EFHC1, implicated in juvenile myoclonic epilepsy, functions at the cilium and synapse to modulate dopamine signaling
Figures
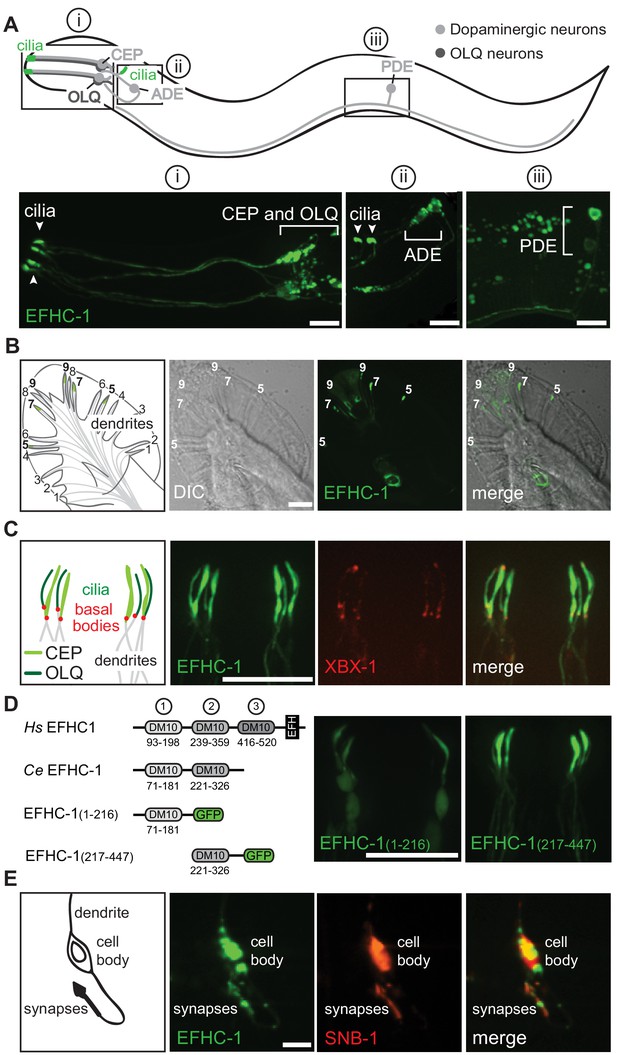
EFHC-1 localizes to cilia and synapses of mechanosensory dopaminergic neurons and to the distal regions of male-specific dopaminergic ray neurons.
(A) EFHC-1::GFP protein marker is expressed in all classes of dopaminergic neurons shared by both hermaphrodites and males (CEP, ADE and PDE), as well as in the mechanosensory OLQ neurons. Localization is enriched in cilia (arrowheads). Scale bar, 10 μm. (B) EFHC-1::GFP is also expressed in dopaminergic ray RnA neurons in the male tail (rays 5, 7 and 9) distal to dendrites. Scale bar, 10 μm. (C) In CEP and OLQ cilia, EFHC-1::GFP localization surrounds the axoneme marked by the intraflagellar transport protein XBX-1::tdTomato. Scale bar, 10 μm. (D) Human (Hs) EFHC1 is composed of three DM10 domains of unknown function and a C-terminal EF-hand (EFH) domain that is thought to bind calcium. In C. elegans (Ce), only the first two DM10 domains are conserved. A truncation protein harboring the first DM10 domain shows non-specific localization, while a truncation protein containing only the second DM10 domain shows specific ciliary localization in OLQ and CEP neurons. Scale bar, 10 μm. (E) EFHC-1::GFP also localizes to synaptic regions in CEP neurons, partially overlapping with the synaptic protein, SNB-1::tdTomato. Scale bar, 10 μm.
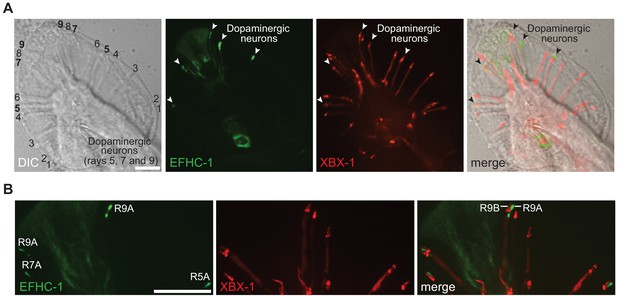
The EFHC-1::GFP fusion protein localizes to the dopaminergic ray neurons in the male tail.
(A) EFHC-1::GFP localizes to regions distal to the dendrites of dopaminergic ray RnA neurons in the male tail (rays 5, 7 and 9). In contrast, XBX-1::tdTomato driven in ciliated cells by the bbs-8 promoter that represents the ciliary axoneme is found in all ray neurons of the male tail. EFHC-1::GFP is also observed in some cell bodies within the male tail, although the identities of these neurons were not determined. Scale bar, 10 μm. (B) XBX-1::tdTomato only localizes to the ciliary axonemes of RnB neurons but appears to localize to dendrites and basal bodies of both RnA and RnB neurons. EFHC-1::GFP is found distal to the basal body in dopaminergic RnA neurons. Scale bar, 10 μm.
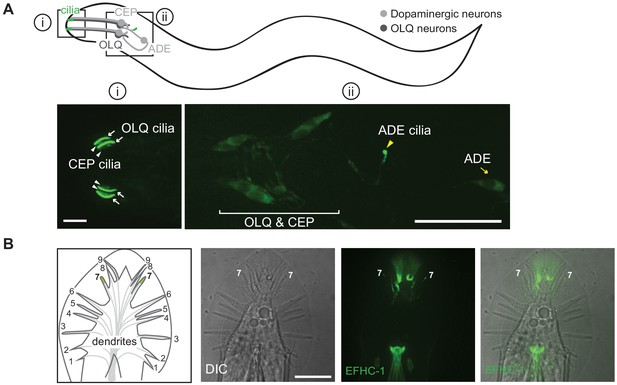
The expression patterns of EFHC-1 in an efhc-1::gfp knock-in strain mirror those seen for the EFHC-1::GFP fusion protein.
(A) Endogenously GFP-tagged EFHC-1 is found in dopaminergic and mechanosensory neurons (CEP, ADE, and OLQ). EFHC-1::GFP predominantly accumulates in cilia (white arrows and yellow arrowheads) of OLQ and ADE neurons, but is weakly present in CEP cilia (white arrowheads). Scale bar, 4 μm (i) and 18 μm (ii). (B) EFHC-1::GFP is highly enriched in the distal end of the dendrite in the male tail (ray 7), but it is not observed in the additional dopaminergic ray neurons (possibly due to lower expression levels that are below detection abilities). Scale bar, 18 μm.
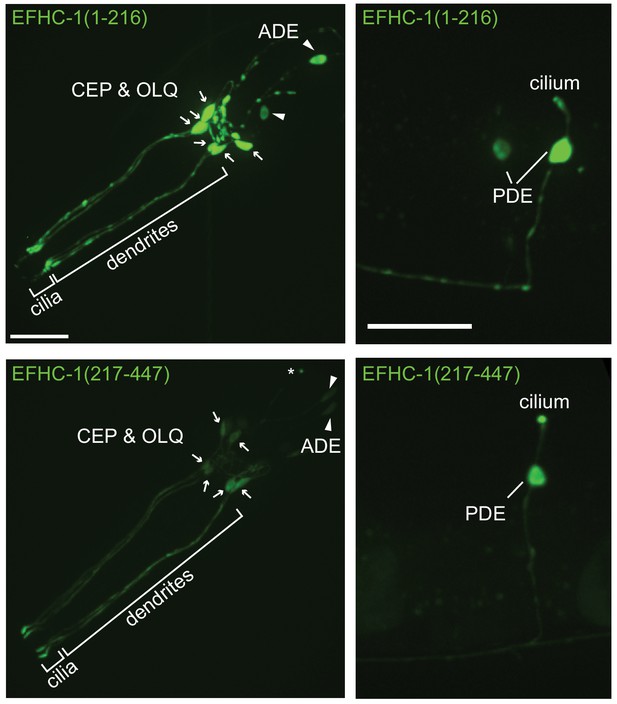
An EFHC-1 truncation with only the first DM10 domain shows non-specific localization throughout OLQ and dopaminergic neurons, while a protein containing only the second DM10 domain shows specific ciliary localization within these neurons.
EFHC-1 (1–216)::GFP is mainly present at cell bodies and dendrites of OLQ and dopaminergic neurons (CEP, ADE and PDE), while EFHC-1 (217–447)::GFP mostly accumulates within cilia. Arrows point to CEP and OLQ cell bodies and arrowheads represent ADE cell bodies. Asterisk indicates the ADE cilium. Scale bar, 22 μm.
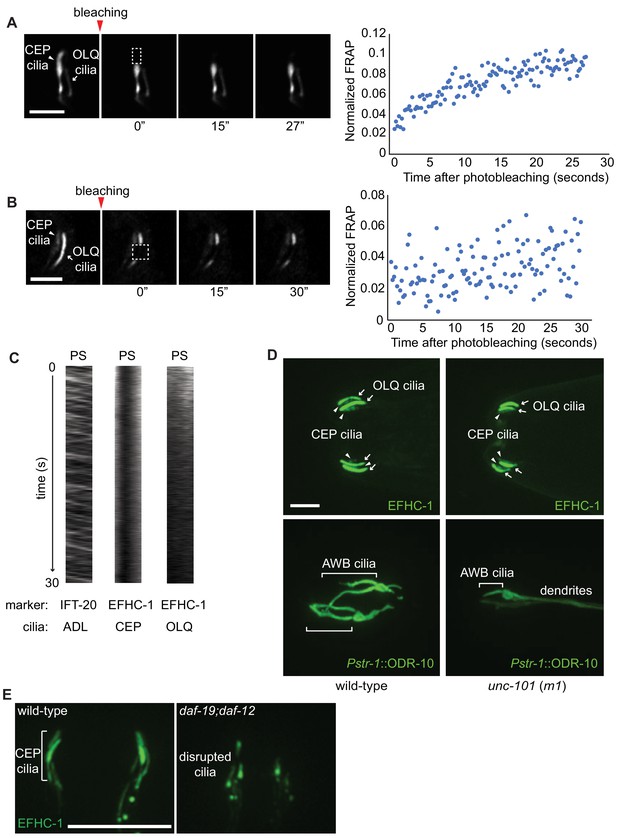
Localization of EFHC-1 within cilia is largely static and independent of typical intraflagellar transport (IFT) mechanisms within cilia, the dendritic/ciliary transport protein, UNC-101, and the ciliary transcription factor, DAF-19.
When distal regions of the CEP cilium in the EFHC-1::GFP overexpression strain (A) and middle regions of the OLQ cilium in the efhc-1::gfp CRISPR-knock-in strain (B) are photobleached, the GFP signal does not fully recover within 30 s. Pre-bleach ratios are normalized to 1. Scale bar, 3 μm. (C) Kymographs of IFT-20::GFP in ADL cilia (left), EFHC-1::GFP in CEP cilia (middle) and EFHC-1::GFP in OLQ cilia (right). PS, proximal segment. Scale bar, 3 μm (D) ODR-10 predominantly localizes to AWB olfactory cilia in wild-type, but it diffuses to dendrites in unc-101 (m1) mutant animals. Ciliary localizations of EFHC-1::GFP in OLQ and CEP neurons (arrows and arrowheads, respectively) are not disrupted in unc-101 mutants. Scale bar, 4 μm. (E) While CEP ciliary structure is disrupted in the absence of the ciliary DAF-19 transcription factor, EFHC-1::GFP is still expressed. Scale bar, 10 μm.
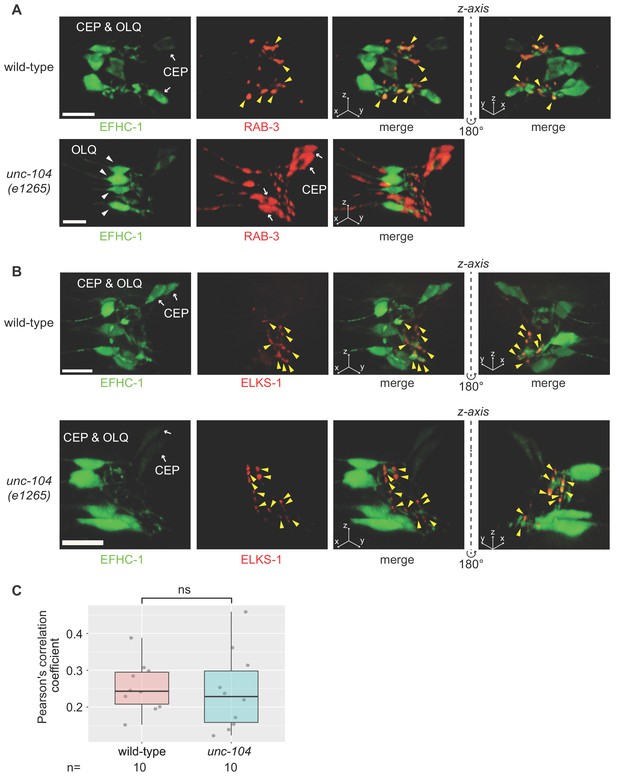
Preynaptic localization of synaptic vesicle protein RAB-3 is dependent on UNC-104, while EFHC-1 and active zone protein ELKS-1 are independent of UNC-104.
(A,B) EFHC-1::GFP in CEP axons colocalizes with synaptic vesicles (marked by Pdat-1::mCherry::RAB-3) and active zones (marked by Pdat-1::ELKS-1::tdTomato). While RAB-3 is mislocalized in unc-104 (e1265) mutant animals, the presynaptic localizations of EFHC-1 and ELKS-1 are not changed. Z-stack images are deconvoluted for three-dimensional reconstruction. Yellow arrowheads represent synaptic vesicles (A) and active zones (B) in presynaptic regions. White arrows, CEP cell bodies; White arrowheads, OLQ cell bodies. Scale bar, 8.12 μm. (C) EFHC-1 shows partial colocalization with ELKS-1 in CEP axons (illustrated using Thresholded Pearson’s correlation coefficients), and this pattern is unchanged in unc-104 mutants. Each dot represents a single axon measured from an individual worm. The statistical significance was calculated using Tukey’s HSD (honest significant difference) test. ns, not significant.
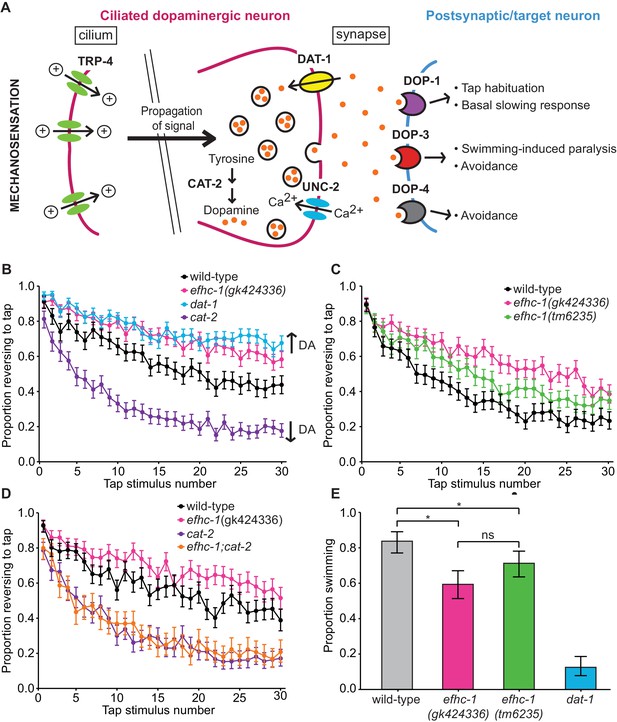
EFHC-1 influences habituation to a mechanical stimulus by modulating dopamine signaling.
(A) The molecular components of dopamine signaling in C. elegans. Food (bacteria) mechanically stimulates the ciliated dopaminergic neurons (dark pink), resulting in the release of dopamine that modulates several behaviors through dopamine receptors on various postsynaptic/target cells (blue). (B) In the presence of food, efhc-1 mutants habituate slower than wild-type animals, and similar to dat-1 mutants that have increased dopamine signaling (DA: dopamine). At each tap, the points represent the number of worms reversing over the total number of worms tracked per strain, and the error bars represent the binomial confidence intervals. Binomial logistic regression followed by Tukey’s HSD (honest significant difference) test was used to determine significance of the habituated level (proportion reversing at tap 30) for each pair of strains. All strains are statistically different from each other (p < 0.001). (C) A second allele of efhc-1, tm6235, also leads to a slow habituation phenotype that is statistically different from wild-type (p < 0.001). All strains are statistically different from each other (p < 0.001). (D) The habituation phenotype seen in efhc-1 mutants is abolished in efhc-1;cat-2 double mutants, suggesting that the slow habituation phenotype of efhc-1 mutants depends on dopamine. All strains are statistically different from each other (p < 0.001) except for cat-2 and efhc-1;cat-2 (p = 0.69). (E) efhc-1 mutants also show defects in the dopamine-dependent swimming-induced paralysis phenotype, with an intermediate level of paralysis after 10 min compared to wild-type and dat-1 mutants. Each bar represents the number of worms swimming after 10 min over the total number of worms per strain, and the error bars represent the binomial confidence intervals (n = 160 for each strain). Binomial logistic regression followed by Tukey’s HSD (honest significant difference) test was used to determine significance for each pair of strains. All strains are statistically different from each other except for the two efhc-1 strains (ns = not significant, p = 0.115). For sample sizes (n) for tap habituation experiments and complete statistical comparisons (p values) see Supplementary files 1 and 2, respectively.
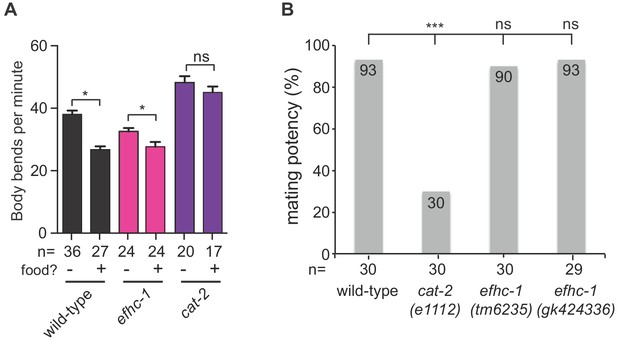
efhc-1 mutants are able to release dopamine in the presence of appropriate behavioral stimuli.
(A) EFHC-1 is not required for the dopamine-dependent basal slowing response, a behavior where animals slow their locomotion in the presence of food. Bars represent mean and error bars represent SEM. (B) Unlike dopamine deficient cat-2 mutant males, both efhc-1 mutant males (tm6235 and gk424336) have similar mating potency to wild-type. The statistical significance was calculated using the fisher’s exact test. ***, p < 0.001; ns, not significant.
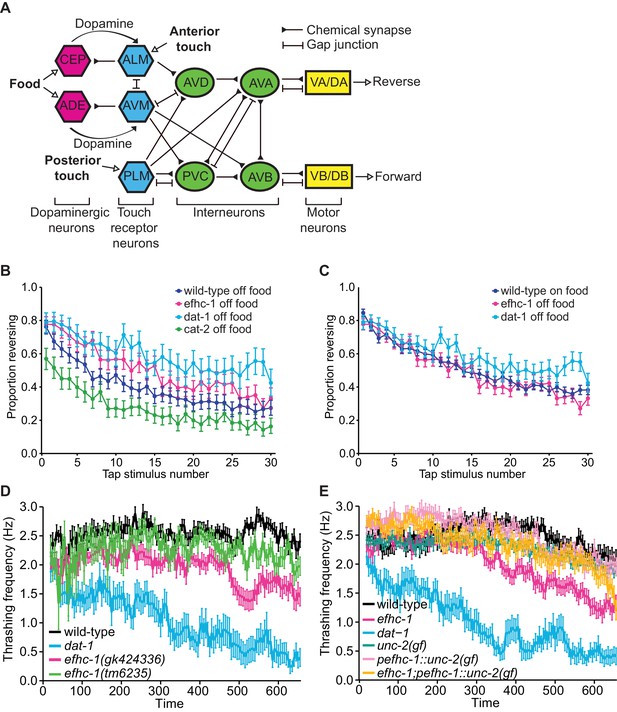
efhc-1 mutants show behavioral phenotypes consistent with increased dopamine signalling.
(A) Neural circuit outlining how dopaminergic neurons influence touch receptor neuron activity (with some connections omitted for clarity). Food (bacteria) mechanically stimulates the dopaminergic CEP and ADE neurons to release dopamine. This dopamine acts extra-synaptically to modulate the sensitivity of the anterior touch receptor neurons (ALM and AVM) to touch. ALM and AVM neurons are activated by anterior touch (or tap) to promote backward movement, while the posterior touch receptor neuron PLM is activated by posterior touch to promote forward movement. This circuit is further fine-tuned by activation of CEP neurons by touch receptor neurons (not shown). (B) efhc-1 mutants show slower habituation compared to wild-type animals off food. At each tap, the points represent the number of worms reversing over the total number of worms tracked per strain, and the error bars represent the binomial confidence intervals. Binomial logistic regression followed by Tukey’s HSD (honest significant difference) test was used to determine significance of the habituated level (proportion reversing at tap 30) for each pair of strains. All strains are statistically different from each other (p < 0.001). (C) efhc-1 mutants off food mimic the habituation seen in wild-type animals on food. All strains are statistically different (p < 0.001) except for wild-type animals on food and efhc-1 mutants off food (p < 0.484). (D) An automated swimming-induced paralysis (SWIP) assay shows intermediate thrashing frequencies for the two efhc-1 alleles compared to dat-1 and wild-type animals. Each point represents the mean of thrashes counted over 5 s (divided by 5), and the error bars represent SEM. (E) efhc-1 mutants show an intermediate thrashing frequency compared to wild-type and dat-1 mutants. An unc-2(gain-of-function) mutation expressed throughout the animals, an unc-2(gain-of-function) mutation driven in EFHC-1-expressing cells, and efhc-1 mutants with the addition of the unc-2(gain-of-function) mutation driven in EFHC-1-expressing cells all show an increased thrashing frequency compared to efhc-1 animals but decreased compared to wild-type animals. For sample sizes (n) for tap habituation experiments and complete statistical comparisons (p values) see Supplementary files 1 and 2, respectively.
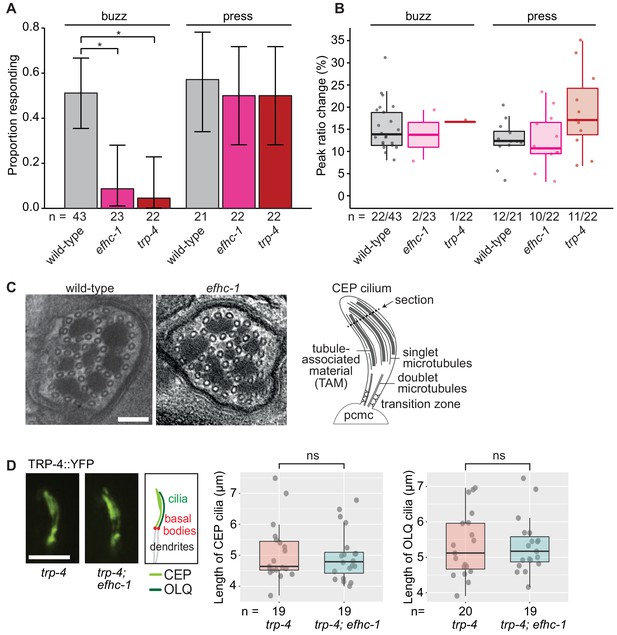
EFHC-1, similar to TRP-4, is required for mechanosensation in non-motile CEP cilia.
(A) Loss of EFHC-1 or TRP-4 impairs activation of CEP neurons upon gentle mechanical stimulation (buzz), resulting in fewer animals responding to the stimulus compared to wild-type animals. In contrast, efhc-1 or trp-4 mutants can respond to stronger stimulation (press). In this panel, n represents the total number of animals assessed. Each bar represents the number of worms responding to mechanical stimulation over the total number of worms assessed per strain, and the error bars represent the binomial confidence intervals. Binomial logistic regression followed by Tukey’s HSD (honest significant difference) test was used to determine significance for each pair of strains. Differences between all strains subjected to the press stimulus are not significant, but differences between wild-type and efhc-1 (p < 0.01) or trp-4 (p < 0.05) mutants subjected to the buzz stimulus are significant. (B) The peak ratio change (%), representing the amplitude of the neuronal response, is similar between strains for animals responding to each stimulus. In this panel, n represents the number of animals responding to the stimulus over the total number of animals assessed. (C) The axonemal structure of CEP neurons by TEM cross-section (refer to schematic) is not abrogated in efhc-1 mutants. Scale bars, 100 nm. (D) The localization of the mechanosensory channel, TRP-4, which is comparable to that of EFHC-1, is unaffected in efhc-1 mutants. Lengths of CEP and OLQ cilia measured in trp-4 and trp-4;efhc-1 mutants are indistinguishable. Each dot represents one cilium. Kruskal-Wallis rank sum test (CEP cilia lengths) and Tukey's Honest Significant Differences (OLQ cilia lengths) were used for calculating the statistical significance. ns, not significant. Scale bar, 3 μm. For complete statistical comparisons (p values) see Supplementary file 2.
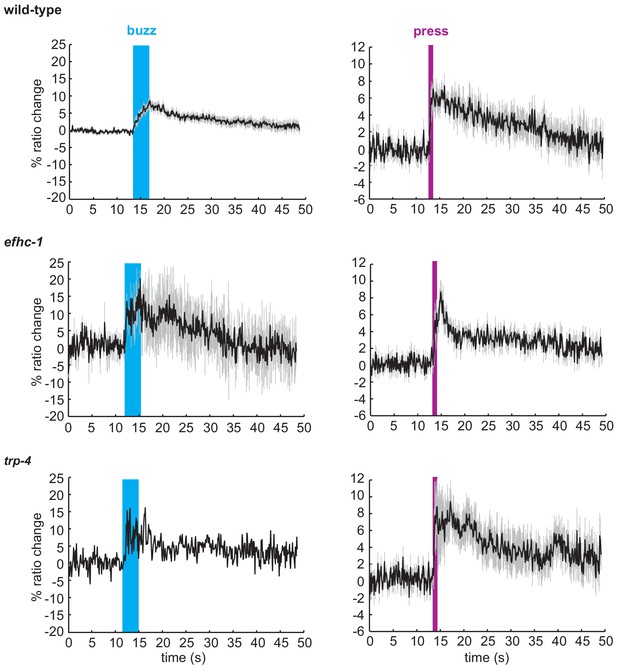
Animals that respond to mechanical stimuli in the CEP neurons show similar patterns of calcium changes.
Average traces of calcium responses in CEP neurons upon the mechanical buzz or press stimuli (black), and SEM values associated with the calcium traces (grey). The duration of the mechanical stimulus is shown in blue (buzz) or purple (press). All worms responding to mechanical stimuli from Figure 3B were included in this analysis.
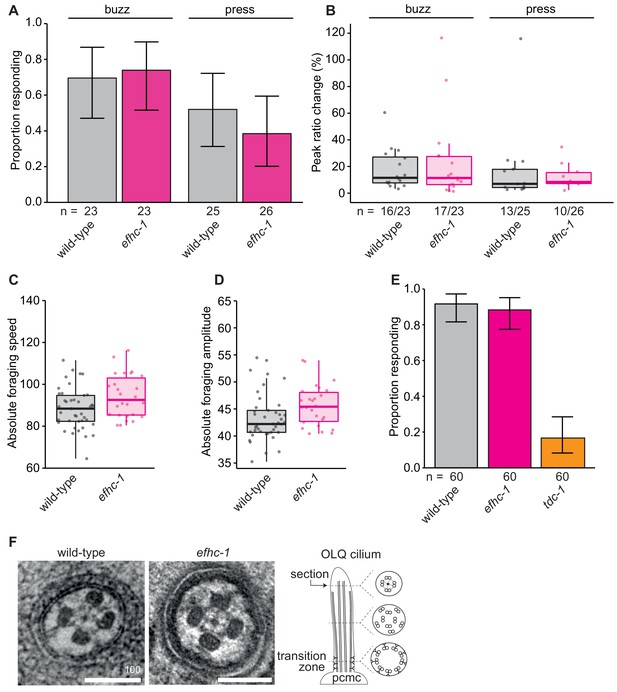
efhc-1 mutants show no defects in OLQ-related mechanosensory or behavioral responses, or OLQ ciliary structure.
(A) Loss of EFHC-1 has no effect on the activation of OLQ neurons upon gentle mechanical stimulation (buzz) (p = 0.943) or a stronger stimulation (press) (p = 0.597). In this panel, n represents the total number of animals assessed. Each bar represents the number of worms responding to mechanical stimulation over the total number of worms assessed per strain, and the error bars represent the binomial confidence intervals. Binomial logistic regression followed by Tukey’s HSD (honest significant difference) test was used to determine significance for each pair of strains. Differences between all strains are not significant. (B) The peak ratio change (%) representing the amplitude of the neuronal response is similar between strains for animals responding to each stimulus. In this panel, n represents the number of animals responding to the stimulus over the total number of animals assessed. (C) efhc-1 mutants showed similar foraging speeds and (D) foraging amplitudes, two behaviors mediated by the OLQ neurons. Here the absolute values of the forward foraging speed/amplitude are depicted. (E) EFHC-1 is also not involved in the suppression of foraging, a behavior in which worms subjected to mechanical stimulation of the anterior body suppress their foraging movements. OLQ neurons play a role in regulating this behavior and mutants like tdc-1 are unable to suppress their foraging movements when reversing. Each bar represents the number of worms that suppressed foraging over the total number of worms assessed per strain, and the error bars represent the binomial confidence intervals. Binomial logistic regression followed by Tukey’s HSD (honest significant difference) test was used to determine significance for each pair of strains, and efhc-1 mutants are not statistically different than wild-type animals. (F) Axonemal structure of OLQ neurons is not abrogated in efhc-1 mutants. Scale bars, 100 nm. For complete statistical comparisons (p values) see Supplementary file 2.
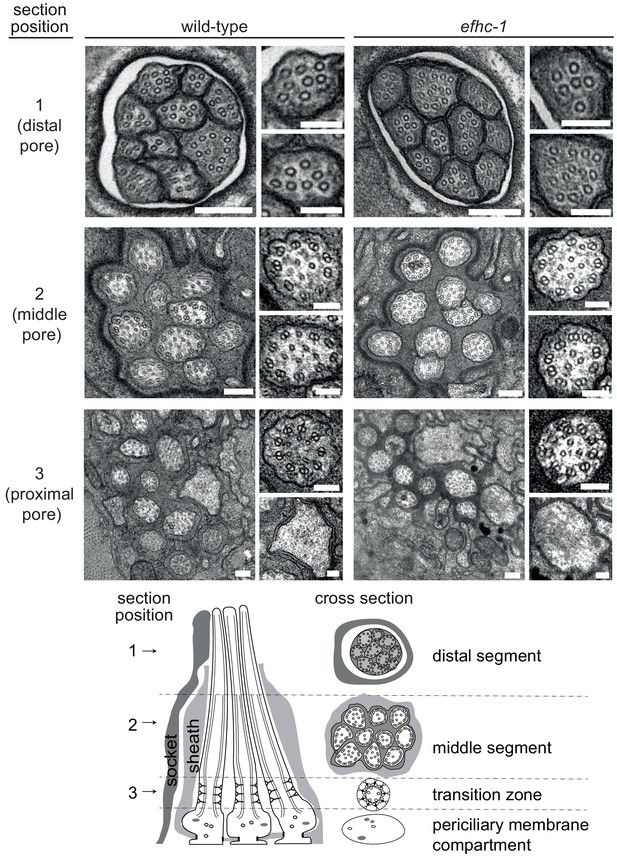
General ciliary structure is retained in efhc-1 mutants.
Transmission electron microscopy (TEM) serial cross-section images of amphid channel cilia in wild-type animals and efhc-1 mutants shows that axonemal structure of conventional cilia is unaffected in efhc-1 mutants. Each amphid channel contains 10 cilia composed of singlet microtubules in the distal pore, doublet microtubules and inner singlet microtubules in the middle pore and a transition zone with Y-link connectors between the microtubules and the membrane in the proximal pore. Large panels show the entire amphid ciliary bundle at low magnification; smaller panels show individual cilia at high magnification. Scale bars: 200 nm (low-magnification images); 100 nm (high-magnification images).
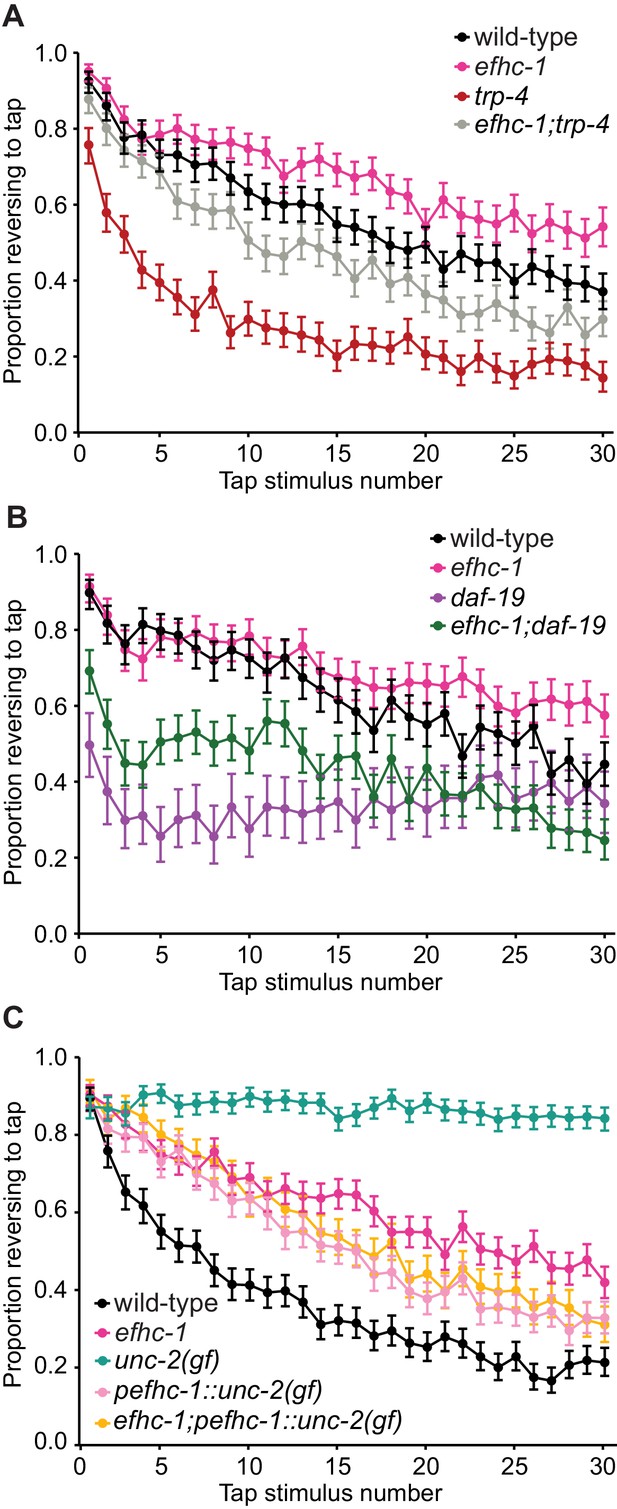
EFHC-1-mediated regulation of dopamine signaling occurs, in part, downstream of cilia.
(A) Double mutants of efhc-1 and trp-4 show intermediate habituation compared to the trp-4 and efhc-1 single mutants. At each tap, the points represent the number of worms reversing over the total number of worms tracked per strain, and the error bars represent the binomial confidence intervals. Binomial logistic regression followed by Tukey’s HSD (honest significant difference) test was used to determine significance of the habituated level (proportion reversing at tap 30) for each pair of strains. All strains are statistically different from each other (p < 0.001). (B) efhc-1 mutants with absent cilia due to a daf-19 mutation show intermediate habituation compared to daf-19 and efhc-1 single mutants. All strains are statistically different from each other (p < 0.001). (C) An unc-2(gain-of-function) mutation driven in EFHC-1-expressing cells shows slow habituation compared to wild-type animals, but faster habituation compared to animals with the unc-2(gain-of-function) mutation expressed throughout. Furthermore, efhc-1 mutants with the addition of an unc-2(gain-of-function) mutation driven in EFHC-1-expressing cells does not exacerbate the habituation phenotype. All strains are statistically different from each other (p < 0.001). For sample sizes (n) for tap habituation experiments and complete statistical comparisons (p values) see Supplementary files 1 and 2, respectively.
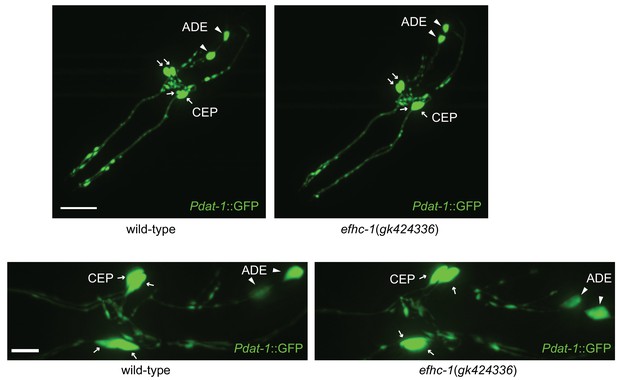
Synaptic structure in efhc-1 mutants is indistinguishable from wild-type.
Soluble GFP protein expressed under the dat-1 promoter marks CEP and ADE neurons including dendrites, axons and cell bodies in head. Z-stack images are taken with 40x oil immersion (top) and 63x water immersion (bottom) objective lens. Arrowheads indicate ADE cell bodies and arrows represent CEP cell bodies. Scale bar, 22 μm (top) and 7 μm (bottom).
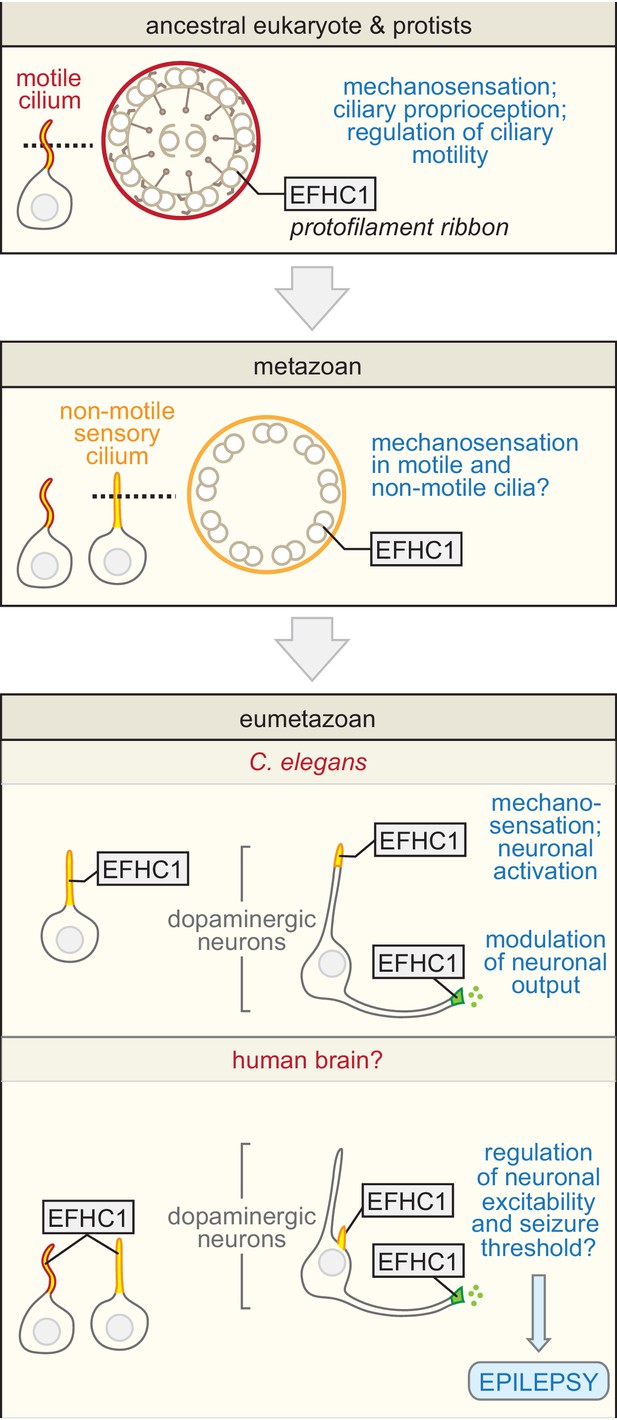
Proposed evolutionary model for the exaptation of EFHC1 motility-associated ciliary functions in non-motile cilia and synapses.
The role of EFHC1 in extant protists (and probably the ancestral eukaryote) is likely to regulate ciliary motility (Ikeda et al., 2003), potentially by acting as a mechanosensor together with a TRP family (or other) calcium channel. In metazoans, this function was retained for motile cilia (Suzuki et al., 2009), and adapted for mechanosensation and potentially other signaling roles in non-motile cilia. EFHC1 evolved in C. elegans, and potentially other neuron-bearing eumetazoans, to also regulate synaptic transmission. In C. elegans, the combined functions for EFHC-1 at cilia (mechanosensory input) and at the synapse (output) help to regulate neuronal excitability and dopamine signaling, both of which may be relevant to epilepsy in human patients with mutations in EFHC1. Motile and non-motile cilia are shown in red and orange, and synapses in green.
Additional files
-
Supplementary file 1
Sample sizes (n) for tap habituation experiments.
- https://doi.org/10.7554/eLife.37271.018
-
Supplementary file 2
p values for statistical comparisons.
- https://doi.org/10.7554/eLife.37271.019
-
Transparent reporting form
- https://doi.org/10.7554/eLife.37271.020





