A homozygous loss-of-function CAMK2A mutation causes growth delay, frequent seizures and severe intellectual disability
Figures
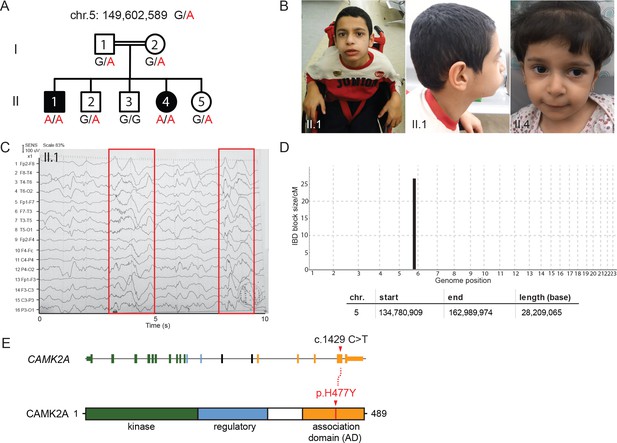
A new syndrome of global neuro-developmental delay with seizures caused by a biallelic mutation in CAMK2A.
(A) Pedigree of a consanguineous Jordanian family with two affected siblings with germline homozygous mutations in CAMK2A. The genotypes of all individuals were verified by Sanger sequencing. (B) Photographs of the two affected siblings with normal head circumferences. (C) EEG graph of patient II.I showing abnormal epileptiform transients (red boxes) (D) Homozygosity mapping delineates one candidate locus on chromosome 5. (E) CAMK2A exonic structure and CAMK2A protein domains. Patients II:1 and II:4 carry biallelic missense mutation p. H477Y located in CAMK2A association domain (AD). Nucleotide change c.1429 C > T refers to position on CAMK2A cDNA.
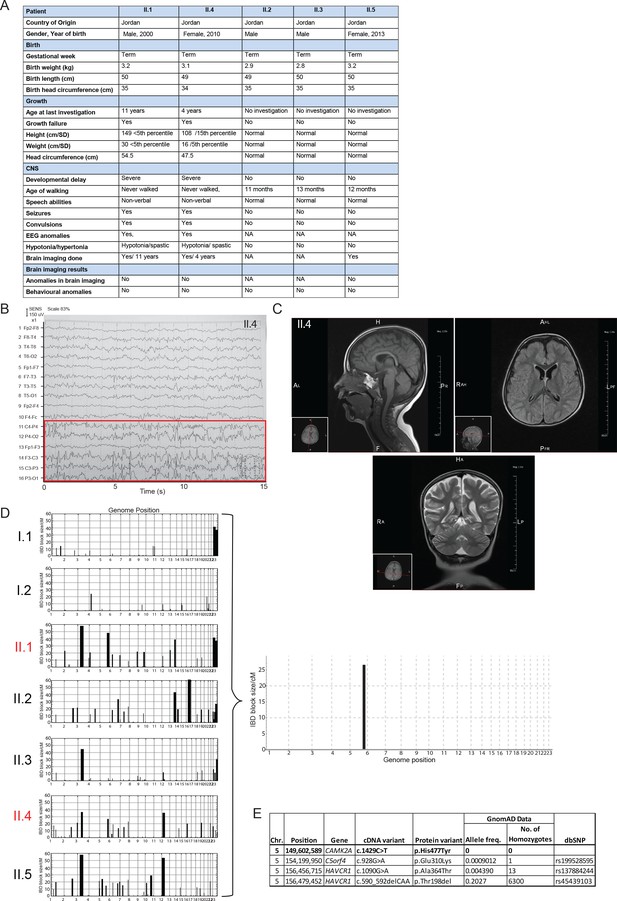
Genetic and clinical findings from the two patients with global developmental delay.
(A) Clinical table detailing the growth parameters and learning deficits of the two affected children. (B) EEG of patient II.4 showing abnormal waveforms (red box). (C) MRI images of patient II.4 showing no gross structural abnormalities in the brain. (D) Graphs showing homozygous regions identified through IBD mapping for each family member prior to filtering (E) Table of 4 homozygous genes that lie within the Chr. 5 IBD region.
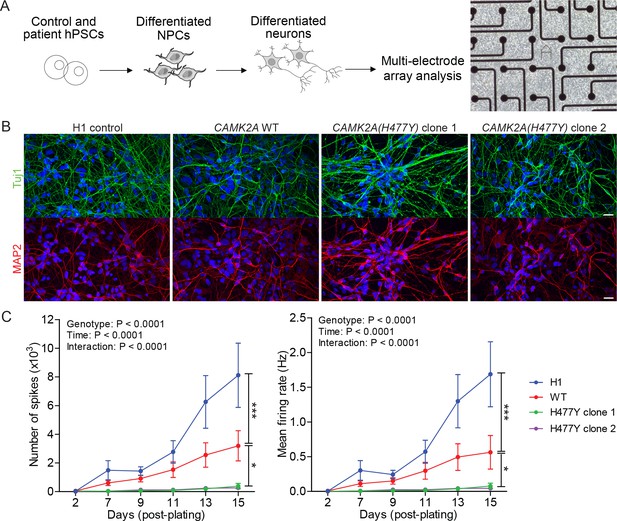
CAMK2A mutant iPSC-derived neurons are functionally less active.
(A) Schematic of the hPSC-derived neuronal activity assay with representative image of iPSC-derived neurons plated on a multi-electrode array (B) Representative confocal images of immunofluorescence staining of neuronal lineage markers TUJ1 (green) and MAP2 (red) show efficient differentiation of iPSCs into neurons. Scale bar represents 20 µm. (C) Graphs showing the number of neuron-evoked spikes and mean firing rate detected by multi-electrode arrays. (n = 7 per line per time-point; Values shown as mean ±SEM; Two-way ANOVA with Tukey post-hoc test; *p<0.05 and ***p<0.001).
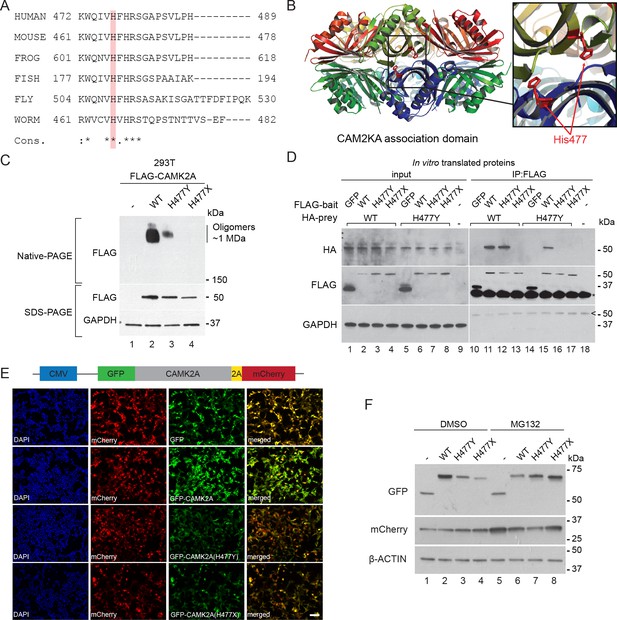
p.H477Y affects CAMK2A oligomerization and protein stability.
(A) Sequence conservation of CAMK2A homologs. Histidine 477 (H477) is highlighted in red. (B) X-ray crystal structure of human CAMK2A AD tetradecamer (PDB: 5IG3). H477 (red) is located at the equatorial dimer interface. (C) Defective oligomerization of the p.H477Y mutant. 293 T cells were transiently transfected with FLAG tagged wild-type CAMK2A and CAMK2AH477Y. A third mutant, CAMK2AH477X which lacks part of the AD (a.a. 478–489) was used as positive control. (D) Defective self-association of the p.H477Y mutant. The indicated FLAG- and HA-tagged CAMK2A wild-type and mutant proteins were synthesized in vitro using rabbit reticulocyte lysate. FLAG-GFP was used a negative control. FLAG-tagged proteins were immunoprecipitated using anti-FLAG M2 agarose resin in the presence of 1% NP40. Co-immunoprecipitated proteins were analyzed by SDS-PAGE. *, IgG light chain. ^, IgG heavy chain. (E) p.H477Y mutation lowers expression of CAMK2A in cells. 293 T cells were transfected with reporter plasmids encoding GFP-tagged wild-type CAMK2A, CAMK2AH477Y and CAMK2AH477X mutants, followed by T2A peptide and mCherry. Representative confocal images show lower expression of mutant GFP- CAMK2AH477Y compared to wild-type. Scale bar represents 100 µm. (F). p.H477Y decreases CAMK2A stability via proteasomal degradation. 293 T cells were transfected as in (E) and treated with 10 µM MG132 or DMSO for 16 hr. 10 µg total cell lysate was used for SDS-PAGE and Western blot.
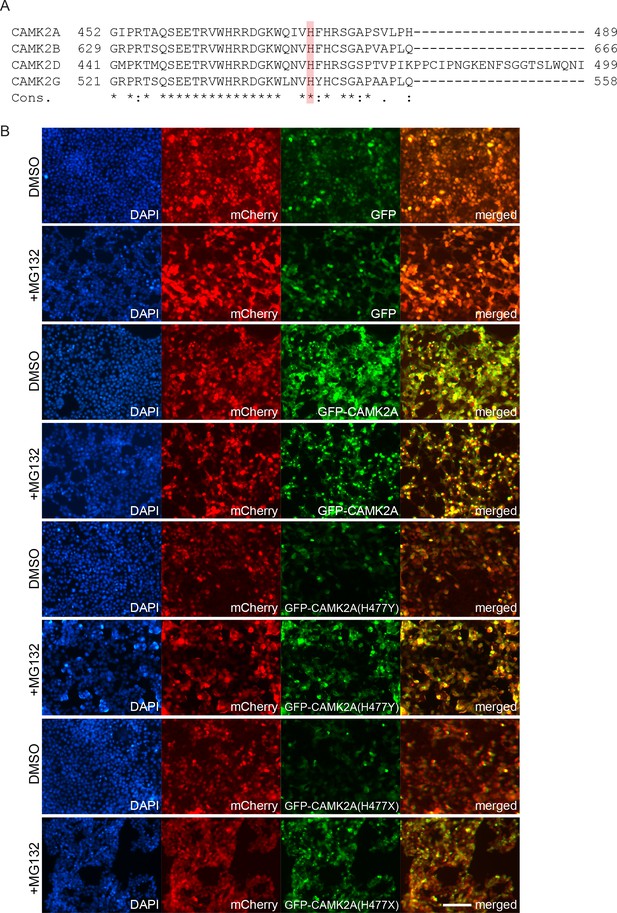
Decreased stability and defective cytoplasmic localization of the CAMK2AH477Y mutant.
(A) Sequence conservation of CAMK2 paralogs. His477 (red) is invariant in all human CAMK2 paralogues with an association domain. (B) p.H477Y mutant is subject to proteasomal degradation. 293 T cells were transfected with plasmids encoding GFP-tagged wild-type CAMK2A, p.H477Y and p.H477X mutants. Cells were incubated with DMSO or 2.5 μM MG132 for 16 hr 1 day post transfection. GFP intensity in the cells is increased in the CAMK2A p.H477Y and p.H477X mutants after MG132 treatment. Scale bar represents 100 µm.
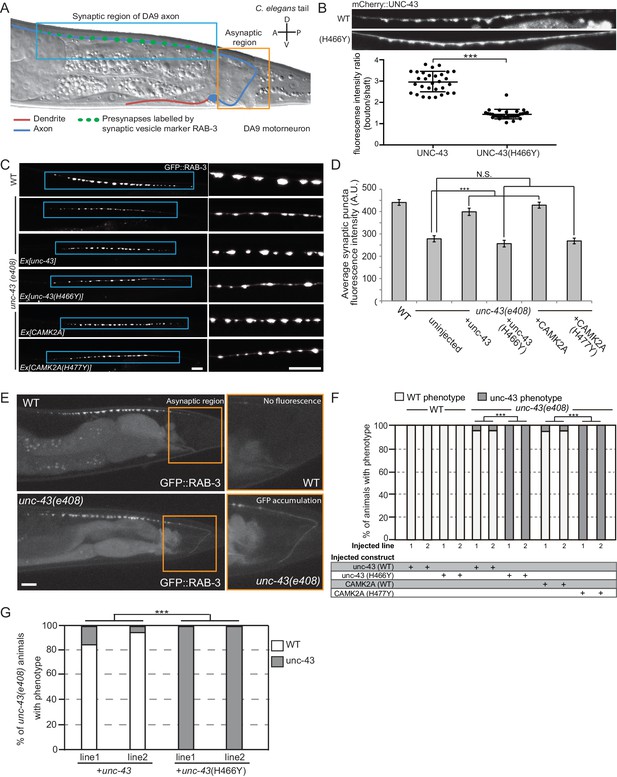
CAMK2AH477Y mutant fails to rescue synaptic defects in unc-43 C. elegans neurons.
(A) Schematic drawing of C. elegans motor neuron, DA9 in the tail region. DA9 extends a dendrite (red) anteriorly and an axon (blue) that extends posteriorly crosses the midline of the animal and anteriorly in the dorsal nerve cord (DNC). It forms approximately 20 en passant synapses within a discrete stretch along the DNC (blue box). DA9 presynaptic vesicles were marked with RAB-3 (GFP::RAB-3). The asynaptic region (yellow box) is devoid of any synaptic vesicle accumulation. (B) The localization of mCherry::UNC-43 and mCherry::UNC-43(H466Y) in DA9 synapses. Note that UNC-43 accumulates at synaptic boutons while UNC-43(H466Y) is diffusely localized. Fluorescent intensity of mCherry::UNC-43 was measured at synaptic boutons and along the axonal shaft. Graph plots the ratio of fluorescence intensity at synaptic boutons compared to the axonal shaft of 30 synapses from three animals. Graph shows the mean and error bars show SEM, ***p-value 6.32e−19, Student’s T-test. (C) Representative confocal images demonstrating presynaptic puncta size changes between WT and unc-43(e408) mutants. unc-43 mutants have smaller presynaptic puncta along the DNC. This defect is rescued by expression of either UNC-43 or CAMK2A in DA9 whilst the mutated UNC-43H466Y and CAMK2AH477Y fail to rescue. (D) Quantification of average puncta intensity from WT and unc-43(e408) animals. Error bars represent SEM with number of synaptic puncta quantified n > 80, N.S. is not significant, ***p-value<0.001 (uninjected vs unc-43 p-value 5.0e8, uninjected vs unc-43H466Y p-value 4.17, uninjected vs CAMK2A p-value 4.25e−12, uninjected vs CAMK2AH477Y p-value 9.40), One-Way ANOVA with Bonferroni posthoc test. (E) Representative confocal images showing mislocalization of GFP::RAB-3 into the asynaptic region (yellow box) in unc-43 DA9 neuron. (F) Rescue of the unc-43(e408) phenotype by DA9 cell-specific expression of UNC-43 or CAMK2A. UNC-43H466Y and CAMK2AH477Y fail to rescue the unc-43 phenotype. Graph shows the percentage of animals with the WT and unc-43 mutant phenotypes. ***p<0.001 (unc-43 vs unc-43H466Y p-value 2.13e−51, CAMK2A vs CAMK2AH477Y p-value 3.77e−50), Fisher Exact test with n = 100 animals scored for each line. (G) Behavioral rescue by expressing wild-type UNC-43 or UNC-43H466Y in unc-43(e408) mutants. The behavior was scored as either wild-type or unc-43. Two independent worm lines were analyzed for each condition. *** p-value 5.29e−41, Fisher Exact test with n = 100 animals scored for each line.
Videos
Video of patient II.1
https://doi.org/10.7554/eLife.32451.005Video of patient II.4
https://doi.org/10.7554/eLife.32451.006Tables
List of homozygous variants identified by Whole Exome Sequencing.
https://doi.org/10.7554/eLife.32451.007| Chr | Position | Gene | cDNA variant | Protein variant |
|---|---|---|---|---|
| 1 | 33,476,435 | AK2 | c.*45–1G > T | |
| 1 | 36,752,343 | THRAP3 | c.512C > T | p.Ser171Phe |
| 1 | 36,932,102 | CSF3R | c.2273C > T | p.Thr758Ile |
| 1 | 39,758,439 | MACF1 | c.1931G > T | p.Gly644Val |
| 1 | 145,365,316 | NBPF10 | c.9941G > A | p.Gly3314Glu |
| 2 | 11,758,842 | GREB1 | c.3841G > A | p.Ala1281Thr |
| 2 | 29,404,617 | CLIP4 | c.1976G > A | p.Arg659Gln |
| 2 | 64,779,195 | AFTPH | c.587G > A | p.Gly196Glu |
| 2 | 238,277,211 | COL6A3 | c.4895G > A | p.Arg1632Gln |
| 2 | 241,987,827 | SNED1 | c.1369G > A | p.Glu457Lys |
| 3 | 38,348,802 | SLC22A14 | c.574G > A | p.Ala192Thr |
| 3 | 44,672,687 | ZNF197 | c.524C > T | p.Ala175Val |
| 3 | 47,452,772 | PTPN23 | c.3484C > T | p.Arg1162Trp |
| 3 | 52,556,184 | STAB1 | c.6403C > G | p.Pro2135Ala |
| 3 | 67,426,232 | SUCLG2 | c.1235T > C | p.Ile412Thr |
| 3 | 197,422,844 | KIAA0226 | c.1366C > T | p.Arg456Trp |
| 4 | 9,174,981 | FAM90A26P | c.83T > G | p.Val28Gly |
| 4 | 9,175,603 | FAM90A26P | c.211C > G | p.Pro71Ala |
| 4 | 10,089,539 | WDR1 | c.743A > G | p.His248Arg |
| 4 | 15,529,151 | CC2D2A | c.1231T > G | p.Ser411Ala |
| 5 | 74,021,852 | GFM2 | c.1820_1825delTTGAGT | p.Glu608_Phe609del |
| 5 | 78,610,479 | JMY | c.2464C > A | p.Pro822Thr |
| 5 | 149,602,589 | CAMK2A | c.1429C > T | p.His477Tyr |
| 5 | 154,199,950 | C5orf4 | c.928G > A | p.Glu310Lys |
| 5 | 156,456,715 | HAVCR1 | c.1090G > A | p.Ala364Thr |
| 5 | 156,479,452 | HAVCR1 | c.590_592delCAA | p.Thr198del |
| 6 | 26,509,392 | BTN1A1 | c.1571G > A | p.Gly524Glu |
| 6 | 27,215,709 | PRSS16 | c.119G > A | p.Ser40Asn |
| 6 | 32,806,007 | TAP2 | c.4C > T | p.Arg2Trp |
| 6 | 33,260,924 | RGL2 | c.1876G > A | p.Gly626Arg |
| 6 | 38,704,952 | DNAH8 | c.221C > A | p.Ala74Asp |
| 6 | 43,412,643 | ABCC10 | c.2807C > T | p.Pro936Leu |
| 6 | 129,932,746 | ARHGAP18 | c.1054C > T | p.Arg352Ter |
| 6 | 131,946,054 | MED23 | c.235C > T | p.Leu79Phe |
| 6 | 151,674,121 | AKAP12 | c.4595_4596insGGC | p.Asp1532delinsGluAla |
| 6 | 168,479,677 | FRMD1 | c.98A > C | p.Glu33Ala |
| 7 | 5,352,665 | TNRC18 | c.7851_7856dupCTCCTC | p.Ser2618_Ser2619dup |
| 7 | 45,123,857 | NACAD | c.1922T > C | p.Val641Ala |
| 7 | 143,884,437 | ARHGEF35 | c.1040C > T | p.Thr347Ile |
| 7 | 149,422,981 | KRBA1 | c.1304C > T | p.Ala435Val |
| 7 | 151,680,130 | GALNTL5 | c.428A > G | p.Tyr143Cys |
| 8 | 12,285,064 | FAM86B1|FAM86B2 | c.310T > C | p.Ser104Pro |
| 8 | 12,285,250 | FAM86B2 | c.808C > T | p.Arg270Trp |
| 8 | 86,574,132 | REXO1L1 | c.1595A > C | p.Asp532Ala |
| 9 | 12,775,863 | LURAP1L | c.149_150insTGGCGG | p.Gly49_Gly50dup |
| 9 | 40,706,047 | FAM75A3 | c.3704A > G | p.His1235Arg |
| 9 | 41,323,425 | FAM75A4 | c.1908C > T | p.Arg637Trp |
| 9 | 41,323,469 | FAM75A4 | c.1864G > A | p.Gly622Asp |
| 9 | 43,822,668 | CNTNAP3B | c.1222C > T | p.Leu408Phe |
| 10 | 51,748,684 | AGAP6 | c.209G > A | p.Arg70Gln |
| 10 | 81,471,741 | FAM22B | c.2137T > C | p.Trp713Arg |
| 11 | 1,651,198 | KRTAP5-5 | c.129_137delAGGCTGTGG | p.Gly44_Gly46del |
| 11 | 12,316,388 | MICALCL | c.1408_1410dupCCT | p.Pro470dup |
| 12 | 7,045,917 | ATN1 | c.1488_1508delGCAGCAGCAGCAGCAGCAGCA | p.Gln496_Gln502del |
| 12 | 7,045,920 | ATN1 | c.1491_1508delGCAGCAGCAGCAGCAGCA | p.Gln497_Gln502del |
| 13 | 99,461,564 | DOCK9 | c.1271_1272insA | p.Leu425LeufsTer? |
| 13 | 114,503,875 | FAM70B | c.500_509 + 72delCCTGCGGGAGG TGAGGGGCACCGGGGACCCCCATATC TACACCTGCGGGAGGTGAGGGGC GCTGGGGACCCCCGTATCTACA | |
| 14 | 105,411,514 | AHNAK2 | c.10274C > T | p.Ala3425Val |
| 14 | 106,994,118 | IGHV3-48 | c.47G > A | p.Gly16Asp |
| 16 | 29,496,359 | c.916T > C | p.Ser306Pro | |
| 16 | 30,772,988 | C16orf93 | c.82G > A | p.Ala28Thr |
| 16 | 70,215,817 | CLEC18C | c.521C > T | p.Ala174Val |
| 17 | 39,211,189 | KRTAP2-2 | c.275G > C | p.Cys92Ser |
| 19 | 1,026,716 | CNN2 | c.56A > C | p.Lys19Thr |
| 19 | 10,084,460 | COL5A3 | c.3584T > C | p.Val1195Ala |
| 19 | 14,517,213 | CD97 | c.1892G > A | p.Ser631Asn |
| 21 | 36,042,462 | CLIC6 | c.776_805delGCGTAGAAGCGGGGGTCCCGGCGGGGGACA | p.Val260_Ser269del |
| 22 | 18,834,773 | c.329C > T | p.Thr110Ile | |
| X | 48,920,059 | CCDC120 | c.110A > G | p.Asp37Gly |
| X | 55,116,478 | PAGE2 | c.25T > A | p.Ser9Thr |
| X | 150,832,702 | PASD1 | c.954_971delCCCAATGGACCAGCAGGA | p.Pro319_Asp324del |
| X | 153,050,158 | SRPK3 | c.1_5delGACAG | p.Thr2LeufsTer57 |
| X | 154,010,046 | MPP1 | c.978A > C | p.Glu326Asp |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (human) | CAMK2A Calcium/Calmodulin Dependent Protein Kinase II Alpha | Uniprot: Isoform B (identifier: Q9UQM7-2) | Q9UQM7-2 | |
| Gene (C. elegans) | unc-43 Calcium/Calmodulin-Dependent Protein Kinase type II | Protein UNC-43 isoform d (Wormbase CDS K11E8.1d) | K11E8.1d | |
| Strain, strain background (C. elegans) | Worm (C. elegans) N2 Bristol Strain;unc-43(e408) | Caenorhabditis Genetics Center (CGC) PMID: 17941711 | 17941711 | |
| Recombinant DNA | pCDH-CMV-MCS-EF1α-Neo | Systems Biosciences (SBI) | CD514B-1 | |
| Recombinant DNA | pSM vector (a derivative of pPD49.26 with additional cloning sites) | Modified for this paper | NA | ADDGENE https://media.addgene.org/cms/files/Vec95.pdf |
| Recombinant DNA | pCDH-CMV-CAMK2A-T2A-mCherry | This paper; based on pCDH-CMV-MCS-EF1α-Neo | NA | |
| Recombinant DNA | pMIG-hOCT4 | Addgene | Plasmid #17225 | |
| Recombinant DNA | MSCV h c-MYC IRES GFP | Addgene | Plasmid #18119 | |
| Recombinant DNA | pMIG-hKLF4 | Addgene | Plasmid #17227 | |
| Recombinant DNA | pMIG-hSOX2 | Addgene | Plasmid #17226 | |
| Cell line (human) | Patient derived iPS neurons | This paper | NA | |
| Cell line (human) | 293T | Lab stock | NA | |
| Chemical compound, drug | MG132 | Sigma-Aldrich | M7449 | |
| Commercial kit | NativeMark PAGE | ThermoFisher | BN1002BOX | |
| Commercial kit | TnT Quick Coupled Transcription/Translation System | Promega | L1170 | |
| Antibody | Anti-FLAG Clone M2 | Sigma-Aldrich | F3165 | |
| Antibody | Anti-HA Clone Y-11 | Santa Cruz Biotechnology | sc-7392 | |
| Antibody | Anti-GADPH | Santa Cruz Biotechnology | sc-47724 | |
| Antibody | Anti-Tuj1 | Covance Research | MMS-435P | |
| Antibody | Anti-MAP2 | Synaptic Systems | 188 004 | |
| Cell culture reagent | 20% Knock Out Serum Replacement | Thermo Fisher | 10828–028 | |
| Cell culture reagent | bFGF | Stemgent | 37316 | |
| Cell culture reagent | Matrigel Basement Membrane Matrix | Corning | 354234 | |
| Cell culture reagent | mTeSR1 | STEMCELL Technologies | 85850 | |
| Cell culture reagent | CytoTune-iPS 2.0 Sendai Reprogramming Kit | ThermoFisher | A16517 | |
| Cell line (human) | H1 embryonic stem cells | Gift from Dr. Lawrence W. Stanton, WiCell | RRID:CVCL_C813 | |
| Antibody | Alexa Fluor 594 secondary Ab | ThermoFisher | Cat# A-11076, RRID:AB_2534120 | |
| Antibody | Alexa Fluor 488 secondary Ab | ThermoFisher | Cat# A-11001, RRID:AB_2534069 | |
| Assay system/kit | Maestro MEA System | Axion Biosystem | - |
Additional files
-
Supplemental file 1
Statistical Data Analysis
- https://doi.org/10.7554/eLife.32451.012
-
Source code 1
IBD linkage program.
- https://doi.org/10.7554/eLife.32451.013
-
Transparent reporting form
- https://doi.org/10.7554/eLife.32451.014





