Clogmia albipunctata) | Cal-otd | This paper | MN122108 | See Materials and methods - RNA in situ hybridization |
| Gene (Clogmia albipunctata) | Cal-nos1 | This paper | MN122109 | See Materials and methods - RNA in situ hybridization |
| Gene (Clogmia albipunctata) | Cal-nos2 | This paper | MN122110 | See Materials and methods - RNA in situ hybridization |
| Gene (Clogmia albipunctata) | Cal-nos3 | This paper | MN122111 | See Materials and methods - RNA in situ hybridization |
| Gene (Clogmia albipunctata) | Cal-nos4 | This paper | MN122112 | See Materials and methods - RNA in situ hybridization |
| Gene (Clogmia albipunctata) | Cal-vas | This paper | MN122113 | See Materials and methods - RNA in situ hybridization |
| Gene (Clogmia albipunctata) | Cal-tud | This paper | MN122114 | See Materials and methods - RNA in situ hybridization |
| Gene (Clogmia albipunctata) | Cal-gcl | This paper | MN122115 | See Materials and methods - RNA in situ hybridization |
| Gene (Lutzomyia longipalpis) | Llo-opaMat | This paper | MN122116 | See Materials and methods - Rapid Amplification of cDNA Ends (RACE) |
| Gene (Lutzomyia longipalpis) | Llo-opaZyg | This paper | MN122117 | See Materials and methods - Rapid Amplification of cDNA Ends (RACE) |
| Gene (Lutzomyia longipalpis) | Llo-osk | This paper | MN122118 | See Materials and methods - RNA in situ hybridization |
| Gene (Chironomus riparius) | Cri-opa | This paper | MN122119 | See Materials and methods - RNA in situ hybridization |
| Gene (Culex quinquefasciatus) | cucoidA | This paper | MN122120 | See Materials and methods - RNA in situ hybridization |
| Gene (Culex quinquefasciatus) | cucoidB | This paper | MN122121 | See Materials and methods - RNA in situ hybridization |
| Gene (Culex quinquefasciatus) | cucoidC | This paper | MN122122 | See Materials and methods - Cloning procedures and mRNA/dsRNA synthesis; RNA in situ hybridization |
| Gene (Culex quinquefasciatus) | Cqu-nos | This paper | MN122123 | See Materials and methods - RNA in situ hybridization |
| Gene (Aedes aegypti) | Aae-cucoidA | This paper | MN122124 | See Materials and methods - Rapid Amplification of cDNA Ends (RACE) |
| Gene (Aedes aegypti) | Aae-cucoidB | This paper | MN122125 | See Materials and methods - Rapid Amplification of cDNA Ends (RACE) |
| Gene (Aedes aegypti) | Aae-cucoidC | vectorbase.org | AAEL013321 | See Materials and methods - Rapid Amplification of cDNA Ends (RACE) |
| Gene (Aedes aegypti) | Aae-nos | vectorbase.org | AAEL012107 | See Materials and methods - RNA in situ hybridization |
| Gene (Anopheles gambiae) | Aga-panMat | This paper | MN122126 | See Materials and methods - Cloning procedures and mRNA/dsRNA synthesis; RNA in situ hybridization; Rapid Amplification of cDNA Ends (RACE) |
| Gene (Anopheles gambiae) | Aga-panZyg | This paper | MN122127 | See Materials and methods - Cloning procedures and mRNA/dsRNA synthesis; RNA in situ hybridization; Rapid Amplification of cDNA Ends (RACE) |
| Gene (Anopheles gambiae) | Aga-cad | This paper | MN122128 | See Materials and methods - RNA in situ hybridization |
| Gene (Nephrotoma suturalis) | Nsu-pan | This paper | MN122129 | See Materials and methods - RNA in situ hybridization |
| Sequence-based reagent | dsRNAs | This paper | see Materials and methods - Cloning procedures and mRNA/dsRNA | |
| Sequence-based reagent | RNA probes | This paper | See Materials and methods - RNA in situ hybridization | |
| Commercial assay or kit | SMARTer RACE 5'/3' Kit | Takara | 634858 | |
| Commercial assay or kit | mMESSAGE mMACHINE SP6 | Thermo Fisher | AM1340 | |
| Commercial assay or kit | QuikChange Lightning Site-Directed Mutagenesis Kit | Agilient | 210515 | |
| Software, algorithm | Geneious | https://www.geneious.com | RRID:SCR_010519 | version 11.1.5 |
| Software, algorithm | GraphPad Prism | https://graphpad.com | RRID:SCR_015807 | version 1.4 |
Additional files
-
Supplementary file 1
Annotation and quantitation of Clogmia transcriptome.
Gene names are those indicated as best-reciprocal-blast hits (see Methods). Contig names correspond to transcriptome assembly. Read counts are given based on non-unique mapping of pre-processed RNA-seq data to annotated transcriptome.
- https://doi.org/10.7554/eLife.46711.026
-
Supplementary file 2
Annotation and quantitation of Lutzomyia transcriptome.
Gene names are those indicated as best-reciprocal-blast hits (see Methods). Contig names correspond to transcriptome assembly. Read counts are given based on non-unique mapping of pre-processed RNA-seq data to annotated transcriptome.
- https://doi.org/10.7554/eLife.46711.027
-
Supplementary file 3
Annotation and quantitation of Culex transcriptome.
Gene names are those indicated as best-reciprocal-blast hits (see Methods). Contig names correspond to transcriptome assembly. Read counts are given based on non-unique mapping of pre-processed RNA-seq data to annotated transcriptome.
- https://doi.org/10.7554/eLife.46711.028
-
Supplementary file 4
Annotation and quantitation of Aedes transcriptome.
Gene names are those indicated as best-reciprocal-blast hits (see Methods). Contig names correspond to transcriptome assembly. Read counts are given based on non-unique mapping of pre-processed RNA-seq data to annotated transcriptome.
- https://doi.org/10.7554/eLife.46711.029
-
Supplementary file 5
Annotation and quantitation of Anopheles transcriptome.
Gene names are those indicated as best-reciprocal-blast hits (see Methods). Contig names correspond to transcriptome assembly. Read counts are given based on non-unique mapping of pre-processed RNA-seq data to annotated transcriptome.
- https://doi.org/10.7554/eLife.46711.030
-
Supplementary file 6
Annotation and quantitation of Nephrotoma transcriptome.
Gene names are those indicated as best-reciprocal-blast hits (see Methods). Contig names correspond to transcriptome assembly. Read counts are given based on non-unique mapping of pre-processed RNA-seq data to annotated transcriptome.
- https://doi.org/10.7554/eLife.46711.031
-
Transparent reporting form
- https://doi.org/10.7554/eLife.46711.032





