Reactivation of RNA metabolism underlies somatic restoration after adult reproductive diapause in C. elegans
Figures
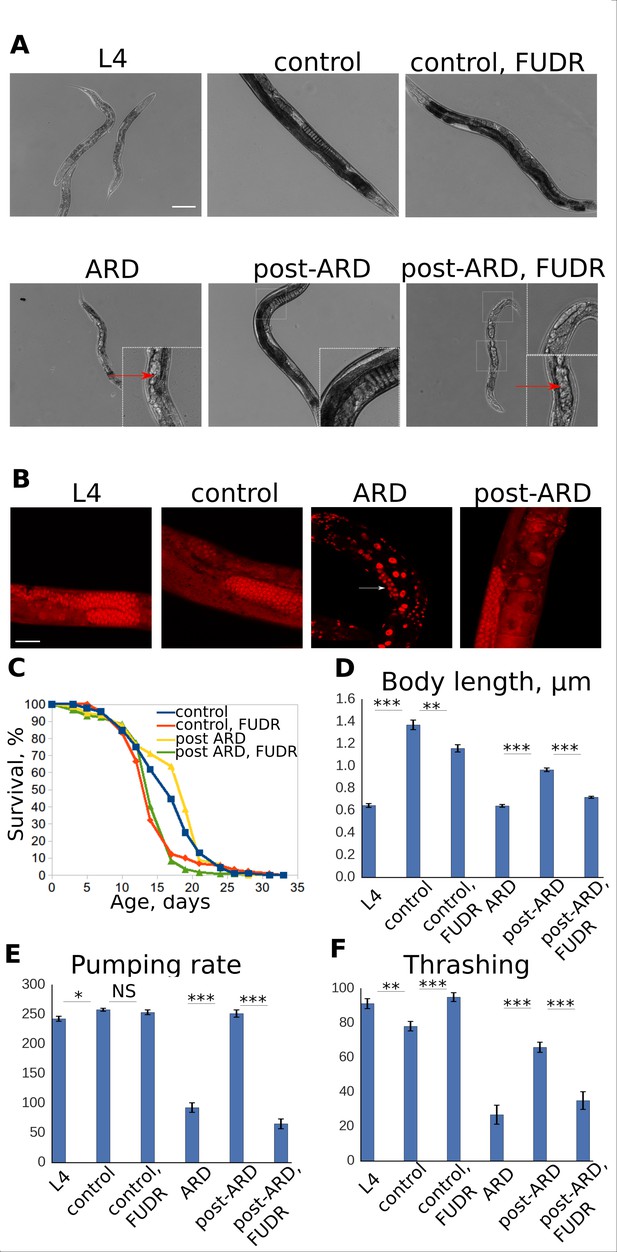
FUDR prevents post-ARD recovery.
(A) Representative DIC images of control, ARD and post-ARD animals. Upper panel: control L4 larva, 4 day old control adult, and 4 day old adult treated with FUDR from L4; lower panel: animal after 3 weeks in ARD (ARD), and animals which were allowed to recover from ARD for 4 days with and without FUDR (post-ARD). Insets show dead embryos and non-recovered germline in post-ARD FUDR condition. Scale bar is 100 µm. (B) DAPI staining of germline in L4 larva, 4 day old adult (control), animal that spent 3 weeks in diapause (ARD), and animal after 4 days of recovery (post-ARD). Reduced pool of GSCs in ARD animal is indicated by an arrow. Scale bar is 25 µm. (C) Survival curves of the control and post-ARD animals with or without FUDR. On day 0 of the experiment, ARD animals after 3 weeks in diapause and control L4 were transferred onto fresh OP50-seeded plates. The next day was considered day1 of adulthood for both conditions. See Figure 1—source data 1. (D) Body length (in micrometers) of L4 larvae, ARD, 4 day old control and post-ARD animals after 4 days of recovery. Shown are mean values ± SEM. See Figure 1—source data 2. (E) Pumping rates of control and post-ARD animals. L4 and 3 weeks old ARD animals were scored about 1 hr after transfer to fresh OP50 plates. The same cohorts (control and post-ARD) were examined again 4 days later. Indicated are rates per minute. Shown are mean values ± SEM. See Figure 1—source data 3. (F) Thrashing rates (per minute) of control and post-ARD animals. Shown are mean values ± SEM. See Figure 1—source data 4. *p<0.05; **p<0.005; ***p<0.0005.
-
Figure 1—source data 1
N2 lifespans.
- https://doi.org/10.7554/eLife.36194.004
-
Figure 1—source data 2
Body length.
- https://doi.org/10.7554/eLife.36194.005
-
Figure 1—source data 3
N2 pumping.
- https://doi.org/10.7554/eLife.36194.006
-
Figure 1—source data 4
N2 thrashing.
- https://doi.org/10.7554/eLife.36194.007
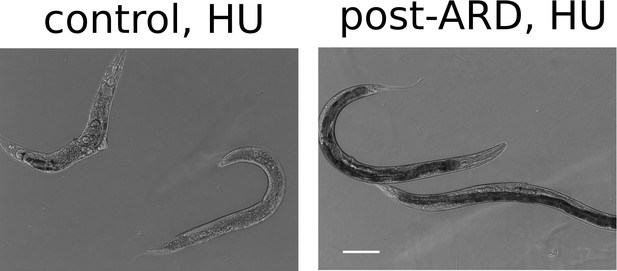
Representative DIC images of control and post-ARD animals treated with hydroxyurea.
L4 or ARD animals were transferred on NGM plates which were supplemented with 40 mM hydroxyurea and seeded with OP50. Animals were examined 4 days later. Post-ARD animals were less sensitive to hydroxyurea than control animals. Scale bar is 100 µm.
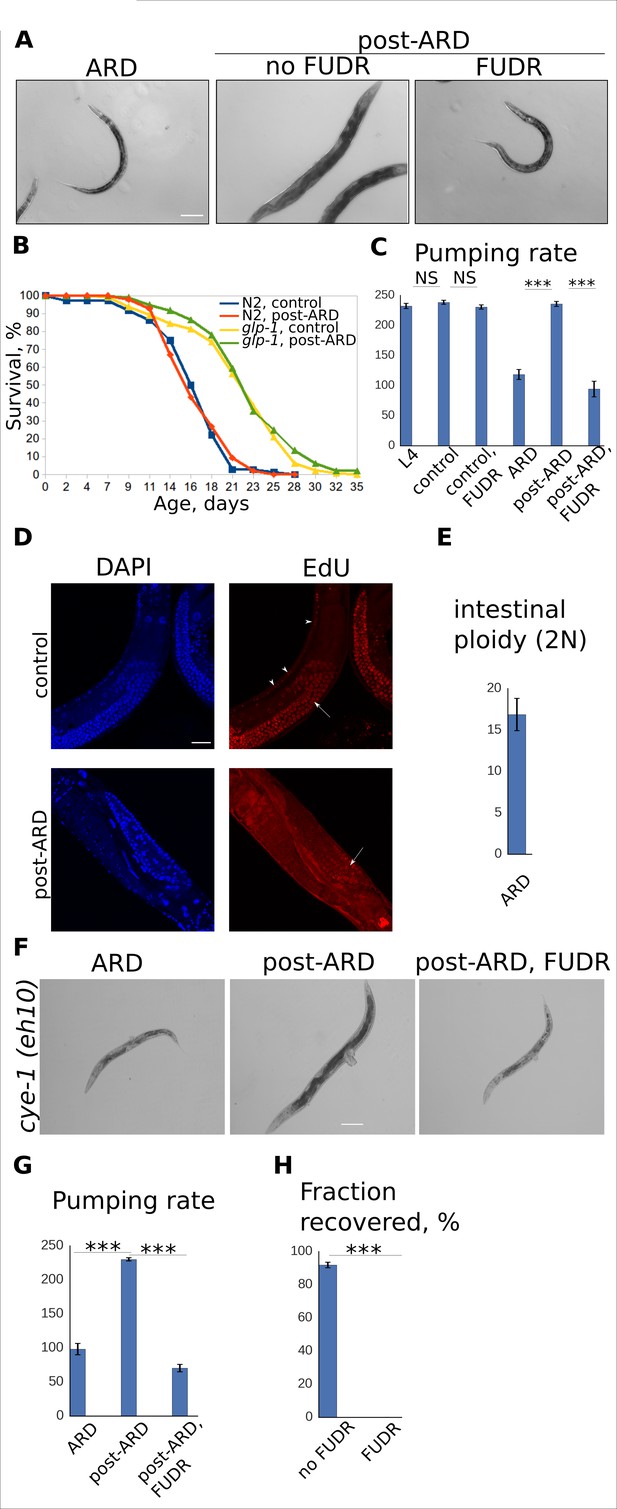
Adult reproductive diapause does not require germline or cell cycle activation.
(A) DIC images of ARD and post-ARD glp-1 (e2141) animals. FUDR prevented morphological improvements of the sterile glp-1 mutants. Scale bar is 100 µm. (B) Survival curves of the wild type (WT) and glp-1 mutants, control and post-ARD. On day 0, two weeks old ARD animals and control L4 were transferred onto fresh OP50. The next day was considered day1 of adulthood for both conditions. Shown are survival plots of recovered animals only. See Figure 2—source data 1. Also see Figure 2—figure supplement 1. (C) Pumping rates (per minute) for control and post-ARD glp-1 (e2141) animals. Shown are mean values ± SEM. See Figure 2—source data 2. (D) EdU labeling of control and post-ARD animals. Control animals were grown on EdU-labeled bacterial cells from egg till day 2 of adulthood. ARD animals were transferred on EdU-labeled cells for recovery and examined 5 days later. Arrows point to EdU-labeled germline cells nuclei, arrowheads point to EdU-labeled somatic nuclei. No somatic cells labeling was observed in post-ARD animals. Scale bar is 25 µm. (E) Ploidy (in diploid equivalents) of intestinal cells in ARD animals. Shown is the mean value ± SEM. See Figure 2—source data 3. (F) DIC images of ARD and post-ARD cye-1 (eh10) animals. FUDR prevented morphological improvements of cye-1 mutants. Scale bar is 100 µm. (G) Pumping rates (per minute) of control and post-ARD cye-1 (eh10) animals. Shown are mean values ± SEM. See Figure 2—source data 4. (H) Fraction of cye-1 (eh10) animals that morphologically recovered after diapause with and without FUDR. Shown are mean values ± SEM. See Figure 2—source data 5. Also see Figure 2—source data 6. *p<0.05; **p<0.005; ***p<0.0005.
-
Figure 2—source data 1
N2 and glp-1 lifespans.
- https://doi.org/10.7554/eLife.36194.010
-
Figure 2—source data 2
glp-1 pumping rate.
- https://doi.org/10.7554/eLife.36194.011
-
Figure 2—source data 3
Intestinal ploidy.
- https://doi.org/10.7554/eLife.36194.012
-
Figure 2—source data 4
cye-1 pumping.
- https://doi.org/10.7554/eLife.36194.013
-
Figure 2—source data 5
cye-1 recovery rate.
- https://doi.org/10.7554/eLife.36194.014
-
Figure 2—source data 6
N2 and glp-1 lifespans.
- https://doi.org/10.7554/eLife.36194.015
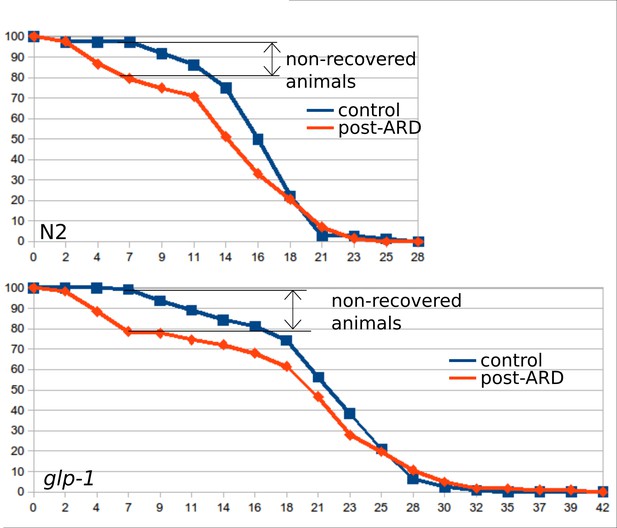
Survival curves of the wild type (WT) and glp-1 mutants, control and post-ARD.
Maintenance at 25°C increased the number of animals that do not recover from the diapause: these animals do not resume growth and do not show visible morphological restoration. These animals tend to die earlier than normally recovering animals. Related Figure 2B shows survival curves in which animals that did not recover morphologically were excluded from analysis. Similar fractions (~20%) of N2 and glp-1 (e2141) animals did not recover after diapause at 25°C. See Figure 2—source data 6.
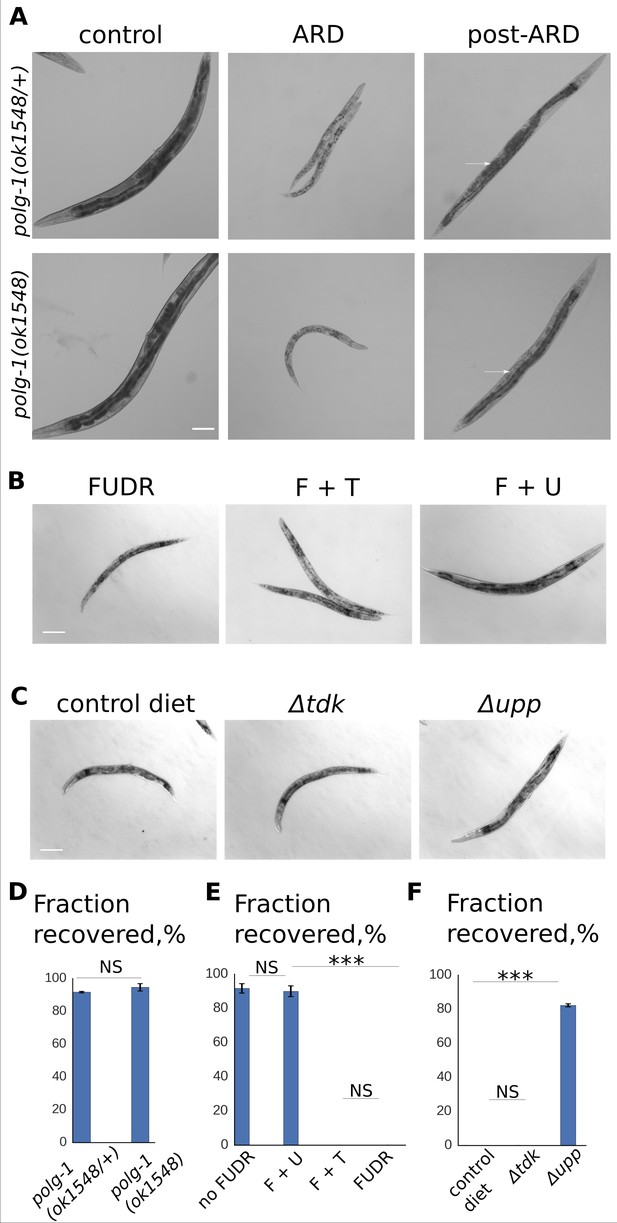
RNA, but not DNA, metabolism is required for post-ARD recovery.
(A) DIC images of polg-1 (ok1548/+) heterozygotes and polg-1 (ok1548) mutants. Animals spent 3 weeks in the diapause and were then allowed to recover for 4 days on fresh OP50 plates. Arrows point to newly produced eggs in case of polg-1 (ok1548/+) heterozygotes and to non-recovered germline in case of polg-1 (ok1548) mutants. Scale bar is 100 µm. (B) DIC images of post-ARD animals recovering on 50 µM FUDR, 50 µM FUDR +1 mM uracil and 50 µM FUDR +1 mM thymine. Uracil, but not thymine rescued growth and recovery of post-ARD animals in presence of FUDR. Scale bar is 100 µm. (C) DIC images of post-ARD animals recovering on NGM plates containing 25 µM FUDR and seeded with different bacterial cells from Keio knockout collection. Control diet – BW25113, Δtdk – JW1226-1, Δupp – JW2483-1. Inhibitory effect of FUDR was reduced by JW2483-1 cells. Scale bar is 100 µm. (D) Fraction of polg-1 (ok1548/+) and polg-1 (ok1548) animals that morphologically recovered after ARD. Shown are mean values ± SEM. See Figure 3—source data 1. (E) Fractions of animals that morphologically recovered after ARD in various conditions (see Figure 3B). Shown are mean values ± SEM. See Figure 3—source data 2. (G) Fractions of animals that morphologically recovered after ARD with different bacterial diets in presence of FUDR (see Figure 3C). Shown are mean values ± SEM. See Figure 3—source data 3. *p<0.05; **p<0.005; ***p<0.0005.
-
Figure 3—source data 1
polg-1 recovery rate.
- https://doi.org/10.7554/eLife.36194.019
-
Figure 3—source data 2
Recovery with uracil.
- https://doi.org/10.7554/eLife.36194.020
-
Figure 3—source data 3
Recovery with bacterial mutants.
- https://doi.org/10.7554/eLife.36194.021
-
Figure 3—source data 4
mtDNA content.
- https://doi.org/10.7554/eLife.36194.022
-
Figure 3—source data 5
Recovery with uridine.
- https://doi.org/10.7554/eLife.36194.023
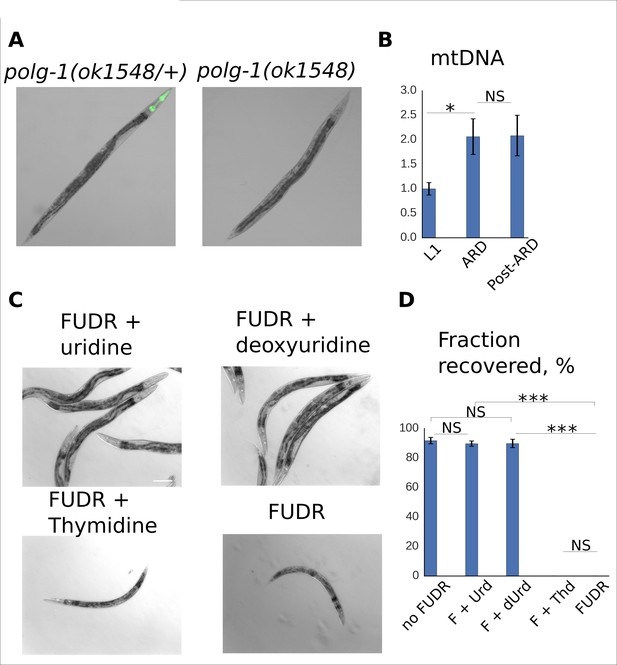
DNA metabolism is dispensable for post-ARD recovery.
(A) VC1224 (polg-1(ok1548/+)) animals were crossed with DR2078 strain carrying a GFP transgene which is expressed in the pharynx. The resulting strain carries a balancer Chromosome II with the fluorescent marker. Polg-1(ok1548/+) heterozygotes carrying the marker can be easily distinguished from polg-1(ok1548) mutants under a fluorescent dissecting scope. Absence of the wild type polg-1 gene in GFP-negative animals was also confirmed by PCR. (B) Relative mtDNA level in L1, ARD and post-ARD germline deficient glp-1 (e2141) animals determined by single-worm qPCR. Values are normalized for mtDNA content of L1 larvae. Shown are mean values ± SEM. See Figure 3—source data 4. (C) DIC images of post-ARD animals recovering on 50 µM FUDR, 50 µM FUDR +1 mM uridine, 50 µM FUDR +1 mM deoxyuridine, 50 µM FUDR +1 mM thymidine. Uridine and deoxyuridine rescued growth and recovery of post-ARD animals in presence of FUDR. Scale bar is 100 µm. (D) Fractions of animals that morphologically recovered after ARD in various conditions (see panel C). Urd = Uridine, dUrd = Deoxyuridine, Thd = Thymidine. Shown are mean values ± SEM. See Figure 3—source data 5. *p<0.05; **p<0.005; ***p<0.0005.
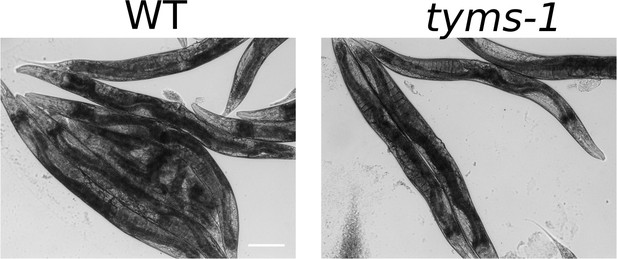
Post-ARD wild type animals and tyms-1 mutants are shown.
Animals were allowed to recover at 25°C, which is a restrictive temperature for tyms-1 mutants. Both strains recovered normally in these conditions. tyms-1 mutant animals did not show any visible abnormalities after ARD, including a normally recovered germline. We confirmed that tyms-1 mutant eggs are inviable at 25°C. Scale bar is 100 µm.
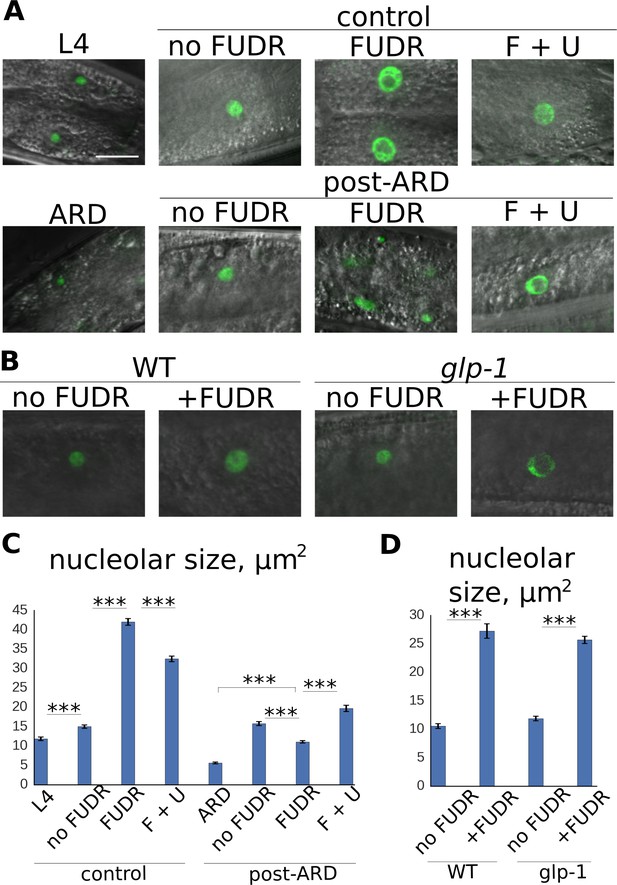
Post-ARD recovery is associated with nucleolar expansion.
(A) Representative images of nucleolar morphology. Shown are nucleoli of intestinal cells visualized with fib-1::GFP. Control L4 and ARD animals were imaged before transfer to fresh OP50 plates containing no FUDR, 50 µM FUDR and 50 µM FUDR +1 mM uracil (F + U). Animals were then analyzed 4 days after transfer. Top panel: control animals at L4 stage and at 4 days of adulthood. Bottom panel: ARD animals before and after 4 days of recovery with or without FUDR. (B) FUDR induces nucleolar stress in post-mitotic tissues independently of the germline. Wild type and sterile glp-1 mutants were grown at restrictive temperature (25°C) until day 1 of adulthood and then transferred onto FUDR-containing plates and analyzed 3 days later. (C) Quantification of nucleolar size from intestinal cells (in μm2) of animals treated as in 4A. Shown are mean values ± SEM. See Figure 4—source data 1. (D) Quantification of nucleolar size from intestinal nucleoli (in μm2) of animals treated as in 4B. Shown are mean values ± SEM. See Figure 4—source data 2. Also see Figure 4—source data . *p<0.05; **p<0.005; ***p<0.0005. Scale bar is 10 µm.
-
Figure 4—source data 1
Post-ARD nucleolar size.
- https://doi.org/10.7554/eLife.36194.026
-
Figure 4—source data 2
- https://doi.org/10.7554/eLife.36194.027
-
Figure 4—source data 3
Nucleolar size, AL condition.
- https://doi.org/10.7554/eLife.36194.028
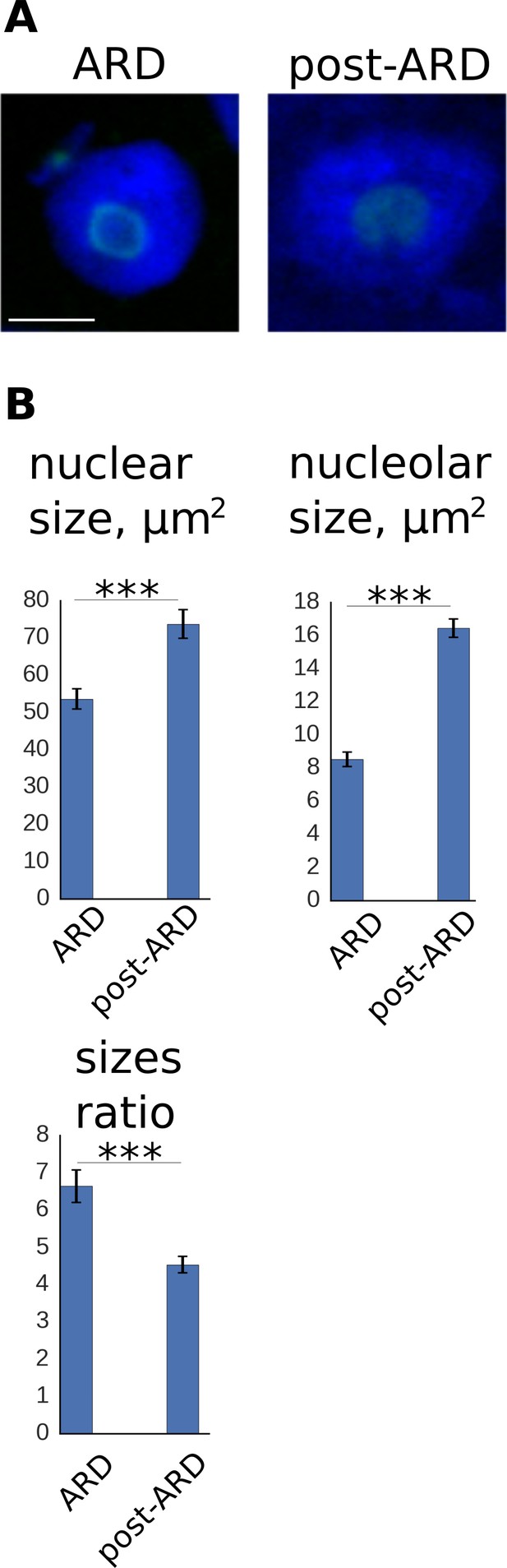
Change of nuclear and nucleolar sizes upon exit from ARD.
(A) Animals expressing fib-1::GFP marker were stained with DAPI for simultaneous examination of nuclei and nucleoli. Shown are nuclei of ARD and post-ARD animals. Scale bar is 5 µm. (B) Quantification of nuclear and nucleolar sizes (in μm2) and their ratio in ARD and post-ARD animals. Both nucleoli and nuclei expand upon ARD recovery. Relatively large nucleolar expansion leads to reduction of nucleus/nucleolus ratio upon exit from ARD. See Figure 4—source data 3.
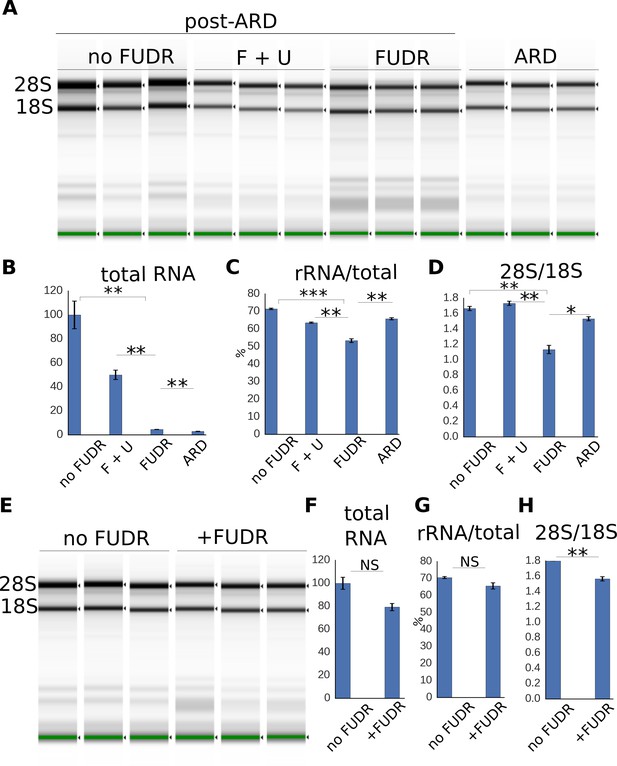
Expansion of the RNA pool during post-ARD recovery.
(A) Total RNA extracted from post-ARD animals recovering in various conditions: no FUDR, 50 µM FUDR +1 mM uracil (F + U), 50 µM FUDR and ARD animals before recovery. Around 500 animals were used for no FUDR and FUDR +uracil sample preps, 3.5-fold more animals (around 1750) were used for FUDR and ARD preps. F + U and no FUDR samples were diluted 5-fold more than FUDR and ARD samples to fit into the quantitative range of TapeStation System. (B,C,D) Quantification of conditions shown in 5A. Normalized total RNA content (B), fraction of rRNA species in the total analyzed RNA, % (C) and ratio of 28S to 18S rRNA abundances (D). Shown are mean values ± SEM. Total RNA content was calculated from the TapeStation and adjusted to reflect dilution factor and number of animals (total RNA per animal). Values are expressed as % of RNA content in no FUDR sample, which was set as 100%. See Figure 5—source data 1. (E) Total RNA extracted from control animals that were transferred on fresh OP50 plates with or without FUDR at L4 stage. Around 300 animals were used per prep. (F, G, H) Quantification of conditions shown in 5E. Total RNA content (F), fraction of rRNA species in the total analyzed RNA, % (G), ratio of 28S to 18S rRNA abundances (H). Total RNA content is presented similarly to Figure 5B. Values are expressed as % of RNA content in no FUDR sample, which is set as 100%. All values are means ± SEM. See Figure 5—source data 1. *p<0.05; **p<0.005; ***p<0.0005.
-
Figure 5—source data 1
RNA analysis.
- https://doi.org/10.7554/eLife.36194.031
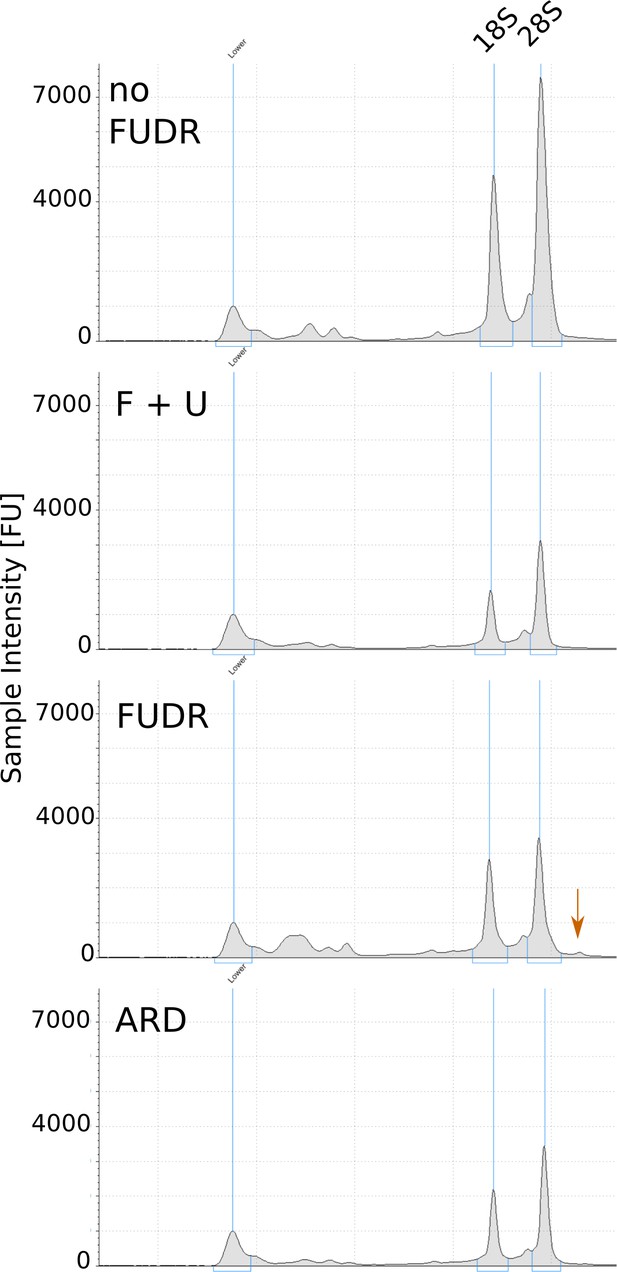
Representative TapeStation electropherograms of total RNA extracted from ARD and post-ARD animals.
Arrow marks high molecular weight species, presumably unprocessed rRNA precursor, accumulating in the presence of FUDR. Samples were prepared as in Figure 5A.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information | ||
|---|---|---|---|---|---|---|
| Material and me strain, strain background (C. elegans) | glp-1 (e2141); glp-1 | CGC | CGC: CB4037 | |||
| Strain, strain background (C. elegans) | CGC | CGC: DR2078 | ||||
| Strain, strain background (C. elegans) | CGC | CGC: VC1224 | ||||
| Strain, strain background (C. elegans) | polg-1 (ok1548); polg-1 (ok1548/+) | This study | Cross of DR2078 and VC1224 | |||
| Strain, strain background (C. elegans) | fib-1::GFP | CGC | CGC: COP262 | |||
| Strain, strain background (C. elegans) | tyms-1 | CGC | CGC: MJ65 | |||
| Strain, strain background (C. elegans) | cye-1; cye-1 (eh10) | CGC | CGC: KM166 | |||
| Strain, strain background (E. coli) | MG1693 (Δthy) | E. coli Genetic Resources at Yale | CGSC#: 6411 | |||
| Strain, strain background (E. coli) | parent strain; control diet; BW25113 | E. coli Genetic Resources at Yale | CGSC#: 7636 | |||
| Strain, strain background (E. coli) | Δtdk; JW1226-1 | E. coli Genetic Resources at Yale | CGSC#: 9112 | |||
| Strain, strain background (E. coli) | Δupp; JW2483-1 | E. coli Genetic Resources at Yale | CGSC#: 9982 | |||
| Chemical compound, drug | 5-Ethynyl-2'- deoxyuridine, EdU | Invitrogen | ThermoFisher: C10339 | |||
| Chemical compound, drug | Uracil, U | Alfa Aesar | Fisher scientific: AAA1557018 | |||
| Chemical compound, drug | Uridine, Urd | Alfa Aesar | Fisher scientific: A1522706 | |||
| Chemical compound, drug | Thymine, T | Acros Organics | Fisher scientific: AC157850050 | |||
| Chemical compound, drugMaterial and me | Thymidine, Thd | Alfa Aesar | Fisher scientific: A1149306 | |||
| Chemical compound, drug | Deoxyuridine, dUrd | Alfa Aesar | Fisher scientific: A1602603 | |||
| Commercial assay or kit | High Sensitivity RNA ScreenTape Ladder | Agilent | Agilent: 5067– 5581 | |||
| Commercial assay or kit | High Sensitivity RNA ScreenTape Sample Buffer | Agilent | Agilent: 5067– 5580 | |||
| Commercial assay or kit | High Sensititive RNA ScreenTape | Agilent | Agilent: 5067– 5579 | |||
| Sequence-based reagent | nd1 forward | IDT | AGCGTCATTTATTG GGAAGAAGAC | |||
| Sequence-based reagent | nd1 reverse | IDT | AAGCTTGTGCTAAT CCCATAAATGT | |||
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.36194.032





