Gdf3 is required for robust Nodal signaling during germ layer formation and left-right patterning
Figures
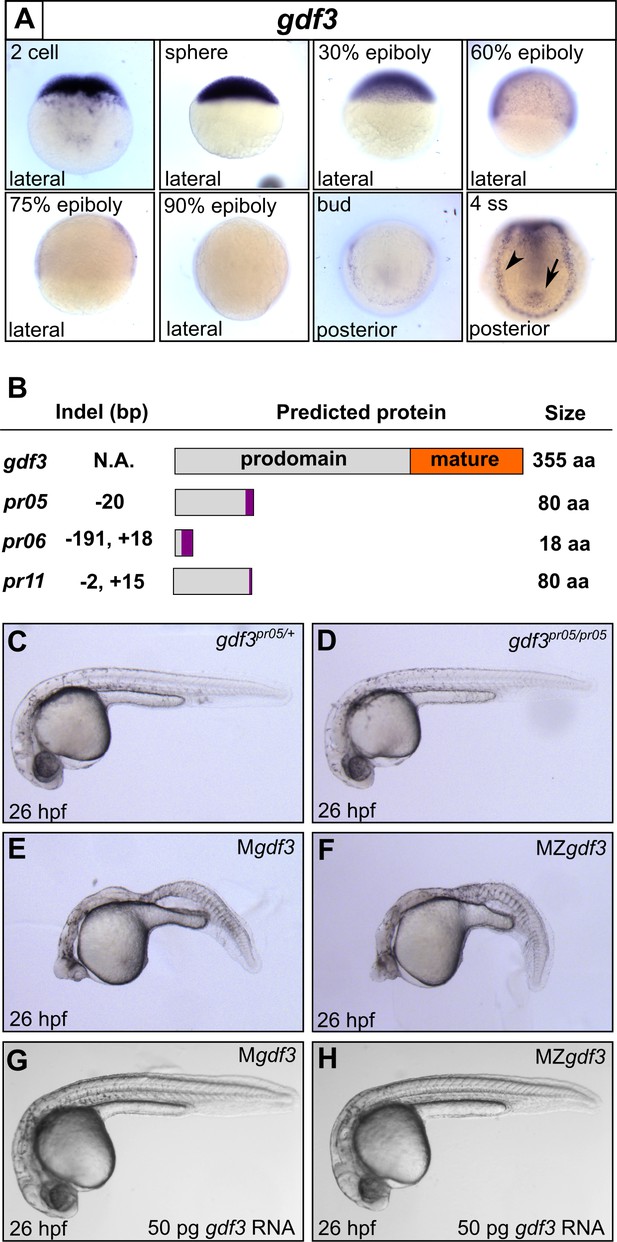
Mutations in maternal gdf3 lead to defects in embryonic patterning.
(A) RNA in situ hybridization shows gdf3 mRNA is broadly expressed throughout early zebrafish development, followed by specific expression in Kupffer’s vesicle (arrow) and the lateral plate mesoderm (arrowhead). Views are indicated. (B) Schematic of gdf3 mutant alleles predicted to form truncated proteins due to early stop codons in the first exon. Purple regions indicate changes in amino acid sequence prior to the premature stop codon in each allele (see Material and Methods for more details). (C–F) Loss of maternally deposited Gdf3 causes patterning defects. Embryos heterozygous (C) or homozygous (D) for the pr05 allele appeared normal at 26 hpf. Heterozygous embryos from homozygous mothers (Mgdf) exhibited defects in mesoderm and endoderm formation (E). Maternal-zygotic gdf3 embryos (MZgdf3) had similar mesoderm and endoderm defects as Mgdf3 embryos (F). gdf3 mRNA injected at the one cell stage rescued defects in both Mgdf3 (G) and MZgdf3 (H) embryos. C-H are lateral views.
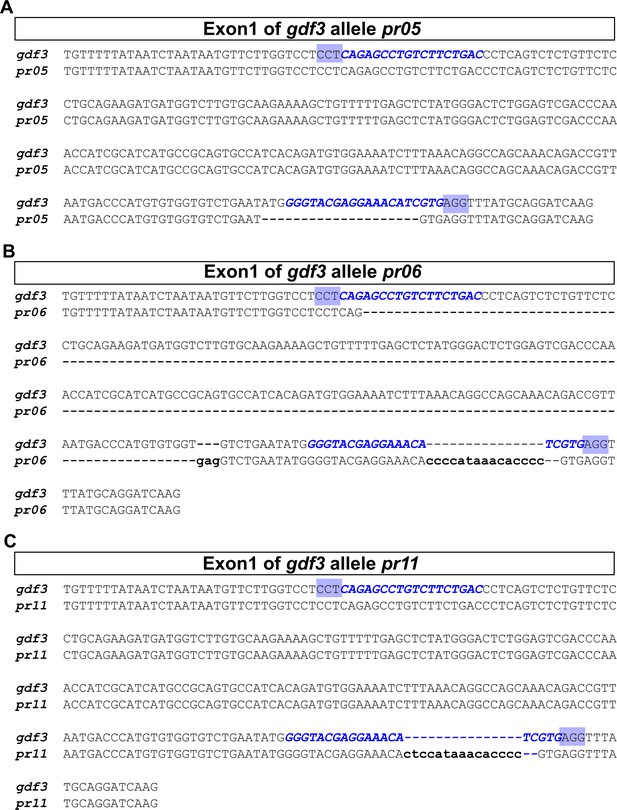
Schematic of gdf3 mutant allele sequences.
The genetic changes generated using CRISPR/Cas9 are diagrammed. (A) gdf3 allele pr05 contains a 20 bp deletion. (B) gdf3 allele pr06 contains a 191 bp deletion and an 18 bp insertion, resulting in a net loss of 173 bp. (C) gdf3 allele pr11 contains a 2 bp deletion and a 15 bp insertion, resulting in a net insertion of 13 bp. The sgRNA target sites (blue text) and the PAM sites (blue highlights) are shown. Deletions are indicated by dashed lines in the mutant alleles. Insertions are represented by lowercase letters in the mutant (pr) sequence and corresponding dashed lines in the wild-type (gdf3) sequence.
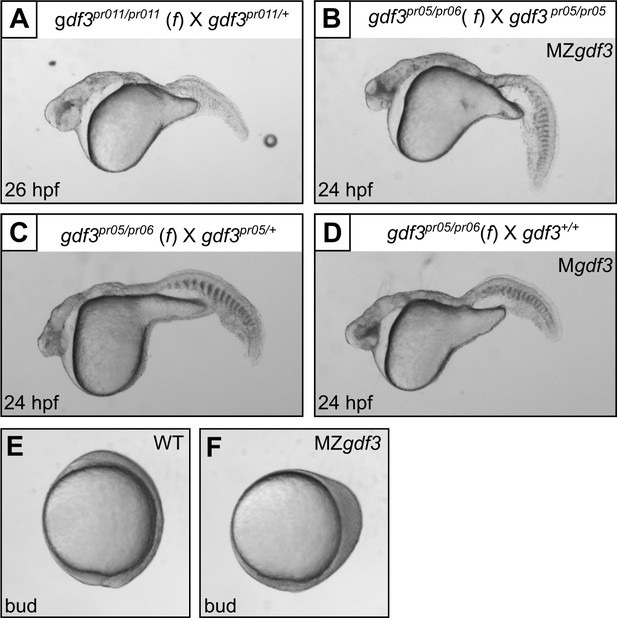
Multiple gdf3 mutant alleles have similar phenotypes.
(A) Embryos from maternal homozygotes for pr11 phenocopied pr5 Mgdf and MZgdf embryos. (B-D) Embryos from maternal compound heterozygotes of the pr05 and pr06 alleles were phenotypic, demonstrating lack of complementation between the alleles. (E) Wild-type (WT) embryo at bud stage. (F) MZgdf3 embryos show abnormal development at the bud stage, with a 90 degree shift of anterior structures vegetally; a phenotype similar to that reported for MZtdgf1 embryos (Gritsman et al., 1999). All views are lateral.
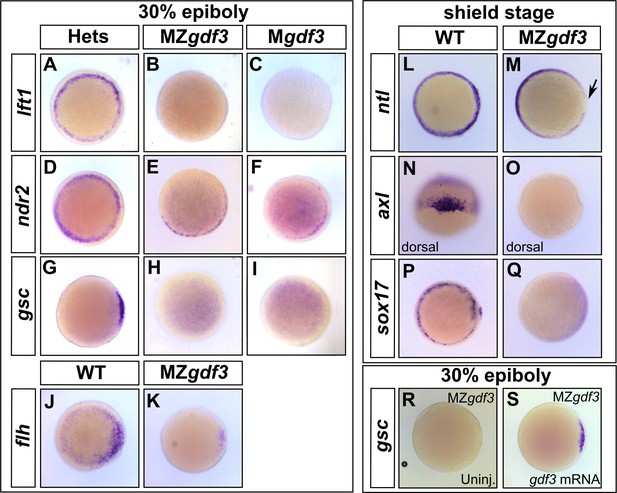
Gdf3 is necessary for the expression of Nodal target genes.
(A–K) RNA in situ hybridization of Nodal target gene expression at 30% epiboly. (A–C) lefty1 (lft1) is normally expressed in the margin (A), but is lost in maternal-zygotic gdf3 mutant embryos (MZgdf3, B) and heterozygous embryos derived from maternal homozygotes (Mgdf3, C). (D–F) cyclops (cyc) is normally expressed in the margin (D). Expression of cyc is reduced in MZgdf3 (E) and Mgdf3 (F) embryos. (G–I) goosecoid (gsc) is normally expressed in the dorsal region (G), but is absent in MZgdf3 (H) and Mgdf3 (I) embryos. (J–K) floating head (flh) is normally expressed in the dorsal region (J) but is diminished in MZgdf3 (K) embryos. (L–Q) RNA in situ hybridization of Nodal target gene expression at the shield stage. (L–M) no tail (ntla) is expressed along the margin in WT embryos (L), but is absent from the dorsal end of MZgdf3 embryos (M, arrow). (N, O) axial (axl) is expressed in the dorsal region of WT embryos (N) but absent in MZgdf3 embryos (O). (P, Q) sox17 is expressed in the margin of WT embryos (P) but absent in MZgdf3 embryos (Q). (R–S) Loss of gsc in MZgdf3 embryos (R) can be rescued by injection of 50 pg gdf3 mRNA. All views are animal, with dorsal to the right and ventral to the left,unless otherwise indicated.
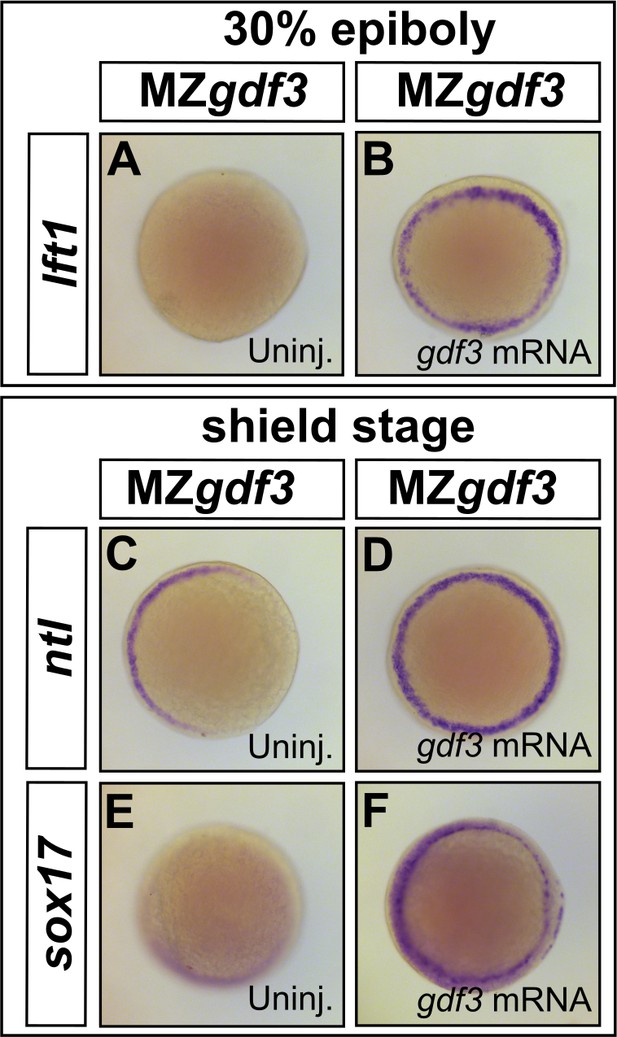
Loss of Nodal target gene expression can be rescued with gdf3 mRNA injection.
(A–F) Loss of lft1, ntl, and sox17 expression in MZgdf3 embryos (A,C, and E) can be rescued by injection of 50 pg gdf3 mRNA (B, D, and F). All views are animal.
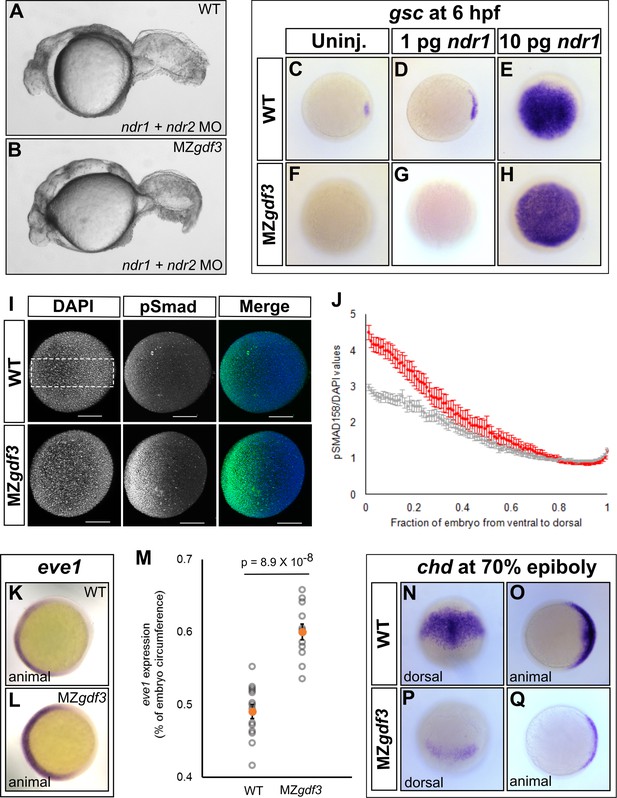
Loss of Gdf3 leads to expansion of Bmp signaling.
(A) Knockdown of nodal related 1 (ndr1) and nodal related 2 (ndr2) in wild-type (WT) embryos causes complete loss of head and trunk mesendoderm and defects in tail patterning. (B) ndr1:ndr2 knockdown in MZgdf3 embryos causes loss of residual tail patterning, producing embryos that more closely resemble those completely lacking Nodal signaling (A). (C-E) Overexpression of 1 pg or 10 pg ndr1 led to increased expression of the Nodal target gene goosecoid (gsc) in WT embryos. (F-H) Overexpression of 10 pg ndr1, but not 1 pg, produced widespread expression of gsc in MZgdf3 mutants. (I) Immunostaining of phosphorylated Smad1/5/8 (green) counterstained with DAPI (blue). Ventral pSmad levels were increased in MZgdf3 mutants. (J) Plotting the normalized pSmad1/5/8 intensity over position on the ventral-dorsal axis confirms that MZgdf3 embryos (n = 14) have increased BMP signaling in the ventral region compared to WT (n = 13). (K-M) RNA in situ hybridization of eve1 showed expansion of expression in the ventral region of MZgdf3 embryos (K) compared to WT (L). (M) Quantitative comparison of WT (n = 14) and MZgdf3 (n = 12) embryos confirmed expansion of the ventral eve1 expression domain in MZgdf3 embryos. (N-Q) RNA in situ hybridization of chordin (chd) revealed its expression is reduced in MZgdf3 embryos. All views are animal unless otherwise indicated. In M, p-value obtained by Student’s t-test (two-sided, homoscedastic). Error bars in J and M, standard error of the mean. Matlab code and data are available as Figure 3—source code 1, 2, and 3 and Figure 3—source data 1 and 2.
-
Figure 3—source code 1
MATLAB script for quantification of the eve1 RNA in situ stain.
- https://doi.org/10.7554/eLife.28635.008
-
Figure 3—source code 2
MATLAB script for quantification of phospho-SMAD1/5/8 immunostain in MZgdf3 mutant embryos.
- https://doi.org/10.7554/eLife.28635.009
-
Figure 3—source code 3
MATLAB script for quantification of phospho-SMAD1/5/8 immunostain in WT embryos.
- https://doi.org/10.7554/eLife.28635.010
-
Figure 3—source data 1
Raw data for quantification of the eve1 RNA in situ stain in WT and MZgdf3 embryos.
- https://doi.org/10.7554/eLife.28635.011
-
Figure 3—source data 2
Raw data for quantification of the pSMAD1/5/8 immunostain in WT and MZgdf3 embryos.
- https://doi.org/10.7554/eLife.28635.012
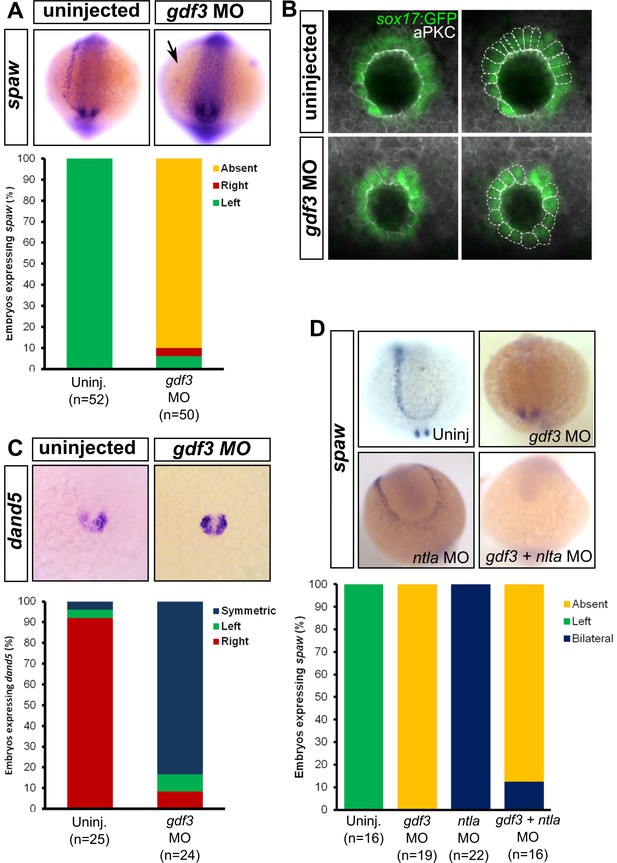
Attenuated production of Gdf3 leads to left-right patterning defects.
(A) spaw expression is altered in gdf3 morphants, with most lacking expression in the lateral plate mesoderm (arrow), as shown by RNA in situ hybridization. (B) Immunostaining of aPKC (white) and GFP driven by the sox17 regulatory region (green) in Kupffer’s Vesicle (KV) at 10 ss. The anterior-posterior asymmetry in cell morphology is lost in gdf3 morphants. Cells are outlined by white dashed lines. (C) RNA in situ hybridization reveals dand5 expression is symmetric around KV in gdf3 morphants at 10 ss. (D) Loss of ntla leads to robust spaw expression in both the left and right LPM and a loss of KV and associated gene expression. spaw expression in ntla + gdf3 double morphants is completely lost, supporting the idea that Gdf3 is required for robust Nodal expression given Nodal is unable to activate in the absence of inhibitors. Raw data provided in Figure 4—source data 1–3.
-
Figure 4—source data 1
Raw data used to generate the spaw RNA in situ results bar graph in Figure 4A.
- https://doi.org/10.7554/eLife.28635.014
-
Figure 4—source data 2
Raw data used to generate the dand5 RNA in situ results bar graph in Figure 4C.
- https://doi.org/10.7554/eLife.28635.015
-
Figure 4—source data 3
Raw data used to generate the spaw RNA in situ results bar graph in Figure 4D.
- https://doi.org/10.7554/eLife.28635.016
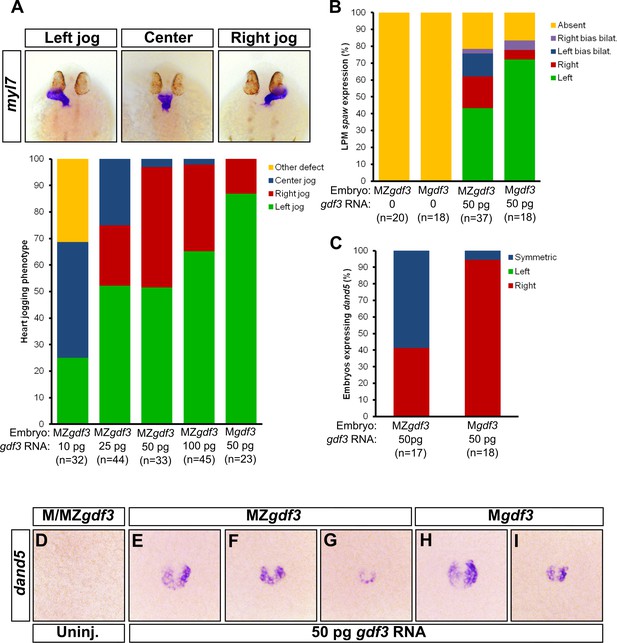
Zygotic Gdf3 can restore proper left-right patterning to embryos rescued from early mesendoderm defects.
(A) Heart jogging defects improved with increasing amounts of injected gdf3 mRNA in MZgdf3 embryos. Zygotic gdf3 (present in Mgdf3 embryos) further rescued heart jogging defects in embryos injected with 50 pg of gdf3 mRNA. (B) M/MZgdf3 embryos lack spaw expression at 12 ss. Embryos injected with 50 pg of gdf3 mRNA rescued spaw expression in the LPM. Proper spaw expression in the left LPM was further restored in the presence of zygotic gdf3. (C) dand5 expression was recovered in MZgdf3 embryos injected with gdf3 mRNA, with most embryos containing symmetric expression of dand5 in KV (F,G). (C) Most Mgdf3 embryos injected with gdf3 mRNA contained proper right-biased asymmetric expression of dand5 (H,I). (D) dand5 expression was absent in all uninjected M/MZgdf3 embryos. Raw data provided in Figure 5—source data 1–3.
-
Figure 5—source data 1
Raw data used to generate the bar graph of heart jog results in Figure 5A.
- https://doi.org/10.7554/eLife.28635.018
-
Figure 5—source data 2
Raw data used to generate the spaw RNA in situ results bar graph in Figure 5B.
- https://doi.org/10.7554/eLife.28635.019
-
Figure 5—source data 3
Raw data used to generate the dand5 RNA in situ results bar graph in Figure 5C.
- https://doi.org/10.7554/eLife.28635.020
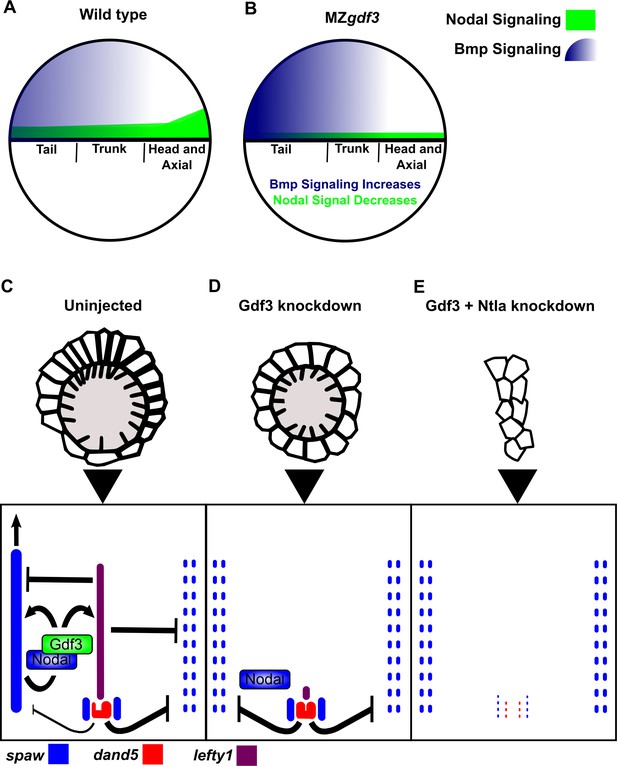
Model of Gdf3 function in early zebrafish development.
(A) In wild-type (WT) embryos, Bmp signaling (blue) is established in a ventral-to-dorsal gradient during blastula and gastrula stages, while Nodal signaling (green) is established along the margin, with strongest expression on the dorsal side. (B) In MZgdf3 embryos, Nodal signaling is weak leading to reduced expression of Nodal target genes including the Bmp inhibitor Chordin. Bmp signaling is increased and leads to an extension of ventral mesoderm markers. However, as Nodal signaling is depleted or lost, the majority of mesoderm and endodermal cell fates fail to be maintained. (C) Proper development of Kupffer’s vesicle includes the generation of asymmetries in cell shape along the anterior-posterior axis of the structure. Anterior cells, closest to the notochord, take on columnar shapes and become more tightly packed, leading to a greater number of cilia in this region. This architecture is required for asymmetric fluid flow in KV and proper dand5 expression. (D) Attenuation of maternal Gdf3 leads to improper cell morphology during KV development, which in turn leads to irregular dand5 expression. Dysregulation of dand5 expression on the left may then inhibit spaw in the left LPM. (E) Removal of Nodal antagonists Lefty1 and Dand5 via ntla morpholino-mediate knockdown does not restore spaw expression in the LPM. This suggests that Gdf3 plays a more direct role in regulating spaw expression, independent of its effects on KV development. Dotted lines (C, D, E) represent lost gene expression.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| gene (Danio rerio) | growth differentiation factor 3 (gdf3) | NA | ZFIN:ZDB-GENE-980526–389 | |
| gene (Danio rerio) | nodal-related 1 (ndr1) | NA | ZFIN:ZDB-GENE-990415–256 | |
| gene (Danio rerio) | nodal-related 2 (ndr2) | NA | ZFIN:ZDB-GENE-990415–181 | |
| gene (Danio rerio) | southpaw (spaw) | NA | ZFIN:ZDB-GENE-030219–1 | |
| gene (Danio rerio) | DAN domain family, member 5 (dand5) | NA | ZFIN:ZDB-GENE-040421–2 | |
| genetic reagent (D. rerio) | gdf3 allele pr05 | this paper | CRISPR/Cas9 generated deletion, predicted null | |
| genetic reagent (D. rerio) | gdf3 allele pr06 | this paper | CRISPR/Cas9 generated indel, predicted null | |
| genetic reagent (ID. rerio) | gdf3 allele pr11 | this paper | CRISPR/Cas9 generated indel, predicted null | |
| antibody | Anti-Digoxigenin-AP | Roche, 11093274910 | 1:3500 | |
| antibody | Anti-phosphorylated SMAD1/5/8 | Cell Signaling Technologies, 9511S | 1:100 | |
| antibody | anti-PKCzeta (aPKC) | Santa Cruz Biotechnology, SC-216 | 1:400 | |
| antibody | Goat anti-Rabbit Alexa Fluor 647 | Invitrogen, A21246 | 1:400 | |
| antibody | Donkey anti-Rabbit Alexa Fluor 647 secondary antibody | Invitrogen, A31573 | 1:400 | |
| recombinant DNA reagent | pCS2-nCas9n | Addgene plasmid # 47929 | ||
| recombinant DNA reagent | pT7-gRNA | Addgene plasmid # 46759 | ||
| recombinant DNA reagent | pCS2(+)-gdf3-WT | this paper | ||
| recombinant DNA reagent | pCS2-3XFlag-sqt | PMID:27101364 | ||
| recombinant DNA reagent | gdf3 in situ probe plasmid | PMID: 8405668 | ||
| recombinant DNA reagent | ndr2 in situ probe plasmid | PMID: 9707578 | ||
| recombinant DNA reagent | flh in situ probe plasmid | PMID: 7477317 | ||
| recombinant DNA reagent | ntla in situ probe plasmid | PMID: 1295726 | ||
| recombinant DNA reagent | gsc in situ probe plasmid | PMID: 8104775 | ||
| recombinant DNA reagent | lft1 in situ probe plasmid | PMID: 10375514 | ||
| recombinant DNA reagent | axial in situ probe plasmid | PMID: 7687227 | ||
| recombinant DNA reagent | sox17 in situ probe plasmid | PMID: 10531029 | ||
| recombinant DNA reagent | spaw in situ probe plasmid | PMID: 12702646 | ||
| recombinant DNA reagent | dand5 in situ probe plasmid | PMID: 15084459 | ||
| sequence-based reagent | gdf3 morpholino | Genetools | 5’-GCTCTGAGGAGGACCAAGAACATTA-3’ | |
| sequence-based reagent | ntla morpholino | Genetools | 5’- GACTTGAGGCAGGCATATTTCCGAT-3’ | |
| sequence-based reagent | ndr1 morpholino | Genetools | 5’-ATGTCAAATCAAGGTAATAATCCAC-3’ | |
| sequence-based reagent | ndr2 morpholino | Genetools | 5’-GCGACTCCGAGCGTGTGCATGATG-3’ | |
| peptide, recombinant protein | T4 PNK | NEB, M0201S | ||
| peptide, recombinant protein | T4 DNA ligase | NEB, M0202S | ||
| peptide, recombinant protein | BsmBI | NEB, R0580S | ||
| peptide, recombinant protein | BglII | NEB, R0144S | ||
| peptide, recombinant protein | SalI | NEB, R0138S | ||
| peptide, recombinant protein | BamHI | NEB, R0136S | ||
| peptide, recombinant protein | XbaI | NEB, R0145S | ||
| commercial assay or kit | Qiaquick PCR Purification Kit | Qiagen, 28104 | ||
| commercial assay or kit | QIAquick Gel Extraction Kit | Qiagen, 28704 | ||
| commercial assay or kit | mMessage mMachine SP6 | ThermoFisher, AM1340 | ||
| commercial assay or kit | MEGAshortscript T7 Transcription kit | ThermoFisher, AM1354 | ||
| commercial assay or kit | QIAprep Spin Miniprep kit | Qiagen, 27106 | ||
| commercial assay or kit | Phusion High- Fidelity PCR Kit | EB, E0553L | ||
| software, algorithm | Imaris | Image analysis and quantification | ||
| software, algorithm | ImageJ | Image analysis and quantification | ||
| software, algorithm | MATLAB | Image analysis and quantification |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.28635.022





