Arid3a regulates nephric tubule regeneration via evolutionarily conserved regeneration signal-response enhancers
Figures
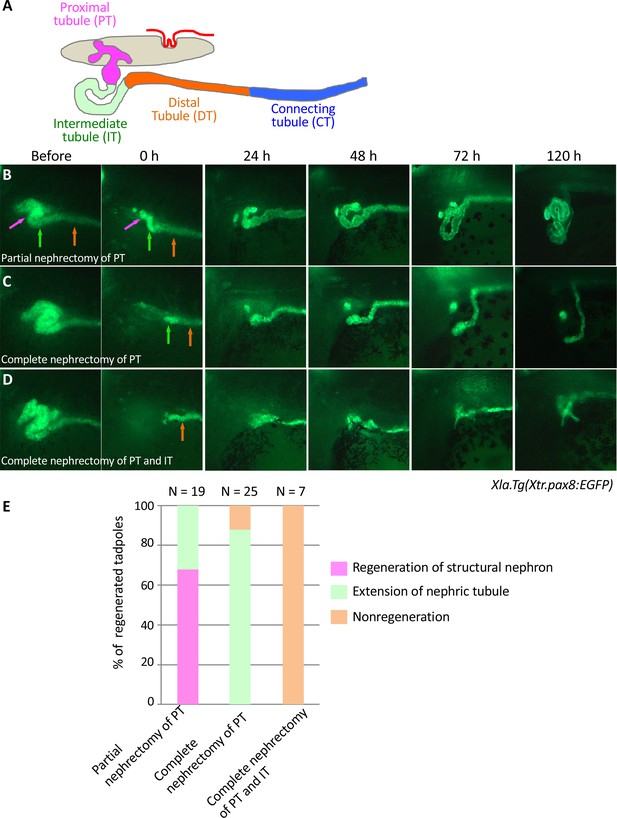
Live imaging of nephric tubules using transgenic X. laevis.
(A) A schematic image of X. laevis nephron. PT: proximal tubule; IT: intermediate tubule; DT: distal tubule; CT: connecting tubule. Magenta arrows: PT; green arrows: IT; orange arrows: DT. (B) Representative regeneration pattern of partially resected proximal tubules. Proximal tubules regenerate a coiled structure. (C) Regeneration pattern of completely resected proximal tubules. The remaining intermediate tubules extend, but no coiled structure is regenerated. (D) Regeneration pattern of completely resected intermediate tubules. No extension of tubules is observed. (E) Statistics of the regeneration pattern. The statistics of three independent experiments are summarized.
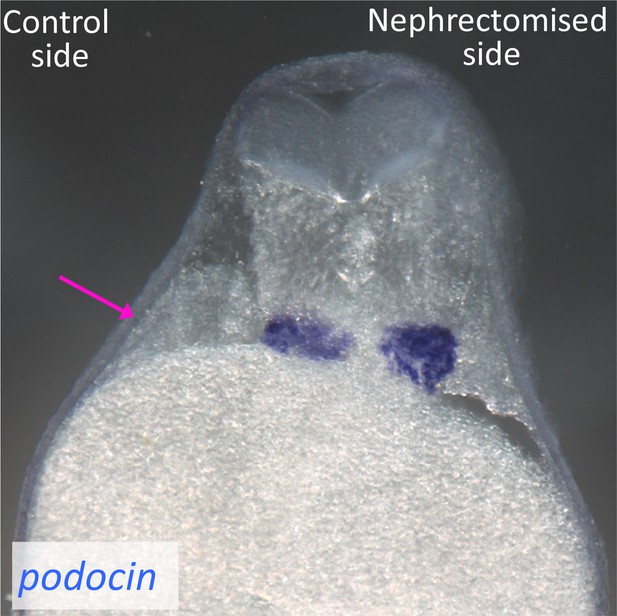
Podocytes are not injured by the surgical removal of nephron tubules.
In situ hybridization of podocin, a podocyte marker, was performed using nephrectomized X. laevis. The image shows a cross section of X. laevis at 48 hr after nephrectomy. The arrow indicates the nephric tubules on the control side.
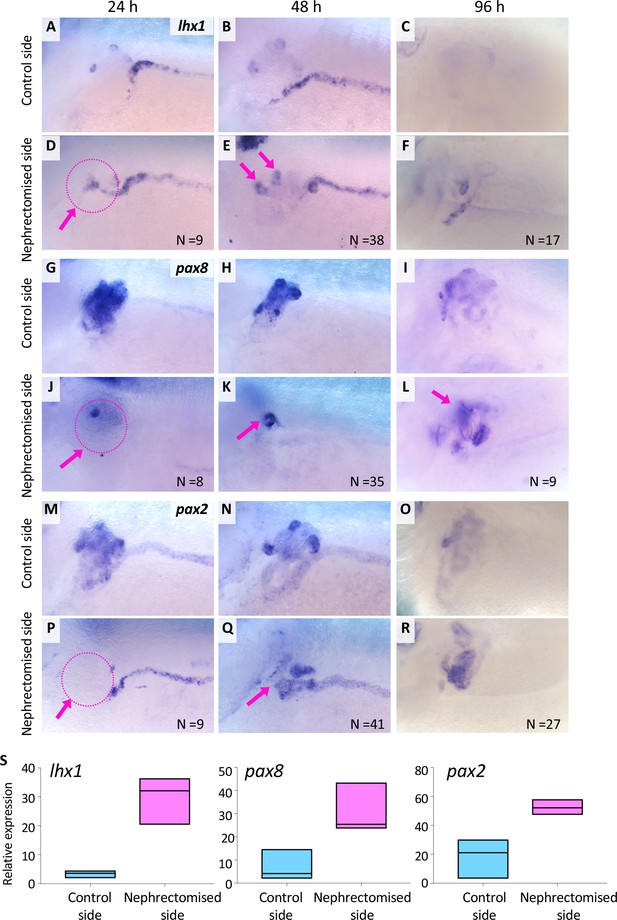
lhx1 expression appears immediately after nephrectomy.
(A–F) Expression of lhx1 on the control side and nephrectomized side at 24, 48, and 96 hr after nephrectomy. N indicates the number of examined embryos. (D) lhx1 expression appears in regenerating nephric tubules within 24 hr (arrows). (E, F) This expression becomes stronger around 48 hr but disappears at 96 hr. (G–L) Expression of pax8. pax8 expression appears in regenerating nephric tubules and is still detected at 96 hr after nephrectomy. (M–R) Expression of pax2 is not observed at 24 hr after nephrectomy (P, arrow). (S) Quantification of expression signals for lhx1, pax8, and pax2. The significance of differences between the control side and the nephrectomized side at 48 hr is calculated by two-tailed paired t-test (lhx1: p=0.0414; pax8: p=0.0102; pax2: p=0.0453). Lines in boxes indicate the median.
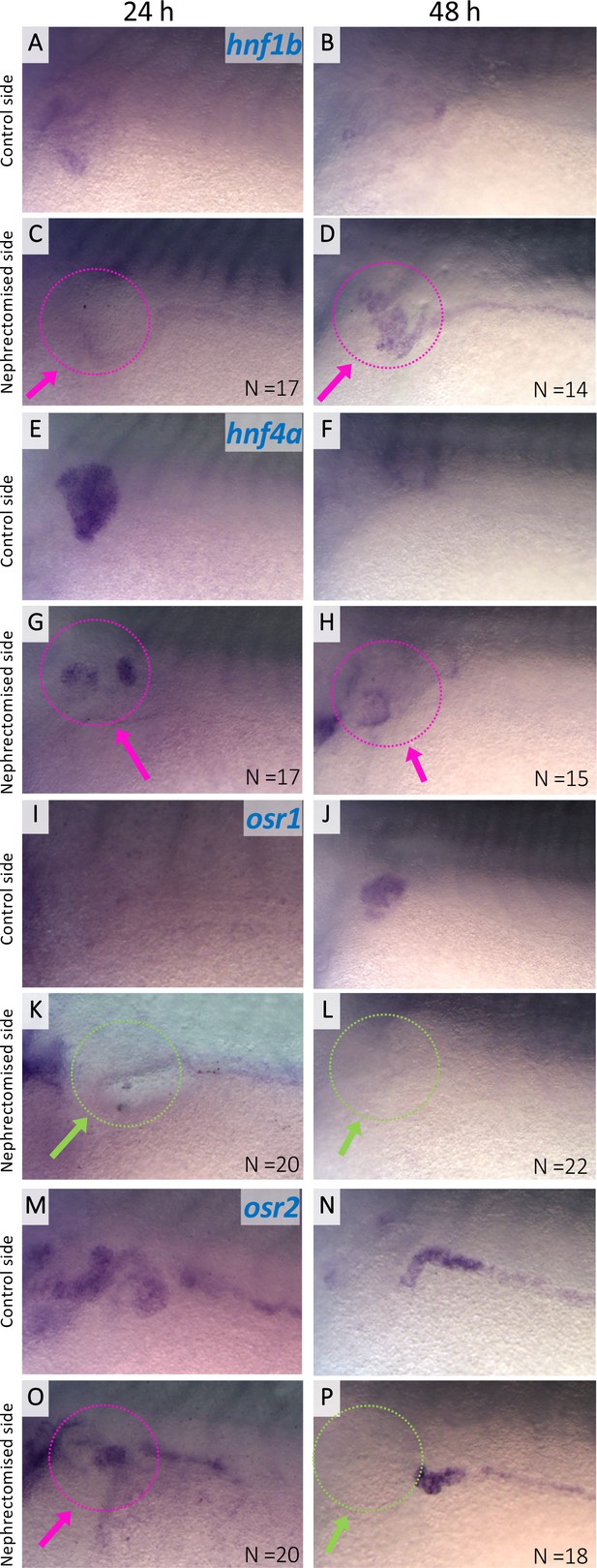
Expression of hnf1b, hnf4a, osr1, and osr2 in regenerating nephric tubules.
Expression of nephric genes at 24 hr and 48 hr after nephrectomy (24 hr: A, C, E, G, I, K, M, and O; 48 hr: B, D, F, H, J, L, N, and P). Control side (A, B, E, F, I, J, M, and N). Nephrectomized side (C, D, G, H, K, L, O, and P). The magenta arrows indicate the expression in regenerating nephric tubules. The green arrows indicate no expression in the regenerating tubules. N indicates the number of examined embryos.
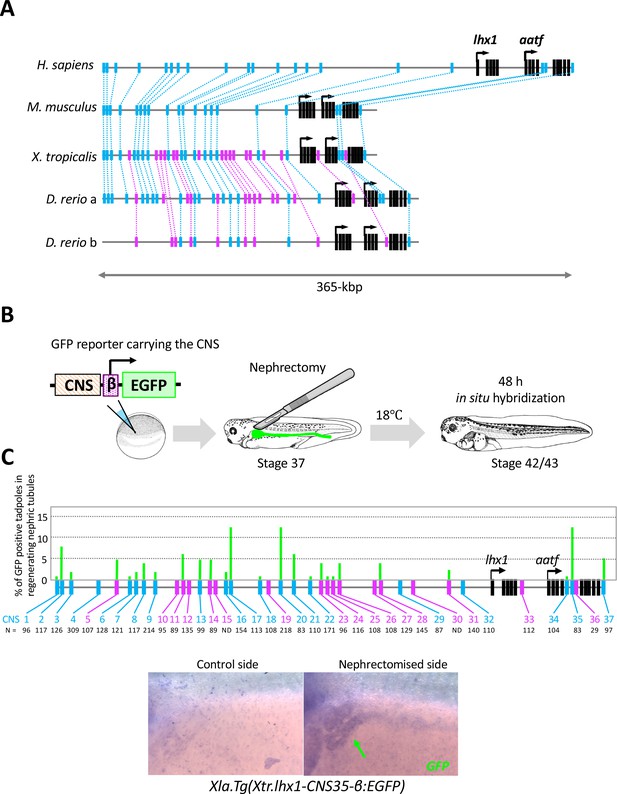
The RSRE for lhx1 is conserved between human and fish.
(A) A diagram of vertebrate lhx1 loci showing the position of CNSs. The magenta boxes indicate CNSs conserved between frog and fish, and the blue boxes indicate CNSs conserved between human and fish. The black boxes indicate the exons. (B) A diagram of the experimental design for mapping RSREs. GFP reporter constructs carrying lhx1-CNSs with the β-actin proximal promoter were subjected to transgenesis. All reporter-injected embryos underwent nephrectomy on the left side at stage 37. Nephrectomized embryos were incubated at 18°C for 48 hr and fixed at stages 42/43. Normally developed embryos were subjected to in situ hybridization in order to examine their GFP expression with maximum sensitivity. (C) A summary of RSRE screening. The green bar indicates % of GFP-positive tadpoles in regenerating nephric tubules. N indicates the number of scored tadpoles. The image shows a representative expression pattern of GFP in regenerating nephric tubules. The green arrow indicates the regenerating nephric tubule.
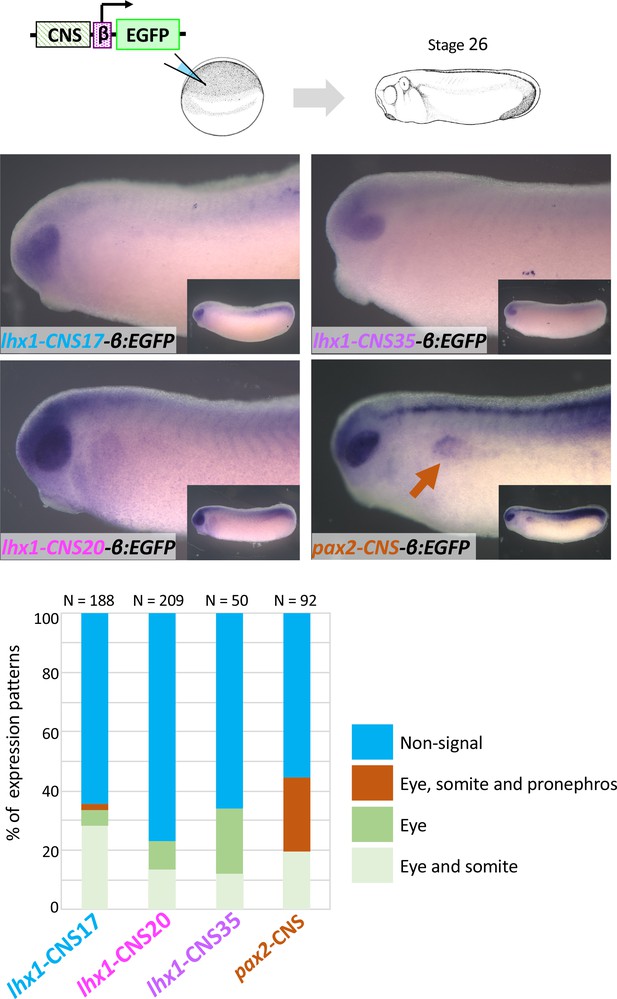
lhx1-CNS17-βEGFP, lhx1-CNS29-βEGFP, lhx1-CNS35-βEGFP and pax2-CNS45-βEGFP were subjected to the transgenic reporter analysis.
Embryos were fixed at stage 26 of which endogenous lhx1 and pax2 expression already begins. An orange arrow indicates the pronephros. N indicates the number of examined embryos. All CNS-carrying reporters were tested at least three times.
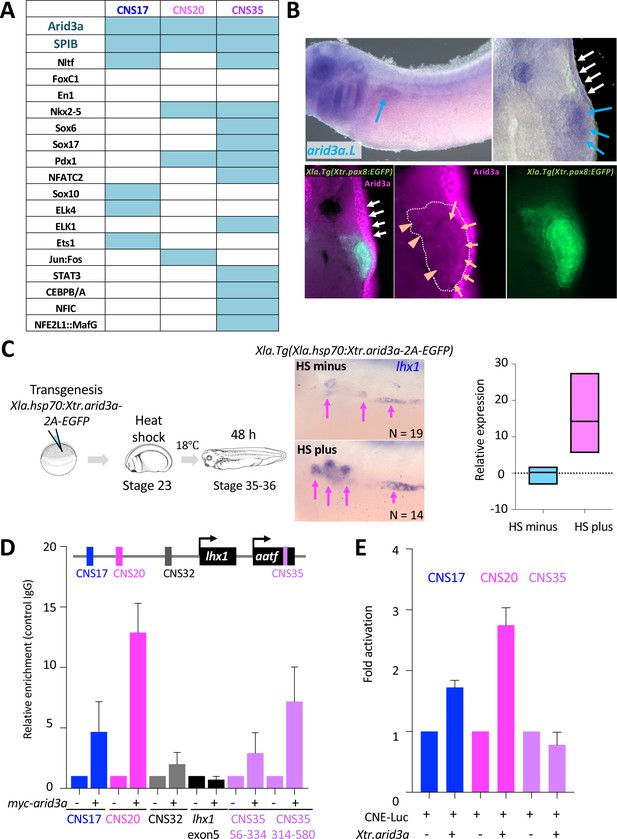
Arid3a is an input transcription factor for RSREs.
(A) A summary of transcription factor binding motifs on RSREs. (B) arid3a is expressed in nephric tubules. A lateral view of embryos at stage 31 and its transverse section. The upper panels show the in situ hybridization of arid3a.L. The blue arrows indicate the nephric tubules and the white arrows indicate the epidermis. The lower panels show immunostaining using anti-Arid3a using Xla.Tg(Xtr.pax8:EGFP) transgenic tadpole. White arrows: epidermis; orange arrows: proximal tubules; orange arrowheads: glomus and/or nephrocoelom. (C) Arid3a induces lhx1 expression. Xla.Tg(Xla.hsp70:Xtr.arid3a-2A-EGFP) transgenic X. laevis at stage 23 were treated at 34°C for 15 min, followed by 15 min at 14°C. These steps were repeated three times, and tadpoles were incubated at 18°C. lhx1 expression was observed 48 hr after the heat shock at stages 35–36. The signal intensity of in situ hybridization was measured and subjected to statistical analysis. The significance of differences between the control side and the nephrectomized side was calculated using two-tailed unpaired t-test (p=0.0131). The magenta arrows indicate lhx1 expression in proximal and intermediate tubules. N indicates the number of examined embryos. (D) Arid3a directly binds to CNS17, CNS20, and CNS35. Myc-tagged Xtr.arid3a mRNA-injected tadpoles were used for ChIP-qPCR. CNS32 and exon 5 were used as negative elements. The significance of differences between the control IgG and the anti-Myc for Arid3a was calculated using two-tailed unpaired Mann–Whitney t-test: CNS17, p=0.0286; CNS20, p=0.0022; CNS32, p=0.3143 (not significant); exon5, p=0.7000 (not significant); CNS35 (56-334), p=0.0079; CNS35 (314-580), p=0.0022. The error bars indicate SEM. (E) Arid3a activates CNS17 and CNS20. The luciferase reporter assay was performed using HEK293T cells. The significance of differences between the control vector and the CNS-containing reporter was calculated using two-tailed unpaired t-test (CNS17, p=0.0033; CNS20, p=0.0256). The error bars indicate SEM.
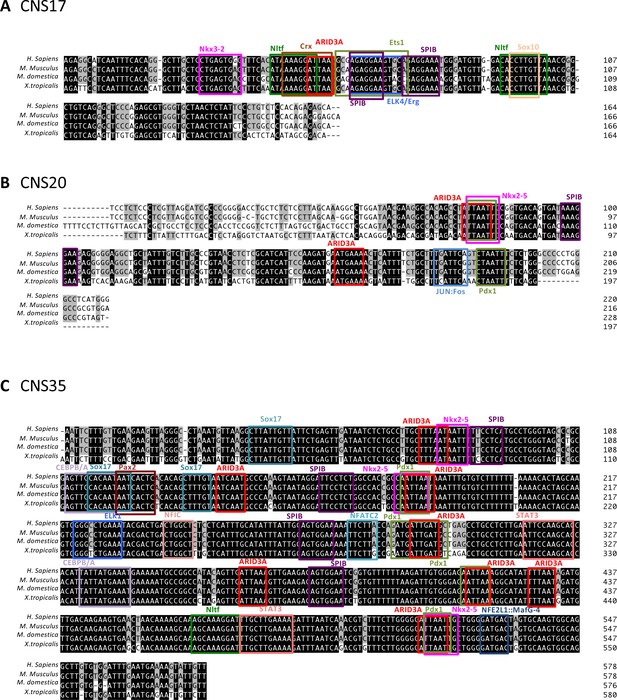
Transcription binding motifs on CNS17, CNS20, and CNS35.
(A–C) Putative transcription binding motifs on CNS17, CNS20, and CNS35. The open-access database JASPAR ver. 5 was used to search for potential transcription factor binding sites on CNS17, CNS20, and CNS35. Then, candidate transcription factors were narrowed down according to their nephric expression using the Expression Atlas. CNSs were aligned using ClustalW mounting in the GENETUX software, and conserved sequences for the candidate transcription factor binding sites were analyzed by phylogenetic footprinting.
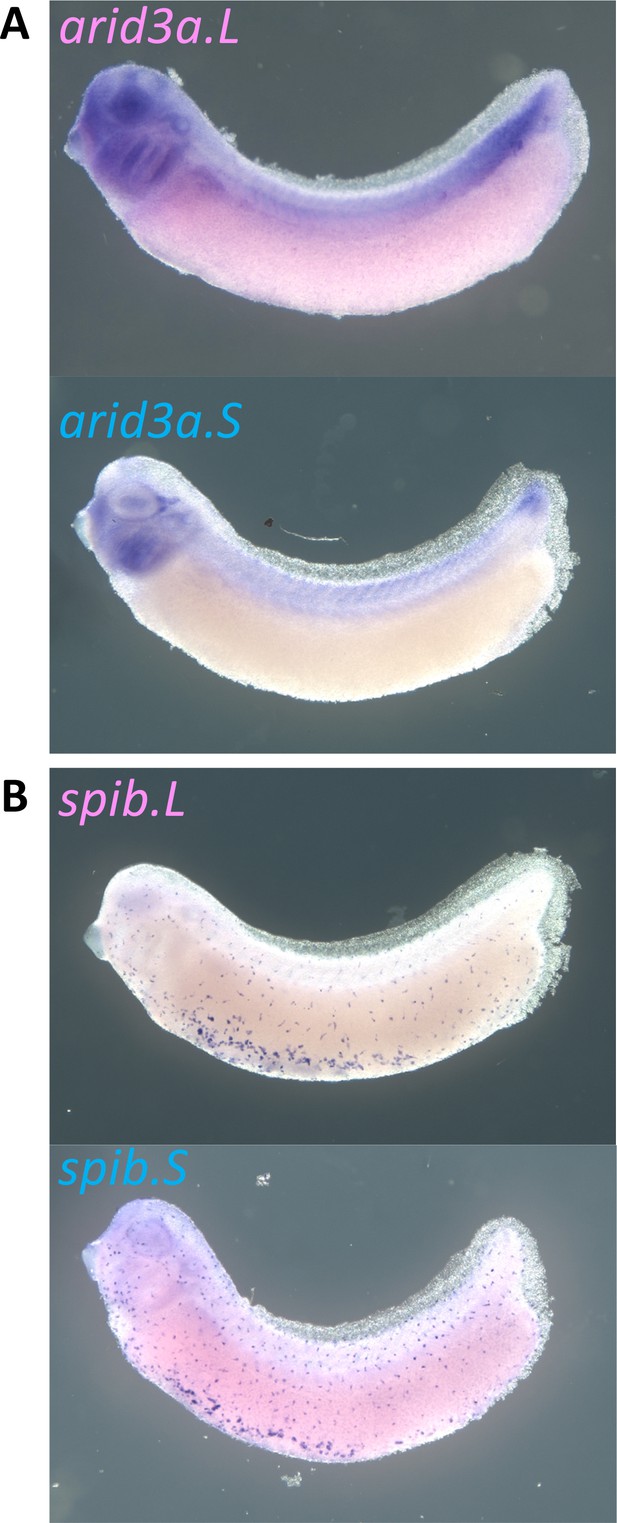
Expression of arid3a.L, arid3a.S, spib.L, and spib.S in X. laevis.
(A) Expression of arid3a.L and Arid3a.S at stage 31. (B) Expression of spib.L and spib.S at stage 31.
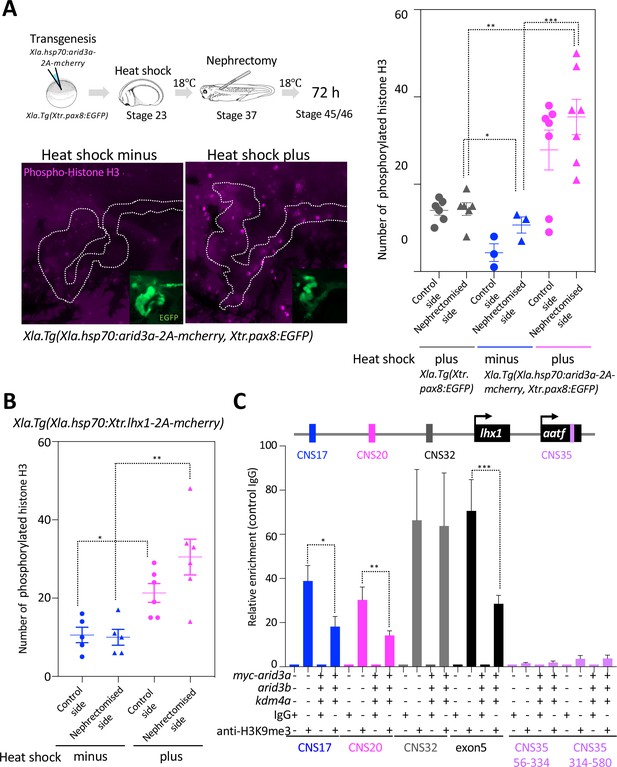
Arid3a promotes cell cycle progression.
(A) The number of phosphorylated histones H3 in the nephrectomized area was increased by the conditionally induced Arid3a. Heat-shocked Xla.Tg(Xla.hsp70:Xtr.arid3a-2A-mcherry, Xtr.pax8:EGFP) was nephrectomized at stage 37, incubated for 72 hr, and then fixed at stages 45/46. The white dotted lines indicate the pax8-expressing cells, and the magenta indicates the phosphorylated histone H3-positive cells. The left graph shows the statistical analysis; one-way analysis of variance (ANOVA) and Tukey’s post hoc test were used. ANOVA p=0.0001; *p = 0.5155 (not significant), **p = 0.001, ***p = 0.0019. The error bars indicate SEM. (B) Lhx1 promotes cell cycle progression in the regenerating area. Heat-shocked Xla.Tg(Xla.hsp70:Xtr.lhx1-2A-mcherry) was nephrectomized at stage 37 and then incubated for 72 hr. One-way ANOVA and Tukey’s post hoc test were used. ANOVA: p=0.0005, *p = 0.0015, and **p = 0.0011. (C) Arid3a with Kdm4a and Arid3b reduced the H3K9me3 levels on RSREs. ChIP analysis was performed using X. laevis. Significant differences were calculated by two-tailed unpaired t-test. The p-values from comparisons between the control and arid3a-, arid3b-, and kdm4a-injected embryos were as follows: *p = 0.0389, **p = 0.0313, and ***p = 0.0456. The error bars indicate SEM.
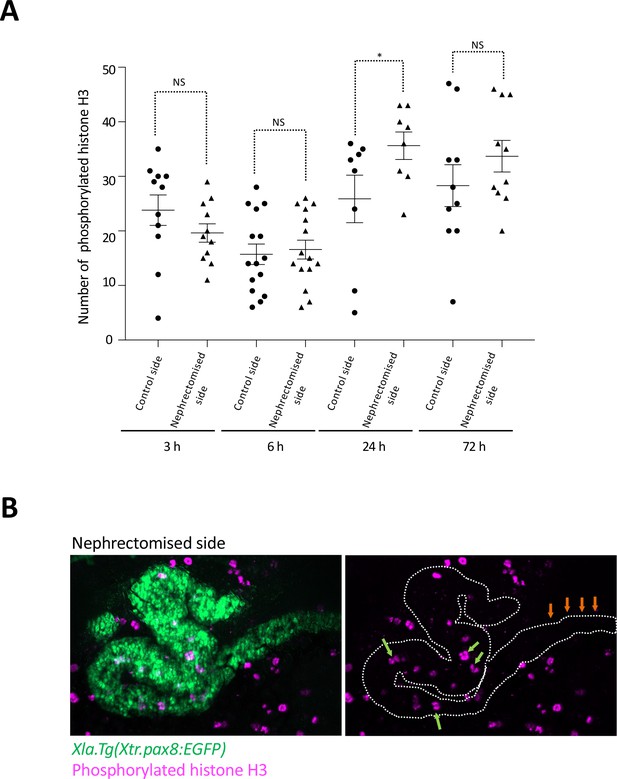
Cell cycle progression in regenerating nephric tubules frequently occurs in the remaining proximal tubule and intermediate tubule.
(A) Cell cycle progression in regenerating nephric tubules. The significance of differences between the control side and the nephrectomized side at 3, 6, 24, and 72 hr is calculated using two-tailed paired t-test (*p = 0.0078, NS: not significant). (B) Immunostaining of phosphorylated histone H3; Xlat.Tg(Xtr.pax8:EGFP) transgenic X. laevis were nephrectomized and then incubated for 24 hr. The green arrows indicate phosphorylated histone H3-positive cells in the remaining proximal tubule and intermediate tubule. The orange arrows indicate the distal tubule.
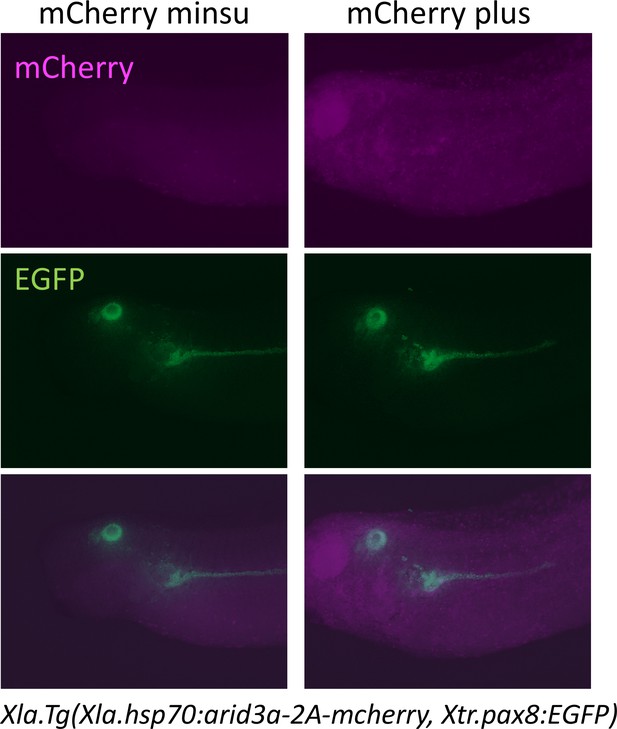
Heat shock induces the expression of mCherry.
Xla.Tg(Xla.hsp70:Xtr.arid3a-2A-mcherry, Xtr.pax8:EGFP) transgenic X. laevis at stage 23 were treated at 34°C for 15 min, followed by 15 min at 14°C. These steps were repeated three times, and tadpoles were incubated at 18°C. mCherry expression was observed at stages 36/37.
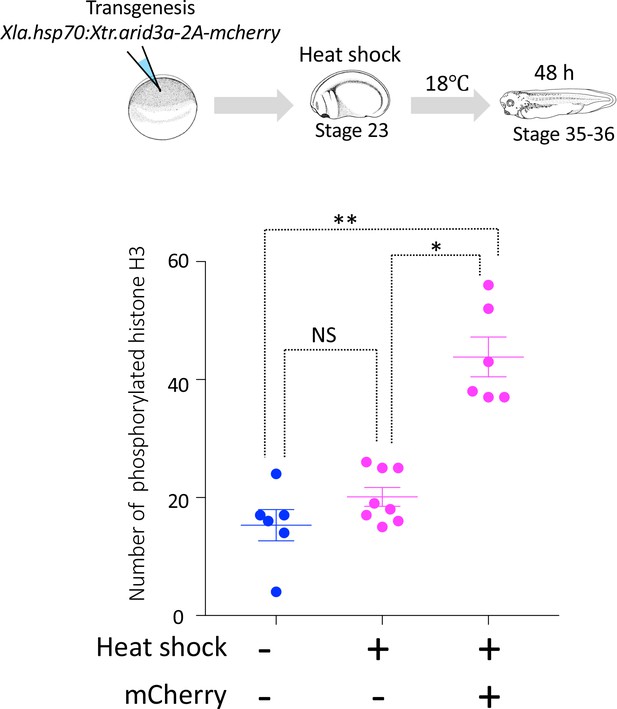
Cell cycle progression in heat-shock-untreated and heat-shock-treated tadpoles.
Xla.Tg(Xla.hsp70:Xtr.arid3a-2A-mcherry) transgenic X. laevis at stage 23 were treated at 34°C for 15 min, followed by 15 min at 14°C. These steps were repeated three times, and tadpoles were incubated at 18°C. mCherry expression was observed at stages 36/37. The significance of differences between the non-heat-shocked tadpole and heat-shocked tadpole was calculated by two-tailed paired t-test (*p = 0.0003, **p = 0.0022, NS: not significant). The error bars indicate SEM.
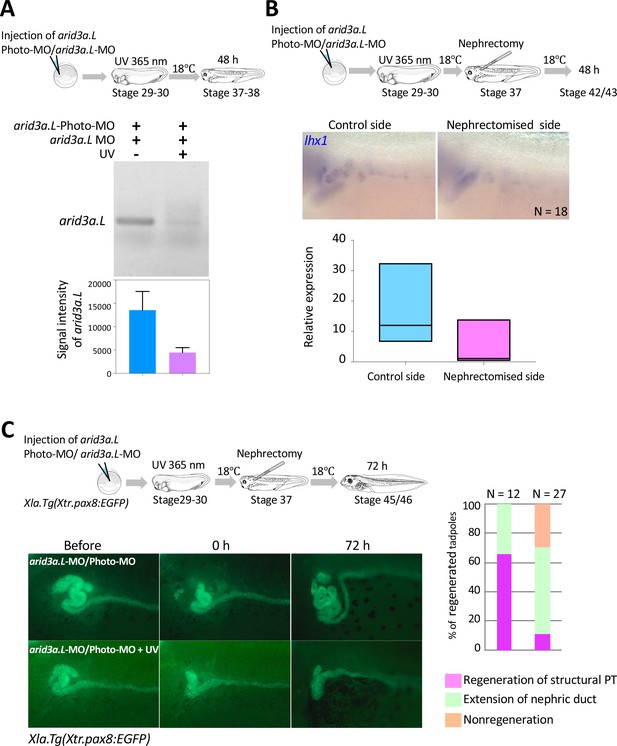
Arid3a is required for the regeneration of proximal tubules in X.laevis.
(A) Conditional knockdown of Arid3a using arid3a.L-photo-morpholinos (arid3a.L-Photo-MO). The upper panel shows the experimental design of conditional gene knockdown experiment using Photo-MO. arid3a.L-antisense-splicing-blocking MO (arid3a.L-MO) inactivated by Photo-MO is injected at the one-cell stage, subjected to UV exposure at stages 29–30, and then sacrificed for RT-PCR analyses. The lower panel shows the statistical analysis. The significance of differences between the UV-untreated and UV-treated embryos was calculated by two-tailed unpaired t-test (p=0.0217). The error bars indicate SEM. (B) Conditional knockdown of Arid3a during nephric regeneration causes the reduction of lhx1 expression. The upper panel shows the experimental design. arid3a.L-MO inactivated by Photo-MO is injected at the one-cell stage, followed by UV exposure at stages 29–30, nephrectomy at stages 36–37, and subsequently incubation for 48 hr. The lower panel shows the quantification of lhx1 expression signals. The analysis indicates that there was no significant difference between the control side and the nephrectomized side (two-tailed paired t-test, p=0.0748). N indicates the number of examined embryos. The lines in boxes indicate the median. (C) Arid3a is required for the regeneration of nephric tubules. The upper panel shows the experimental design. arid3a.L-MO inactivated by Photo-MO is injected at the one-cell stage, and UV exposure is performed at stages 29–30, followed by nephrectomy at stages 36–37 and subsequently incubation for 72 hr. The left panel shows a summary of the statistics of three independent experiments.
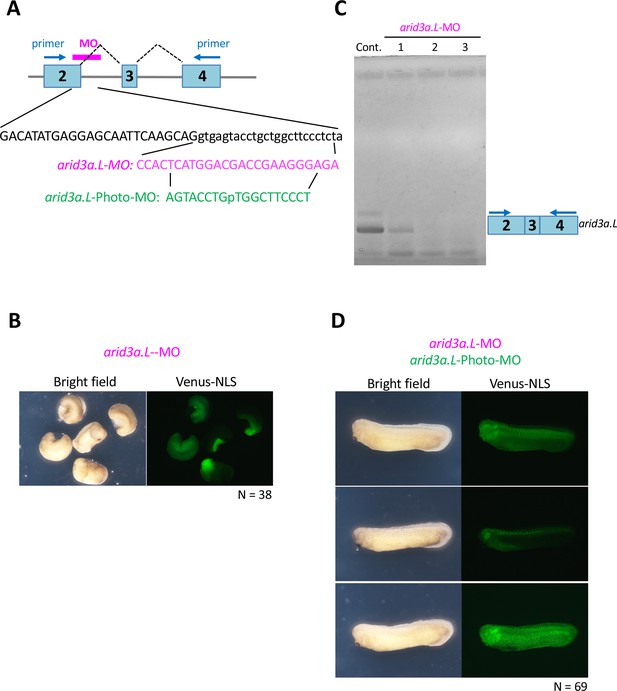
Arid3a-photo-morpholino blocks the effect of Arid3a-antisense morpholino.
(A) Sequences of arid3a.L-antisense-MO (arid3a.L-MO) and arid3a.L-Photo-MO. The upper panel shows a schematic illustration of arid3a-antisense-MO. Sequences show the target region of arid3a.L-MO and its sense arid3a.L-Photo-MO. (B) arid3a.L-MO causes developmental defects in early-stage embryos. arid3a.L-MO with venus-NLS mRNA is injected at the one-cell stage. (C) The arid3a.L-MO blocks the splicing of the arid3a transcript. Total RNA was purified from control and arid3a.L-MO-injected embryos, and RT-PCR analysis was performed. Lanes 1–3: three independent experiments. (D) Embryos injected with arid3a.L-MO inactivated by arid3a.L-Photo-MO developed normally.
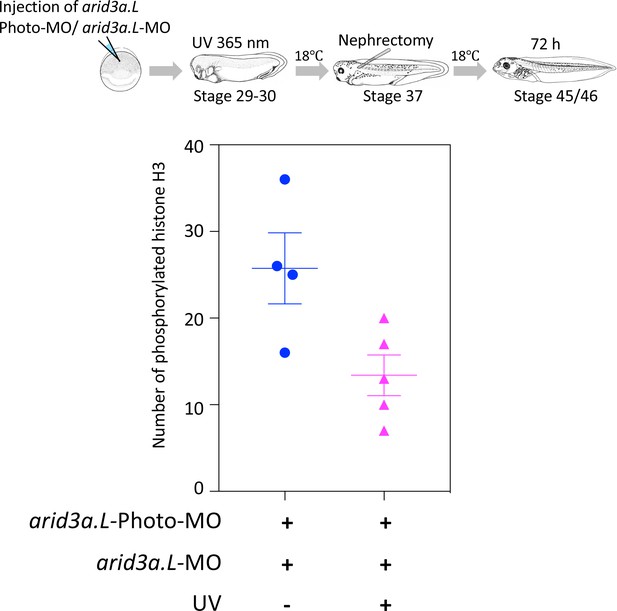
Conditional knockdown of Arid3a during nephric regeneration causes the reduction of cell cycle progression.
The upper panel shows the experimental design. arid3a.L-antisense-MO inactivated by arid3a.L-Photo-MO is injected at the one-cell stage, and UV exposure is performed at stage 29, followed by nephrectomy at stages 37/38 and then incubation for 72 hr. The fixed nephrectomized X. laevis were subjected to immunostaining with phosphorylated histone H3. The lower panel shows the statistical analysis. The significance of differences between the UV-untreated and UV-treated embryos was calculated by two-tailed unpaired t-test (p=0.0278). The error bars indicate SEM.
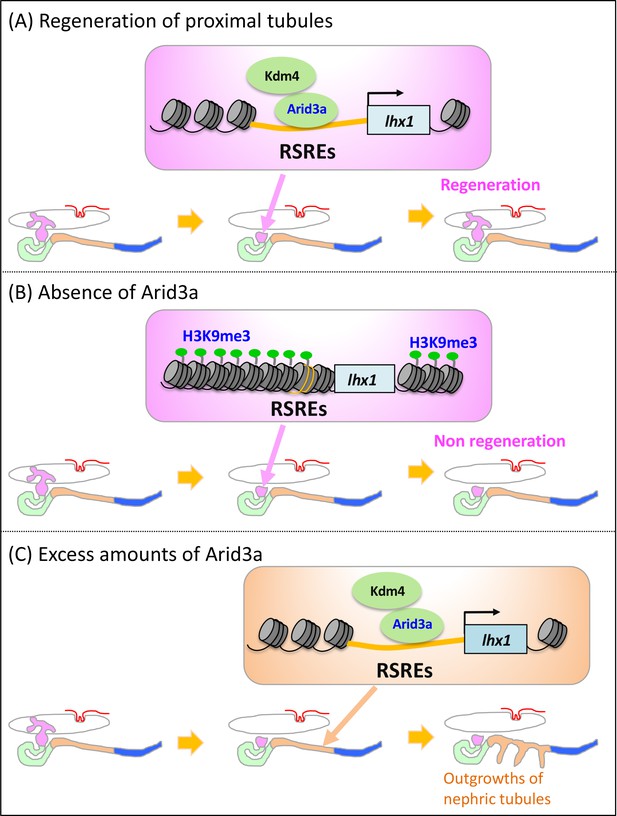
Model illustrating the Arid3a function in the regeneration of proximal nephric tubules.
(A) Arid3a binds to RSREs on lhx1 and changes the H3K9me3 levels. This chromatin modification allows the induction of lhx1 expression. (B) In the absence of Arid3a, proximal tubules fail to regenerate a complete nephron structure. (C) Excess amounts of Arid3a cause the outgrowth of nephric tubules from the distal nephric duct.
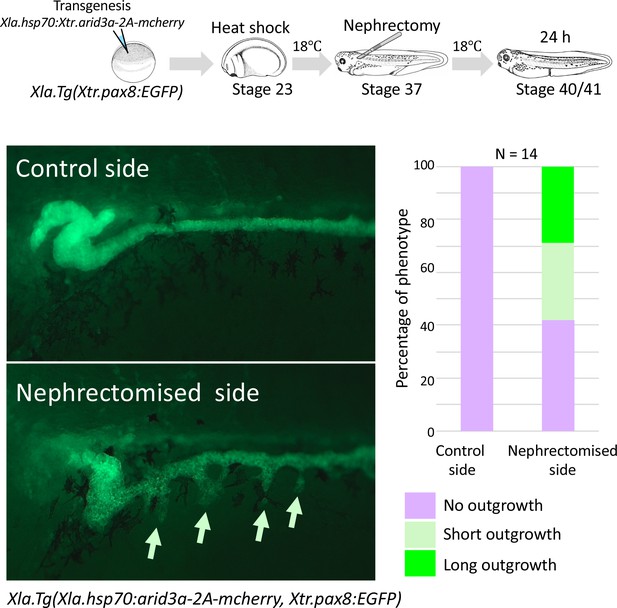
Conditionally induced Arid3a causes the outgrowth of nephric tubules in regenerating nephrons.
The upper panel shows the experimental design. Xla.Tg(Xtr.arid3a-2A-mcherry) transgenic X. laevis prepared using Xla.Tg(Xtr.pax8:EGFP) were induced by heat shock at stage 23. Nephrectomy was performed at stage 37, and embryos were then incubated for 24 hr.

A) A diagram of the experimental design.
B) RT-PCR analysis was performed. Lanes 1: Marker, Lane2: UV untreated embryos, Lane3: UV treated embryos. UV treatment and RT-PCR were performed at three times independently. C) The significance of differences between the UVuntreated and UV-treated wiled type embryos was calculated by two-tailed unpaired t-test (p = 0.4857, not significant). The error bars indicate SEM.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Xenopus tropicalis) | arid3a | This paper | RefSeq: NM_001011106.1 | |
| Gene (Xenopus tropicalis) | arid3b | This paper | RefSeq: XM_002938881.4 | |
| Gene (Xenopus tropicalis) | lhx1 | This paper | RefSeq: NM_001100228.1 | |
| Genetic reagent (Xenopus laevis) | Xla.Tg(Xtr.pax8:EGFP) | Ochi, H., et al., 2012; doi: 10.1038/ncomms1851. | ||
| Cell line (Homo sapiens) | 293T | RIKEN BRC CELL BANK | RCB2202, RRID:SCR_003163 | |
| Transfected construct | pGL4.23 | Promega | E8411 | |
| Transfected construct | pGL-lhx1-CNS17-Luc | This paper | New regent. The CNS17 fragments from IS-lhx1-CNS17-β-GFP vector introduced into the SacI and EcoRV sites of pGL4.23 vector. | |
| Transfected construct | pGL-lhx1-CNS20-Luc | This paper | New regent. The CNS20 fragments from IS-lhx1-CNS17- β-GFP wereintroduced into the SacI and EcoRV sites of pGL4.23 vector. | |
| Transfected construct | pGL-lhx1-CNS35-Luc | This paper | New regent. The CNS35 fragments from IS-lhx1-CNS17- β-GFP vector were introduced into the SacI and EcoRV sites of pGL4.23 vector. | |
| Antibody | Anti-phospho histone H3 (Ser10) antibody | Milipore | 06–570 | (1:1000) |
| Antibody | Anti-Arid3a antibody | DSHB | PCRP-ARID3A-1E9, RRID:AB_2618410 | (1:10) |
| Antibody | Alexa 488 -conjugated goat anti-rabbit IgG | Invitrogen | A11001 | (1:1000) |
| Antibody | Alexa 568-conjugated goat anti-mouse IgG | Invitrogen, | A11011 | (1:1000) |
| Antibody | Anti-H3K9 (tri-methyl K9) antibody | Abcam, | ab8898 | (1:750) |
| Antibody | Mouse monoclonal c-Myc (9E10) antibody | Santa Cruz Biotechnology Inc. | sc-40, RRID:AB_291323 | (1:750) |
| Recombinant DNA reagent | Xenopus laevis hsp70 promoter | Wheeler, G. N., et al. 2000; doi.org/10.1016/S0960-9822 (00)00596–0 | ||
| Recombinant DNA reagent | IS-β-GFP reporter | Ogino, H., et al., 2008; doi: 10.1242/dev.009548 | ||
| Recombinant DNA reagent | pCS-myc-arid3a | New regent. The PCR product was introduced into the XhoI and XbaI sites of the pCS2 + MT plasmid. | ||
| Recombinant DNA reagent | pCS-his-arid3b | New regent. The PCR product was introduced into the ClaI and XbaI site of the pCS vector. | ||
| Recombinant DNA reagent | hsp70-myc-arid3a-2A-mcherry | This paper | New regent. The PCR amplified myc-arid3a and 2A-mcherry were introduced into the ClaI and XbaI sites of the IS-hsp70-cloning vector. | |
| Recombinant DNA reagent | hsp70-myc-arid3a-2A-EGFP | This paper | New regent. The PCR amplified myc-arid3a and 2A-EGFP were introduced into the ClaI and XbaI sites of the IS-hsp70-cloning vector. | |
| Recombinant DNA reagent | hsp70-lhx1-2A-EGFP | This paper | New regent. The PCR amplifiedlhx1 was introduced into the EcoRI sites of the pCS2 + MT plasmid. The PCR amplified myc-lhx1 and 2A-EGFP were introduced into the EcoRI and XbaI sites of the IS-hsp70-cloning vector. | |
| Recombinant DNA reagent (Xenopus laevis) | arid3a.L | This paper | Xelaev18006256m | New regent. The PCR product was introduced into the EcoRI and XhoI sites of the pBluescript II SK plasmid. |
| Recombinant DNA reagent (Xenopus laevis) | arid3a.S | This paper | Xelaev18009788m | New regent. The PCR product was introduced into the EcoRI and XhoI sites of the pBluescript II SK plasmid. |
| Recombinant DNA reagent (Xenopus laevis) | spib. L | This paper | Xelaev18036193m.g | New regent. The PCR product was introduced into the EcoRI and XhoI sites of the pBluescript II SK plasmid. |
| Recombinant DNA reagent (Xenopus laevis) | spib.S | This paper | Xelaev18037903m.g | New regent. The PCR product was introduced into the EcoRI and XhoI sites of the pBluescript II SK plasmid. |
| Recombinant DNA reagent (Xenopus laevis) | hnf4a | This paper | Xelaev17043619m, Xelaev17043619m | New regent. The PCR product was introduced into the BamHI and HindIII sites of the pBluescript II SK plasmid. |
| Recombinant DNA reagent (Xenopus laevis) | hnf1b | This paper | Xelaev18012186m.g, Xelaev18014991m.g | New regent. The PCR product was introduced into the BamHI and HindIII sites of the pBluescript II SK plasmid. |
| Recombinant DNA reagent (Xenopus laevis) | osr1 | This paper | Xelaev14054577m.g, Xelaev14010174m.g | New regent. The PCR product was introduced into the XhoI and BamHI sites of the pBluescript II SK plasmid. |
| Recombinant DNA reagent (Xenopus laevis) | osr2 | This paper | Xelaev14045820m.g, Xelaev14031017m.g | New regent. The PCR product was introduced into the XhoI and BamHI sites of the pBluescript II SK plasmid. |
| Recombinant DNA reagent (Xenopus laevis) | six2 | This paper | Xelaev16000858m, Xelaev16036496m | New regent. The PCR product was introduced into the HindIII and XhoI sites of the pBluescript II SK plasmid. |
| Recombinant DNA reagent (Xenopus laevis) | lhx1 | This paper | Xelaev16044871m.g | New regent. The PCR product was introduced into the SmaI and HindIII sites of the pBluescript II SK plasmid. |
| Recombinant DNA reagent (Xenopus laevis) | pax2 | Heller and Brändli, 1997: doi.org/10.1016/S0925- 4773(97)00158-5 | ||
| Recombinant DNA reagent (Xenopus laevis) | pax8 | Heller and Brändli, 1999: doi.org/10.1002/(SICI)1520- 6408(1999)24:3/4 < 208::AID- DVG4 > 3.0.CO;2 J | ||
| Recombinant DNA reagent (Mus musculus) | kdm4a | Mammalian Gene Collection (MGC) Clones | 4207552 | BC028866 |
| Sequence-based reagent (Xenopus tropicalis) | PCR primers for CNS | This paper | ||
| Sequence-based reagent (Xenopus laevis) | ChIP-qPCR primers | This paper | ||
| Sequence-based reagent (Xenopus laevis) | Photo-Morpholino oligonucleotide for arid3a,L | This paper | Gene Tools, LLC | AGAGGGAAGCCAGCAGGTACTCACC |
| Sequence-based reagent (Xenopus laevis) | Morpholino oligonucleotide for arid3a,L | This paper | Gene Tools, LLC | AGTACCTGpTGGCTTCCCT |
| Sequence-based reagent (Xenopus laevis) | PT-PCR primers for arid3a.L | This paper | ||
| Sequence-based reagent (Homo sapiens) | hg19 chr17-34994909–35360679 | hg19 | UCSC Genome Browser, RRID:SCR_005780 | |
| Sequence-based reagent (Mus musculus) | mm10 chr11-83838963–85151744 | mm10 | UCSC Genome Browser, RRID:SCR_005780 | |
| Sequence-based reagent (Monodelphis domestica) | monDom5 chr2-185210169–185976291 | monDom5 | UCSC Genome Browser, RRID:SCR_005780 | |
| Sequence-based reagent (Xenopus tropicalis) | xenTro3 GL173152-472286-845619 | xenTro3 | Xenbase, RRID:SCR_003280 | |
| Sequence-based reagent (Danio rerio) | danRer10-chr15_27468859–28180541 | danRer10 | UCSC Genome Browser, RRID:SCR_005780 | |
| Sequence-based reagent (Danio rerio) | danRer10- chr5_55422952–55633560 | danRer10 | UCSC Genome Browser, RRID:SCR_005780 | |
| Software, algorithm | GraphPad Prism 7.0 | GraphPad Software | RRID:SCR_002798 | |
| Software, algorithm | Adobe Photoshop | Adobe | RRID:SCR_014199 | |
| Software, algorithm | MultiPipMaker | Schwartz, S., et al., 2000: doi: 10.1101/gr.10.4.577 | RRID:SCR_011806 | |
| Software, algorithm | JASPAR ver. 5 | Mathelier, A., et al., 2014: doi: 10.1093/nar/gkt997. | RRID:SCR_003030 | |
| Commercial assay or kit | ISOGEN | NIPPON GENE | Code No. 317–02503 | |
| Commercial assay or kit | Dual-Luciferase Reporter Assay System | Promega | E1910 | |
| Commercial assay or kit | Dynabeads Protein A | Dynabeads | 10001D | |
| Chemical compound, drug | jetPEI (transfection) | Polyplus-transfection SA | 101–10N |
Primer sequences for the RT-PCR and quantitative RT-PCR.
https://doi.org/10.7554/eLife.43186.020| Xtr.arid3a_full-length-F | ATGAAGCTGCAAGCGGTG |
| Xtr.arid3a_full-length-R | TCAGGGAGAAGGATTGTTAG |
| Xtr.arid3b_full-length-F | CGATGCCGCCACCATGCACCATCACCACCATCATCACCACCATCACT |
| Xtr.arid3b_full-length-R | CTAGAGTGATGGTGGTGATGATGGTGGTGATGGTGCATGGTGGCGGCAT |
| Xla.CNS17-qPCR-F | CTGAGTGAGTTTCAAATAAAAGGATTAAG |
| Xla.CNS17-qPCR-R | GCTATGTAGAGTGGAATAGAGTTAGAATGA |
| Xla.CNS20-qPCR-F | AATACTCACACAGGGAAGACAGC |
| Xla.CNS20-qPCR-R | AAGGCCAAAATTACTTTTCATTTATCTTA |
| Xla.CNS32-qPCR-F | GGGAATTAACCCCCATGGGAA |
| Xla.CNS32-qPCR-F | TTTGCCTCCCTCCTGATCTATAGG |
| Xla.exon5-qPCR-F | CCAGGTTCCATGCACTCTATG |
| Xla.exon5-qPCR-R | TTTCTGGTGGGTGTGACAAA |
| Xla.CNS35-qPCR-56–334 F | AGTTTATAATCTCTGCCGTGCT |
| Xla.CNS35-qPCR-56–334 R | TGTGCTGCTTGGAATTCAAG |
| Xla.CNS35-qPCR-314–580 F | CTTGAATTCCAAGCAGCACAT |
| Xla.CNS35-qPCR-314–580 R | CCTCAAGAACAATTCTCATTTAAATCCAC |
| Xla.arid3a-L-exon2-RT-PCR-F | CCCAAGCAATCTAGTCAACAGACATTTCC |
| Xla.arid3a-L-exon4-RT-PCR-R | GCTGCACTGGTGATTGAAGTTGGTAG |
| Xla.lhx1-F | TCTACTGTAAAAACGACTTCTTCAGG |
| Xla.lhx1-R | CCATTGACTGATAGAGAAGAAAAGG |
| Xla.six2.L-F | CGAAGCCAAAGAGAGGTACG |
| Xla.six2.L-R | TTGGGATCCTTCAACTCTGG |
| Xla.six2.S-F | ACCCGTTGTCCTCTTCAATG |
| Xla.six2.S-R | TGACCTGCTGAATGCAAGT |
| Xla.osr1-F | TCCTTCCTACAAGCCTTCAATGGAC |
| Xla.osr1-R | CTGAACAGAACACAATCATGTACAAGGAATTC |
| Xla.osr2-F | GGGAAGATGGGCAGCAAAGCT |
| Xla.osr2-R | TAGAAGTCCTGTCTGGGGCTGTG |
| Xla.hnf1b-F | TGGCTATGGATGCCTATAGTACTGGCC |
| Xla.hnf1b-R | TGCTGATGCTGCTAGTATCTGTGACAAC |
| Xla.hnf4a-F | CGGCTTTCTGTGAACTTCCACTGG |
| Xla.hnf4a-R | CTACATAGCTTCCTGTTTGGTGATGGTC |
Additional files
-
Supplementary file 1
The primer sequences used in this study.
- https://doi.org/10.7554/eLife.43186.021
-
Transparent reporting form
- https://doi.org/10.7554/eLife.43186.022





