Recurrent loss of HMGCS2 shows that ketogenesis is not essential for the evolution of large mammalian brains
Figures
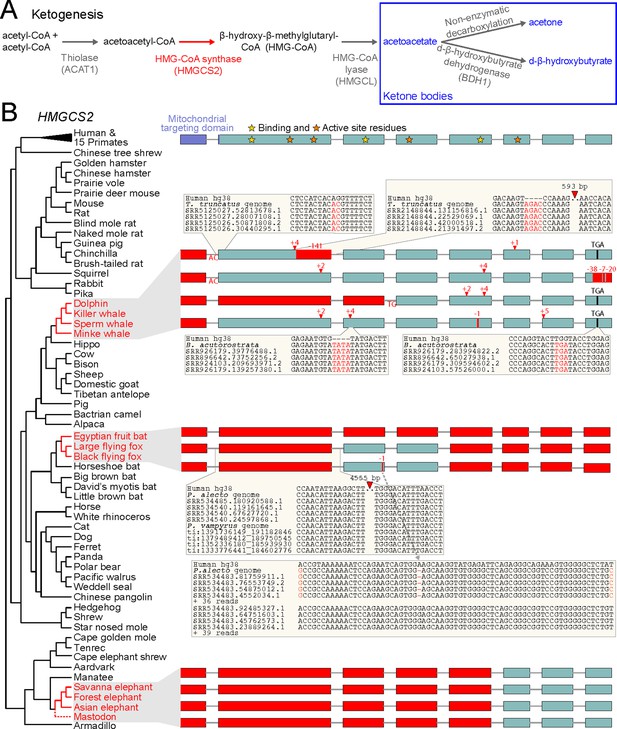
Evolution of ketogenesis in placental mammals.
(A) Biosynthesis of ketone bodies (blue font). With the exception of the mitochondrial HMG-CoA synthase (HMGCS2, red font), the other two enzymes required for acetoacetate production also have roles in amino acid metabolism and are thus pleiotropic. BDH1 is only required for converting acetoacetate into d-β-hydroxybutyrate. (B) Recurrent loss of HMGCS2 in three independent lineages (red font). All species in black font have an intact HMGCS2 reading frame. Boxes are coding exons proportional to their size, introns are shown as horizontal lines. Red boxes are exon deletions. In-frame stop codon, frameshifting insertion/deletion and splice site-disrupting mutations are indicated. With the exception of the heterozygous 1 bp deletion in the black flying fox that has a read support of ~50:50 for the derived and ancestral allele and reveals two distinct haplotypes (inset), all shown mutations are supported by at least 30 reads with no support for the ancestral allele (Supplementary file 1). Insets exemplify the validation of inactivating mutations by showing the local genomic context and four reads.
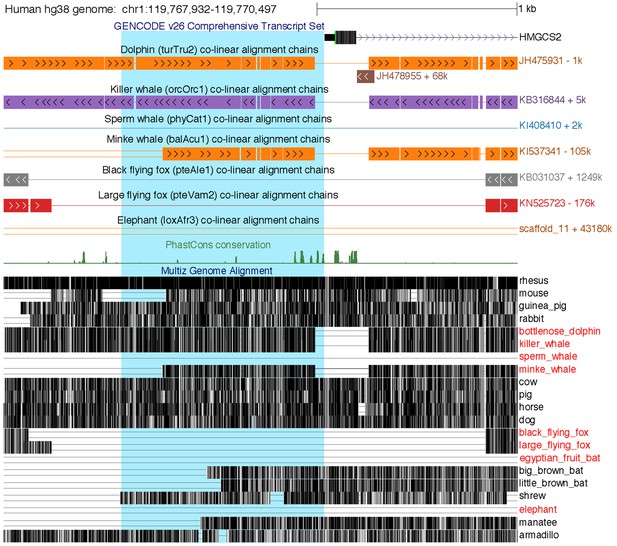
Loss of the HMGCS2 promoter region in the sperm whale, pteropodids, and the elephant.
The figure shows the 2.5 kb genomic region around HMGCS2 exon 1 in the UCSC human (hg38 assembly) genome browser (Casper et al., 2018). The 1 kb region upstream of the transcription start site, highlighted in blue background, is deleted in the sperm whale and also lost in fruit bats and elephant, as shown by the co-linear pairwise alignment chains (blocks are aligning regions, a single line represents the shared deletion, double lines represent regions that do not align between the human and the query genome).
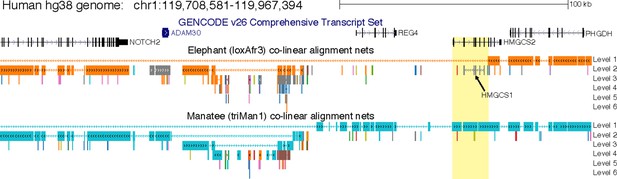
Deletion of a large part of HMGCS2 in elephant.
Screenshot of the human UCSC genome browser shows the locus of the NOTCH2, ADAM30, REG4, HMGCS2 and PHGDH genes. Co-linear pairwise alignment chains to the African savanna elephant (loxAfr3 assembly) and the manatee (presumably its closest sister species [Poulakakis and Stamatakis, 2010]) show that several rearrangements, including probably two nested inversions, happened in this locus. Importantly, while NOTCH2, ADAM30 and PHGDH align to both species, the elephant exhibits a large deletion that includes REG4 and most of HMGCS2 (yellow background, see Figure 1B). Alignment chains to the African savanna elephant loxAfr4 assembly are virtually identical and confirm the partial HMGCS2 deletion. The grey elephant chain (arrow) that only aligns exons but not introns of HMGCS2 is the elephant HMGCS1 gene. As described in the main text, searching unassembled reads of 2 individuals of the African savanna elephant (whose genome has been sequenced) and read data of the African forest elephant, the Asian elephant and the mastodon did not reveal any evidence for the presence of the deleted HMGCS2 exons or the REG4 gene, suggesting that this genomic region was already deleted in ancestor of elephants and mastodons.
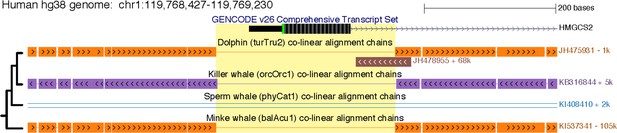
A shared ~ 270 bp deletion removed HMGCS2 exon one in cetaceans.
The figure shows exon one in the UCSC human (hg38 assembly) genome browser. This exon is deleted in all cetaceans (yellow background) and has the same breakpoints, as shown by the co-linear pairwise alignment chains (blocks are aligning regions, a single line represents the shared deletion). This region was subsequently removed by another large deletion in the sperm whale lineage. The phylogenetic relationships of the four species are indicated on the left side.

Large deletions in the HMGCS2 locus in pteropodid fruit bats.
(A) Screenshot of the human UCSC genome browser shows the HMGCS2 locus with its flanking genes and co-linear pairwise alignment chains to three pteropodid bats. Chains that align the paralog HMGCS1 or a processed HMGCS1 pseudogene in flying foxes are labeled. Please note that we show in this figure the transcript ENST00000369406, which includes an alternative exon three that is skipped in the transcript shown in Figure 1B (ENST00000544913). Yellow background highlights the deletion of exon 1, a separate deletion that removed exon 2 (shown in detail in the inset in Figure 1B) and a deletion that removed four consecutive exons downstream. At least the second deletion exhibits shared breakpoints between both flying foxes, showing that loss of HMGCS2 already occurred in the ancestor of both species. The phylogenetic relationships of the three species are indicated on the left side. (B) To validate the deletion of HMGCS2 in the Egyptian fruit bat, we used an independent PacBio assembly. The locus around the neighboring PHGDH and REG4 gene is covered by several large PacBio reads. Blastx results only reveal the presence of PHGDH and REG4 but no evidence of HMGCS2. Consistent with HMGCS2 deletion, the PHGDH and REG4 are separated by ~ 50 kb in human but only ~ 6 kb in the Egyptian fruit bat.
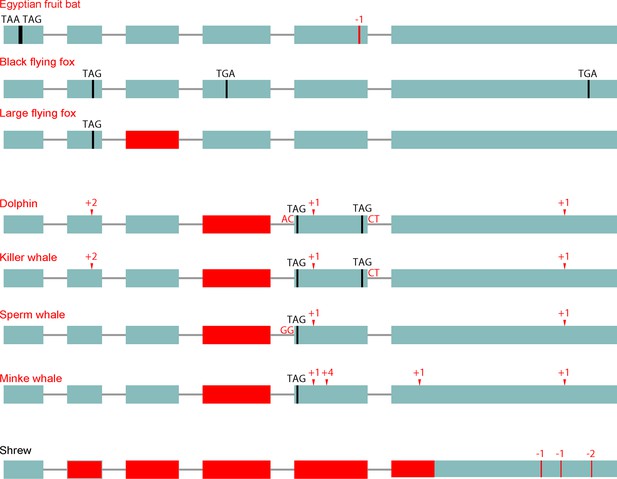
Loss of the d-β-hydroxybutyrate producing BDH1 gene.
BDH1 converts the ketone body acetoacetate into the ketone body d-β-hydroxybutyrate (Figure 1A). BDH1 is a non-essential gene as a knockout in mouse results in the absence of d-β-hydroxybutyrate and a compensating increase in acetoacetate without major problems (Otsuka et al., 2017). Furthermore, Bdh1 knockout mice survive a 48 hr fast. We found that pteropodids and cetaceans but not the elephant exhibit several gene-inactivating mutations in this gene (visualization as in Figure 1B). RELAX analysis shows that BDH1 also evolved under relaxed selection in pteropodids and cetaceans (Supplementary file 2). In addition, we found that this gene has several inactivating mutations and a large deletion in the shrew. Importantly, ACAT1, HMGCS2 and HMGCL are intact in the shrew, suggesting that acetoacetate and acetone are the primary ketone bodies in this species. None of the other analyzed placental mammals had inactivating mutations in BDH1.
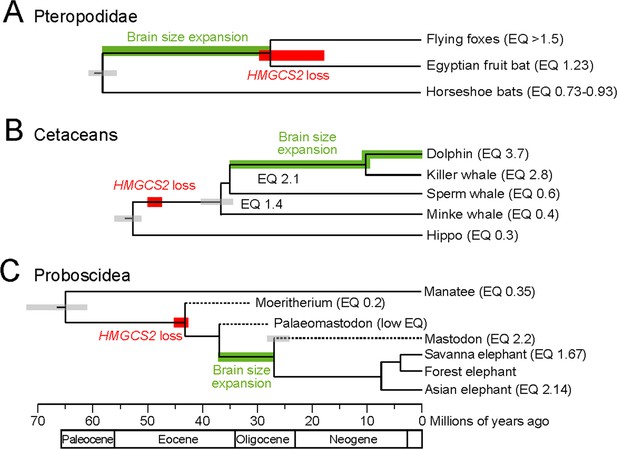
HMGCS2 loss and brain size evolution.
(A) In pteropodids, molecular dating estimates that the loss of HMGCS2 happened 29–18 Mya and thus may overlap the split of the flying foxes and the Egyptian fruit bat. It is not possible to resolve whether gene loss happened before or after the split as HMGCS2 is completely deleted in the Egyptian fruit bat. While horseshoe bats and other insectivorous bat lineages have brains not larger than expected for their body size (encephalization quotient (EQ) <1), brain size has increased in the lineage leading to the fruit bats that have EQ values > 1 (Stephan et al., 1981). Thus, brain size expansion presumably predates the loss of ketogenesis. (B) HMGCS2 was already lost in the cetacean ancestor before the split of toothed and baleen whales ~ 36 Mya, as inferred from shared inactivating mutations in exons 1, 2 and 8. Molecular dating further estimates that the loss of this gene happened early on the cetacean branch 50–47 Mya. The cetacean ancestor had a brain slightly larger than expected for its body size with an EQ of 1.4. While EQ values increased and decreased in several cetacean lineages, brain size has greatly expanded in dolphins, reaching an EQ of 3.7 (Montgomery et al., 2013). Thus, brain size expansion in dolphins occurred after the loss of ketogenesis. (C) Early proboscids such as Moeritherium, an extinct lineage that split from other proboscids ~ 43 Mya, had brains about 20% of the size expected for a mammal of the same body size, and thus an EQ of 0.2 (Shoshani et al., 2006). Exact EQ values of Palaeomastodons are not known; however, fossils have a small braincase, which indicates a low EQ (Sanders et al., 2010; Benoit, 2015). In contrast, mastodons that diverged from elephants ~ 27 Mya had brains about twice as large as expected from their body size (EQ 2.2), similar to extant elephants (Shoshani et al., 2006). This suggests that brain size expansion happened in a period between 37 and 27 Mya. Molecular dating indicates that HMGCS2 loss happened between 45 and 42 Mya, suggesting that the loss of ketogenesis precedes brain size expansion in the elephant lineage. Divergence times of extinct proboscid lineages were taken from (Shoshani and Tassy, 2013) and (Rohland et al., 2007). Supporting Information.
-
Figure 2—source data 1
Sequence alignment.
This file contains the HMGCS2 sequence alignment (fasta format) including the sequences of the African forest elephant, the Asian elephant and the American mastodon. This alignment was used to date the loss of HMGCS2.
- https://doi.org/10.7554/eLife.38906.010
Tables
Additional files
-
Supplementary file 1
Number of unassembled sequence reads supporting gene-inactivating mutations in cetaceans and fruit bats.
Please note that sequencing errors can change a real stop codon to a sense codon, which happened in two cases; however, given that > 60 reads confirm the stop codon, these single erroneous reads to not support the presence of the ancestral allele. The heterozygous 1 bp deletion in the black flying fox (last row) is discussed in the main text.
- https://doi.org/10.7554/eLife.38906.011
-
Supplementary file 2
RELAX analysis of genes in the ketogenesis pathway.
The estimate for ‘All’ and Elephantimorpha was computed from an alignment including the Asian elephant, the African forest elephant and the mastodon.
- https://doi.org/10.7554/eLife.38906.012
-
Supplementary file 3
Dating the loss of HMGCS2.
The table lists divergence times and estimates for how long HMGCS2 was functional along the mixed branch leading to the three loss lineages. Using the method of (Meredith et al., 2009; Gaudry et al., 2017), we calculated a point estimate for when HMGCS2 was lost and an upper/lower bound of this estimate. (A) Elephant lineage. We estimated HMGCS2 loss dates both from an alignment that includes sequences of Elephantimorpha and from an alignment that includes only the African savanna elephant. (B) Fruit bat lineage. (C) Cetacean lineage.
- https://doi.org/10.7554/eLife.38906.013
-
Supplementary file 4
Species and genome assemblies that were analyzed in this study.
- https://doi.org/10.7554/eLife.38906.014
-
Supplementary file 5
Sources of unassembled genomic sequencing reads used for the validation of inactivating mutations in HMGCS2 loss species
- https://doi.org/10.7554/eLife.38906.015
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38906.016





