A bacterial riboswitch class for the thiamin precursor HMP-PP employs a terminator-embedded aptamer
Figures
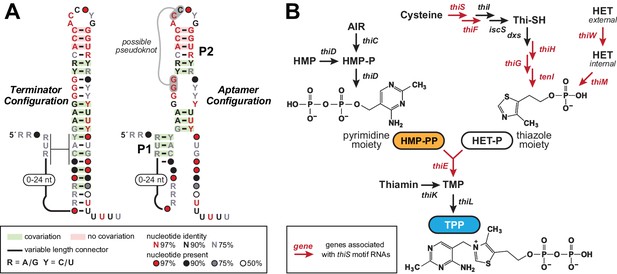
The thiS motif consensus models and gene associations.
(A) Conserved nucleotide sequences and two possible secondary structure models based on 400 representatives of thiS motif RNAs. (Left) The terminator configuration exhibits features (strong stem followed by a run of U nucleotides) that are consistent with a typical bacterial intrinsic transcription terminator stem (‘OFF’ state). (Right) Another secondary structure model (‘ON’ state) for thiS motif RNAs, consistent with the pattern of conserved base-pairing potential, involves the formation of a distinct P1 stem and a possible pseudoknot, whereas the base-pairing of P2 is identical to the tip of the terminator stem configuration. (B) Typical biosynthetic pathways for the pyrimidine and thiazole moieties of thiamin, and their eventual conversion into the enzyme cofactor thiamin pyrophosphate (TPP). Genes coding for the enzymes involved in HET transport and catalysis of various biosynthetic steps are annotated with colors that reflect their association with thiS motif representatives. Note that a carboxylated derivative of HET-P, called cHET-P, has been proposed to be the main thiazole-based biosynthetic intermediate for TPP in some bacteria (Begley et al., 2012).
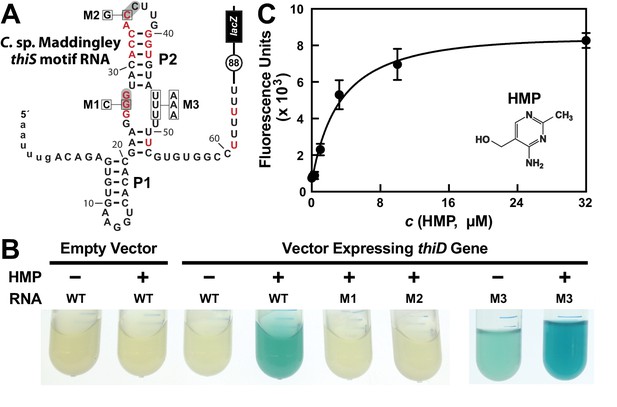
HMP-PP triggers reporter gene expression.
(A) Sequence and predicted secondary structure of the WT thiS motif RNA associated with the thiS gene of C. sp. Maddingley fused to a β-galactosidase reporter gene (lacZ). The structural model is depicted in its presumed ‘ON’ state, where the shaded nucleotides highlight possible base-pairs of the predicted pseudoknot. The encircled number represents the additional nucleotides linking the end of the intrinsic terminator element (6 U nucleotides) and the coding region joined to the lacZ reporter gene sequence. Red nucleotides are >97% conserved as depicted in the thiS consensus model (Figure 1A). The nucleotides are numbered beginning with the predicted natural transcription start site, and the lowercase letters at the 5′ end identify additional nucleotides included in the reporter-fusion construct. Boxed nucleotides identify positions that are altered in the mutant constructs indicated. (B) Photos of reporter gene assay liquid cultures of B. subtilis cells carrying the protein expression plasmid pDG148-Stu either lacking (empty vector) or carrying a native thiD gene. Reporter constructs carrying the WT riboswitch construct or mutations M1 through M3 as depicted in A were grown in LB either without (‒) or with (+) HMP supplementation. (C) HMP concentration dependent reporter gene expression of WT B. subtilis cells carrying the riboswitch-lacZ fusion construct depicted in A. Cells were grown in minimal (GMM) liquid medium supplemented with various concentrations of HMP as indicated. Data points are the average of three measurements and are representative of experiments performed on multiple days. Error bars indicate the standard deviation of the measurements, which when not visible are smaller than the corresponding data points.
-
Figure 2—source data 1
Fluorescence values for reporter gene expression data for Figure 2C.
- https://doi.org/10.7554/eLife.45210.006
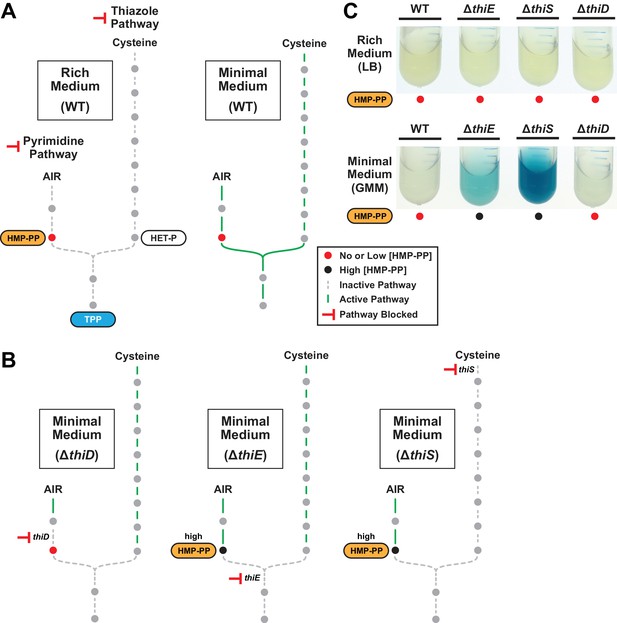
The effects of thiamin biosynthetic pathway knockouts on expression of a thiS riboswitch-reporter construct.
(A) Schematic representation of the expected metabolic activities of the pyrimidine and thiazole pathways for TPP biosynthesis in rich (LB, left) and minimal (GMM, right) media. Solid green lines indicate pathways with flux, whereas dashed gray lines represent pathways with low or no flux. Shaded circles represent molecules in the pathway. (B) Schematic representation of the expected metabolic flux of thiamin knockout strains grown in minimal medium. Other annotations are as described for panel A. (C) Reporter gene expression of WT B. subtilis cells and cells lacking the coding region for various thiamin biosynthesis genes grown in rich or minimal liquid media.
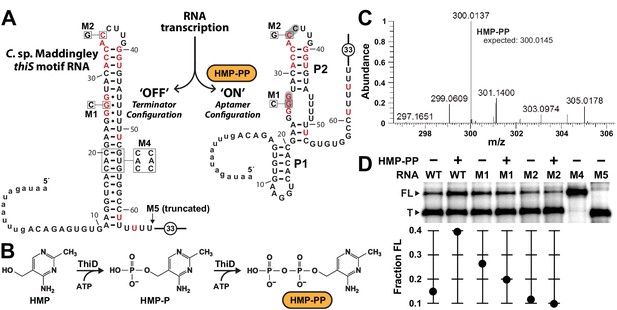
Pyrophosphorylated HMP triggers riboswitch-mediated transcription elongation in vitro.
(A) Sequence and secondary structure models for the ‘OFF’ and ‘ON’ states of an HMP-PP riboswitch construct from C. sp. Maddingley used for transcription termination assays. In the absence of ligand (‘OFF’ state, left), an intrinsic transcription terminator stem is expected to form and cause transcription termination near nucleotide number 66. In the ligand-bound structure (‘ON’ state, right), a distinct P1 stem and a P2 stem (partial formation of the terminator) are expected to form, thereby blocking formation of the complete terminator stem and leading to transcription read-through. (B) Pathway for the enzymatic pyrophosphorylation of HMP using 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate kinase (ThiD), carried out as previously described (Hanes et al., 2007). (C) Plots of the mass-to-charge ratios (m/z) of HMP and HMP-PP prepared by enzymatic pyrophosphorylation. Relative abundance (peak heights) are normalized to that of HMP-PP. (D) (Top) PAGE analysis of single-round transcription termination assays of WT and various mutant HMP-PP riboswitch constructs in the absence or presence of ligand. ‘FL’ and ‘T’ denote the full length and terminated transcripts, respectively. Transcription reactions in lanes denoted ‘(‒) HMP-PP’ were supplemented with the processed reaction mixture including all the reagents (except ThiD protein) required for the preparation of HMP-PP. Thus, these lanes include ~0.5 mM HMP. (Bottom) Values for the fraction of full-length RNA transcript relative to the total transcription yield are plotted for each reaction.
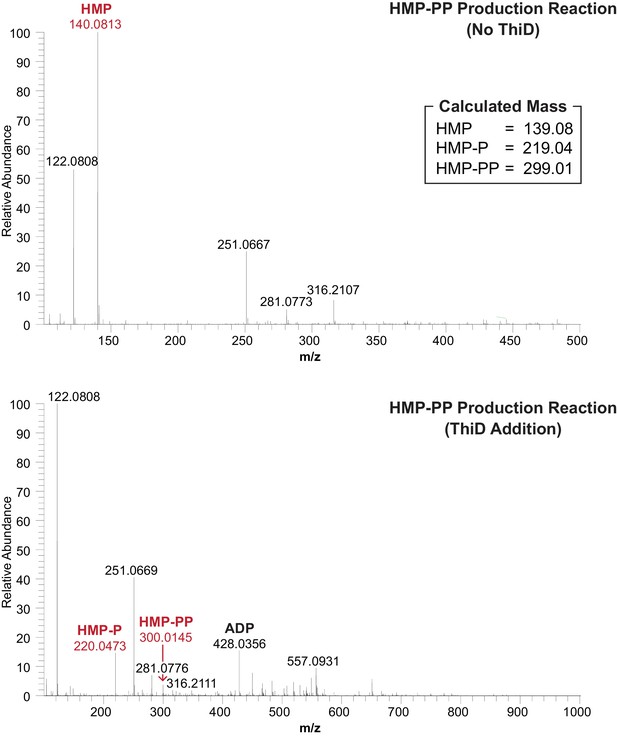
Mass spectrum analyses of the HMP-PP enzymatic biosynthesis reactions before (top) and after (bottom) the addition of ThiD protein.
https://doi.org/10.7554/eLife.45210.008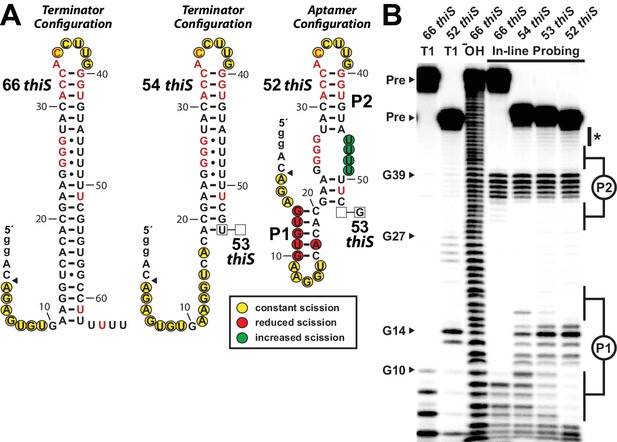
Substantial conformational rearrangement of an HMP-PP riboswitch is brought about by single-nucleotide differences at the 3′ terminus.
(A) Sequences and secondary structure models of RNA constructs used to assess conformational determination between terminator and aptamer structures. Nucleotide differences between constructs are boxed, and empty boxes indicate the nucleotide is absent. The secondary structure models were predicted based on the in-line probing data derived from B. Note that the 53 thiS RNA secondary structure model adopts both the terminator (predominant form for 54 thiS) and aptamer (predominant form for 52 thiS) conformations with near equal probability. Yellow circles identify nucleotide linkages that undergo robust spontaneous strand scission. Some remain unstructured regardless of the length of the construct, suggesting these nucleotides always remain unstructured. Red circles identify linkages that exhibit reduced spontaneous scission compared to larger constructs, suggesting the corresponding nucleotides become more structured in the 52 thiS construct. Green circles identify linkages undergoing increased strand scission, suggesting these nucleotides become less structured in 52 thiS construct. The arrowhead identifies the start of the interpretable data in B. (B) PAGE analysis of 5′ 32P-radiolabeled RNA constructs subjected to in-line probing analysis in the absence of HMP-PP. T1 and ‒ OH indicate RNase T1 partial digestion (cleavage after G nucleotides) and alkaline-mediated partial digestion (cleaves after each nucleotide), respectively. Bands corresponding to precursor RNA (Pre) and enzymatic cleavage after certain G residues are annotated. The asterisk identifies bands corresponding to nucleotides 46–49 of 52 thiS, which appear to increase in intensity compared to the 66 thiS construct.
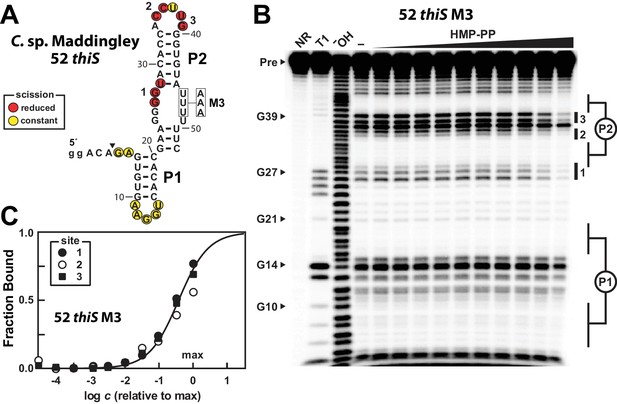
A modified representative HMP-PP riboswitch aptamer binds HMP-PP.
(A) Sequence and secondary structure model of the 52 thiS RNA construct derived from the thiS gene of C. sp. Maddingley, and its M3 mutant (U46A, U47A, U48A). Both the WT and M3 constructs are known to retain gene control activity in response to ligand (Figure 2). Nucleotides at the 5′ end denoted in lowercase letters were added to enhance the efficiency of transcription in vitro. The characteristics of sites of spontaneous RNA cleavage resulting from in-line probing reactions in the absence or presence of HMP-PP, as derived from the image in B, are indicated. (B) PAGE analysis of 5′ 32P-radiolabeled 52 thiS M3 RNA that was subjected to in-line probing assays in the absence (‒) or presence of HMP-PP. The ligand HMP-PP was enzymatically produced using ThiD protein and used in a range of concentrations relative to the maximum HMP-PP concentration (precise concentration unknown, see Materials and methods). The lane denoted "‒" was supplemented with the processed reaction mixture including all the reagents (except ThiD protein) required for the preparation of HMP-PP. Thus, this lane includes ~0.5 mM HMP. NR, T1 and ‒OH indicate no reaction, RNase T1 partial digestion (cleavage after G nucleotides), and alkaline-mediated partial digestion (cleaves after each nucleotide), respectively. Bands corresponding to precursor RNA (Pre) and enzymatic cleavage after certain G residues are annotated. Notable sites of ligand-mediated structural modulation are numbered 1 through 3. (C) Plot depicting the fraction of RNA bound to ligand versus the logarithm of the concentration of HMP-PP relative to the maximum (max) ligand concentration tested. The data points were generated by quantifying band intensity changes at sites 1, 2 and 3 in B with correction for loading and yield differences, including the general reduction in RNA cleavage observed in the final lane containing the maximum HMP-PP concentration tested. The line presents a theoretical 1:1 binding curve (Hill coefficient of 1) and is overlaid on the data points for comparison.
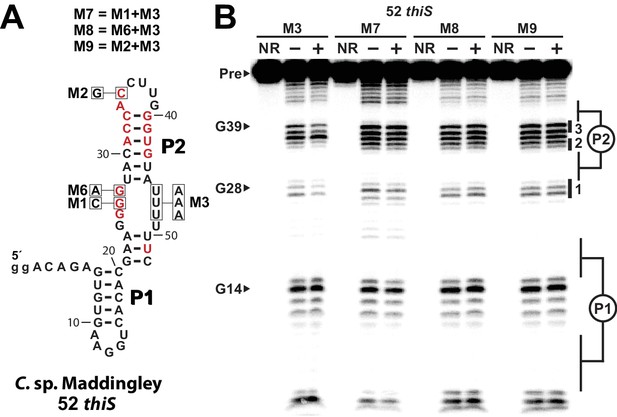
Mutational analysis of an HMP-PP aptamer.
(A) Sequence and secondary structure model of the 52 thiS parent sequence and the mutations used to form the variant constructs. Nucleotides in red identify the highly conserved (>97%) positions as determined by comparative sequence analysis (Figure 1A). Nucleotide positions that were mutated in each RNA construct are boxed and annotated with the mutant nucleotide identity. The mutant constructs M6 through M8 were obtained by including M1, M6, and M2 mutations in the 52 thiS M3 RNA construct, which originally exhibits direct binding of HMP-PP (Figure 5). (B) PAGE analysis of in-line probing assays performed with 52 thiS M3 RNA, or with each mutant RNA construct (M7 through M9) as indicated in the absence (‒) or presence (+) of HMP-PP. Additional annotations are as described in the legend to Figure 5.
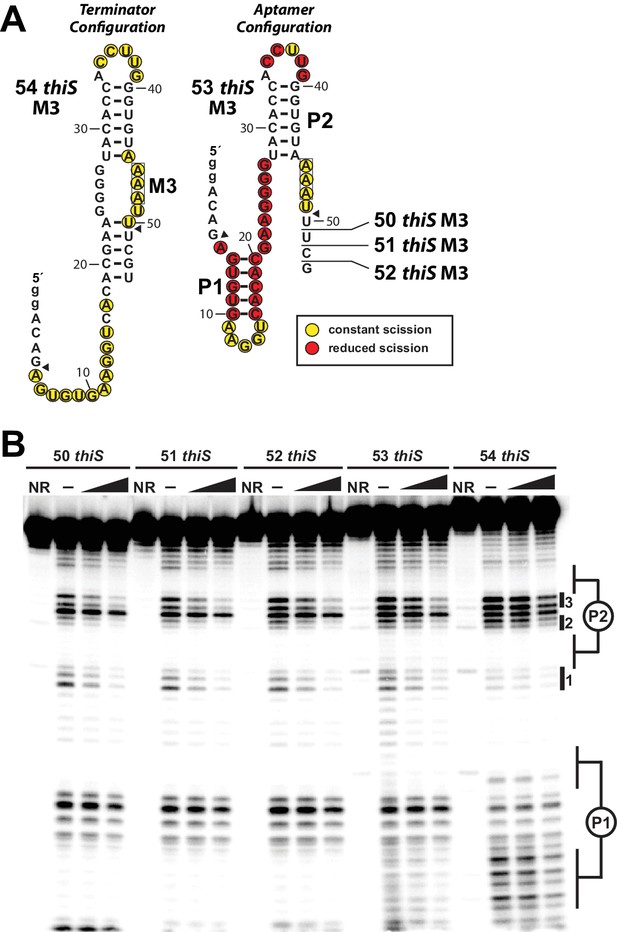
HMP-PP mediated conformational rearrangement of construct 53 thiS M3 and related RNAs.
(A) Sequences and secondary structure models for five progressively shorter thiS M3 RNA constructs. The 54 thiS M3 construct (left) is identical to the 54 thiS construct depicted in the main text (Figure 4A), except that three U nucleotides spanning positions 46–48 have been converted to A nucleotides (M3). The remaining constructs are each, consecutively, shortened one nucleotide at their 3′ termini, to yield constructs 53 thiS M3 through 50 thiS M3. Scission annotations for 54 thiS M3 are derived from the data presented in B in the absence of HMP-PP. Scission annotations for 53 thiS M3 are derived from the data presented in B, and reflect the difference in banding pattern between the (‒) and max HMP-PP lanes. (B) PAGE analysis of 5′ 32P-radiolabeled M3 mutants of 50 thiS through 54 thiS RNA constructs, subjected to in-line probing analysis in absence or presence of HMP-PP. The two concentrations of ligand used are the 1 and 1/3 of the maximum concentration (max) of HMP-PP as described in the main text (see legend to Figure 5). NR indicates no reaction and (‒) indicates no HMP-PP added. Regions of the banding pattern corresponding to the locations of stems (P1 and P2) and notable sites of modulation (1 through 3) are denoted as observed previously (Figure 4B, Figure 5B).
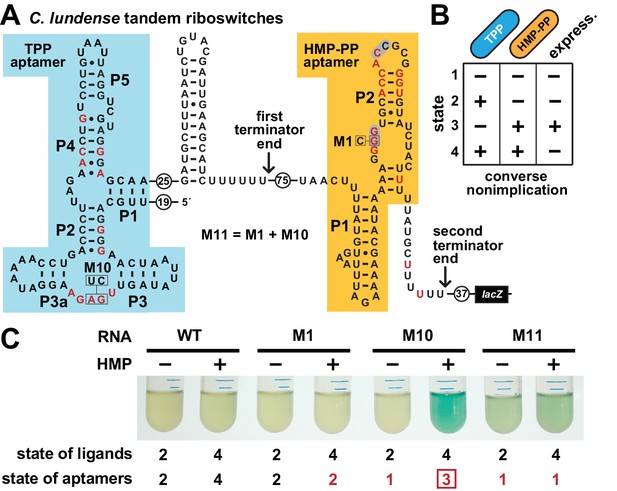
Tandem TPP and HMP-PP riboswitches form a two-input Boolean logic gate to regulate gene expression.
(A) Sequence and secondary structure of a tandem riboswitch arrangement derived from the thiW gene from C. lundense DSM 17049 that is responsive to TPP and HMP-PP ligands. Red letters identify positions matching the most highly conserved nucleotides in the consensus sequence models for TPP (McCown et al., 2017) and HMP-PP (Figure 1A) aptamers. (B) Predicted truth table for tandem TPP and HMP-PP riboswitches represents a converse nonimplication logic gate. Minus and plus symbols, respectively, represent low or high gene expression (express.) or ligand concentrations as indicated. (C) Reporter gene expression of a B. subtilis strain expressing ThiD, and carrying either a wild-type (WT) or a mutant (M1, M10 or M11) C. lundense tandem riboswitch-lacZ reporter fusion construct as described in A. Cells were grown in rich (LB) medium containing either no (‒) or 10 μM additional HMP and 200 μg mL−1 X-gal. The numbers listed as ‘state of ligands’ represents the cellular presence or absence of ligands corresponding to the ‘states’ depicted in the truth table in B. The numbers listed as ‘state of aptamers’ represents the expected status (free or ligand bound) of the aptamers in the riboswitch, again as depicted in the truth table. Red numbers identify states artificially created by riboswitch mutations. The box identifies state 3, which is created by the addition of HMP to construct M10.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Salmonella typhimurium) | thiD | NA | GenBank: AZI82914.1 | |
| Strain, strain background (Bacillus subtilis) | B. subtilis strain 168 | Bacillus Genetic Stock Center | 1A1 | |
| Strain, strain background (Escherichia coli) | BL21 (DE3) | New England Biolabs | C2527I | |
| Genetic reagent (Bacillus subtilis) | ΔthiS | Bacillus Genetic Stock Center | 11680 | |
| Genetic reagent (Bacillus subtilis) | ΔthiD | Bacillus Genetic Stock Center | 11710 | |
| Genetic reagent (Bacillus subtilis) | ΔthiE | Bacillus Genetic Stock Center | 38290 | |
| Recombinant DNA reagent | pDG1661-thiS-lacZ | This paper | HMP-PP riboswitch β-galactosidase reporter construct | |
| Recombinant DNA reagent | pDG1661-tandem-lacZ | This paper | Tandem TPP and HMP-PP riboswitch β-galactosidase reporter construct | |
| Recombinant DNA reagent | pDG148-Stu-thiD | This paper | pDG148-Stu expression vector for ThiD protein | |
| Recombinant DNA reagent | pETDuet:His-ThiD | This paper | pETDuet expression vector for N-terminal HisX6 tagged ThiD protein | |
| Commercial assay or kit | E. coli RNA polymerase holoenzyme | New England Biolabs | M0551S | |
| Chemical compound, drug | (4-amino-2-methylpyrimidin- 5-yl)methanol | Enamine Ltd | EN300-135390 | |
| Chemical compound, drug | X-Gal | MilliporeSigma | 3117073001 | |
| Chemical compound, drug | 4-Methylumbelliferyl β-D-galactopyranoside | MilliporeSigma | M1633 | |
| Chemical compound, drug | [γ-32P]-ATP | PerkinElmer | ||
| Chemical compound, drug | [α-32P]-ATP | PerkinElmer | ||
| Software, algorithm | GraphPad Prism (RRID:SCR_002798) | GraphPad Prism (https://graphpad.com) | Version 7 |
Additional files
-
Supplementary file 1
This file provides the coordinates for each HMP-PP riboswitch representative, and includes the genes associated with each representative when such annotations are available.
- https://doi.org/10.7554/eLife.45210.014
-
Supplementary file 2
This file presents sequence alignments in Stockholm format for individual HMP-PP riboswitch aptamers and for tandem TPP and HMP-PP aptamers.
- https://doi.org/10.7554/eLife.45210.015
-
Supplementary file 3
This file includes four supplementary data figures and a table listing the DNA constructs used in the study.
- https://doi.org/10.7554/eLife.45210.016
-
Transparent reporting form
- https://doi.org/10.7554/eLife.45210.017





