Essential role for SUN5 in anchoring sperm head to the tail
Figures
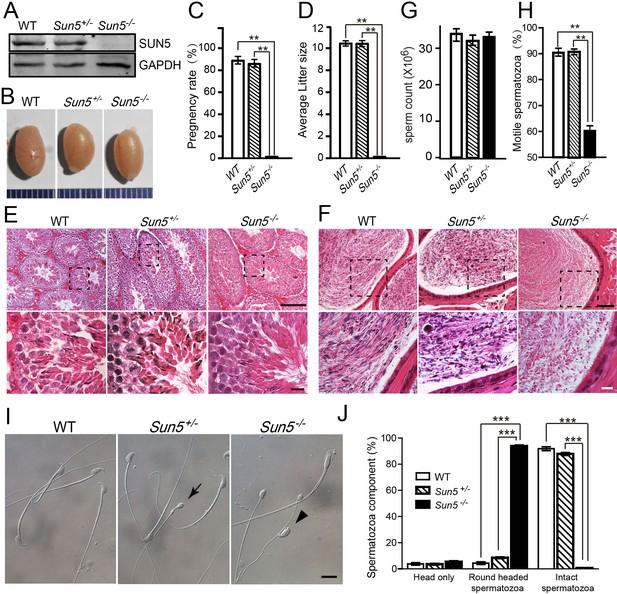
Ablation of SUN5 leads to male infertility and sperm malformation.
(A) Immunoblotting of SUN5 in WT, Sun5+/− and Sun5−/− testes. (B) The size of the testes was not altered in the Sun5+/−and Sun5−/− mice. (C) The pregnancy rate of WT (92.46 ± 3.39%), Sun5+/− (88.33 ± 3.73%) and Sun5−/− (0) male mice (n = 6). p(WT VS Sun5-/-)= 1.24 × 10−6, p(Sun5+/- VS Sun5-/-)= 1.37 × 10−7, p(WT VS Sun5+/-)= 0.29. (D) The average litter size of WT (10.65 ± 0.21), Sun5+/− (10.27 ± 0.38) and Sun5−/− (0) male mice (n = 6). p(WT VS Sun5-/-)= 9.85 × 10−27, p(Sun5+/- VS Sun5-/-)= 5.47 × 10−25, p(WT VS Sun5+/-)= 0.39. (E) HE (hematoxylin-eosin) staining of testes from WT, Sun5+/− and Sun5−/− mice, seminiferous tubules shown in the figures were at stage IV-VI. Scale bar: upper panel, 100 μm; lower panel, 10 μm. (F) HE staining of the caudal epididymis from WT, Sun5+/− and Sun5−/− mice. Scale bar: upper panel, 50 μm; lower panel, 10 μm. (G) The sperm concentration of WT (33.92 ± 1.71 × 106), Sun5+/− (31.29 ± 0.93 × 106) and Sun5−/− (33.03 ± 1.67 × 106) male mice (n = 5). p(WT VS Sun5−/−)= 0.65, p(Sun5+/−VS Sun5−/−)= 0.41, p(WT VS Sun5+/−)= 0.17 (H) The percentage of motile spermatozoa in WT (90.20 ± 0.63%), Sun5+/ -(90.40 ± 0.14%) and Sun5−/− (60.2 ± 1.98%) male mice (n = 5). p(WT VS Sun5-/-)= 0.0001, p(Sun5+/- VS Sun5-/-)= 0.0005, p(WT VS Sun5+/-)= 0.92. (n = 5) (I) Caudal epididymal spermatozoa of WT, Sun5+/− and Sun5−/− mice. The arrow indicates the round-headed spermatozoon in Sun5+/− mice, and the arrowhead indicates the tailless head spermatozoon in Sun5−/− mice. Scale bar: 10 μm. (J) The percentage of different spermatozoon components in WT, Sun5+/− and Sun5−/− caudal epididymides (n = 5). The first group of columns show the percentage of isolated sperm heads in WT (3.78 ± 0.90%), Sun5+/− (3.60 ± 0.62%) and Sun5−/− (5.44 ± 0.79%) mice, p(WT VS Sun5-/-)= 0.32, p(Sun5+/- VS Sun5-/-)= 0.07; The 2nd group of columns show the percentage of round headed spermatozoon in WT (4.38 ± 0.96%), Sun5+/− (8.56 ± 0.06%) and Sun5−/− (93.86 ± 0.79%) mice, p(WT VS Sun5-/-)= 5.82 × 10−7, p(Sun5+/- VS Sun5-/-)= 2.73 × 10−7; The 3rd group of columns show the percentage of intact spermatozoon in WT (92.44 ± 1.63%), Sun5+/− (87.84 ± 1.13%) and Sun5−/− (0.7 ± 0.28%) mice, p(WT VS Sun5-/-)= 1.01 × 10−6, p(Sun5+/- VS Sun5-/-)= 7.63 × 10−8. Data represent mean ±SEM.
-
Figure 1—source data 1
Source data for mouse fertility, sperm concentration, sperm motility and spermatozoa components in epididymis.
- https://doi.org/10.7554/eLife.28199.006
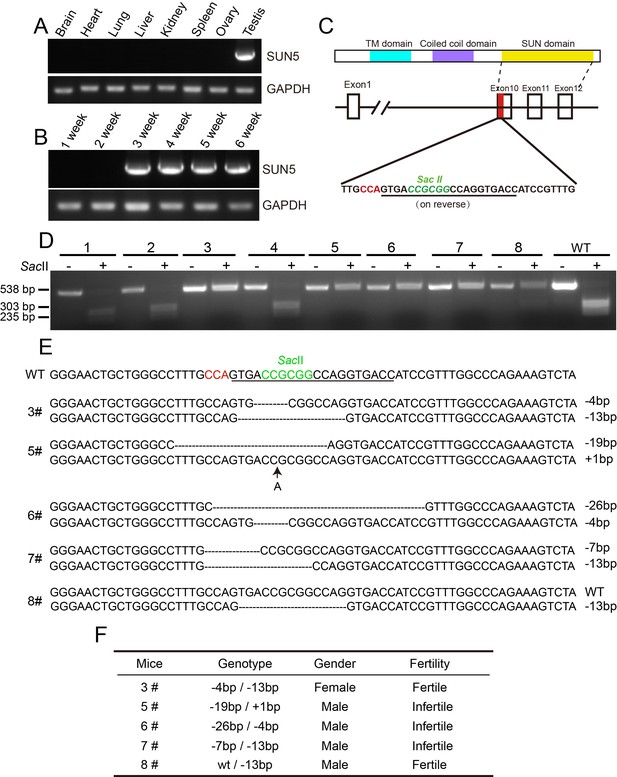
The generation of Sun5 knockout mice.
(A) The expression of Sun5 was restricted to testis. (B) Sun5 expression begins in 3-week-old testes. (C) The knockout strategy of Sun5 in mice. (D) Genotyping of founders to identify Sun5 knockouts. (E) Sequences of the mutated alleles in Sun5 knockout mice. (F) Summary of the fertility of 5 Sun5 knockout founders.
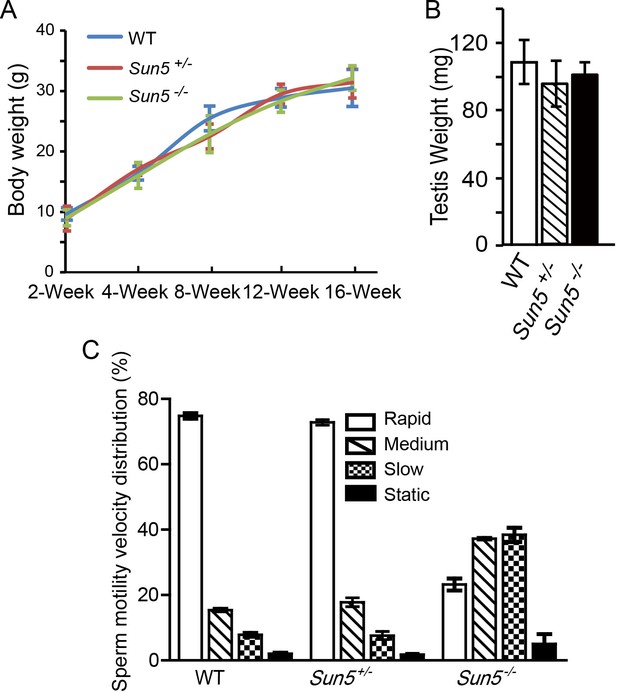
Sun5 knockout does not affect growth and testis development, but influences sperm motility.
(A) Body weight of WT, Sun5+/− and Sun5−/− male mice, showing that Sun5 knockout does not affect mouse growth (n = 5). (B) Testis weight of adult WT, Sun5+/− and Sun5−/− male mice, indicating that testis development was not affected by Sun5 knockout (n = 5). (C) The motility velocity distribution of motile spermatozoa in WT, Sun5+/− and Sun5−/− male mice, most of the Sun5-null motile spermatozoa belonged to medium- and slow- moving groups (n = 5). Data represent mean ±SEM.
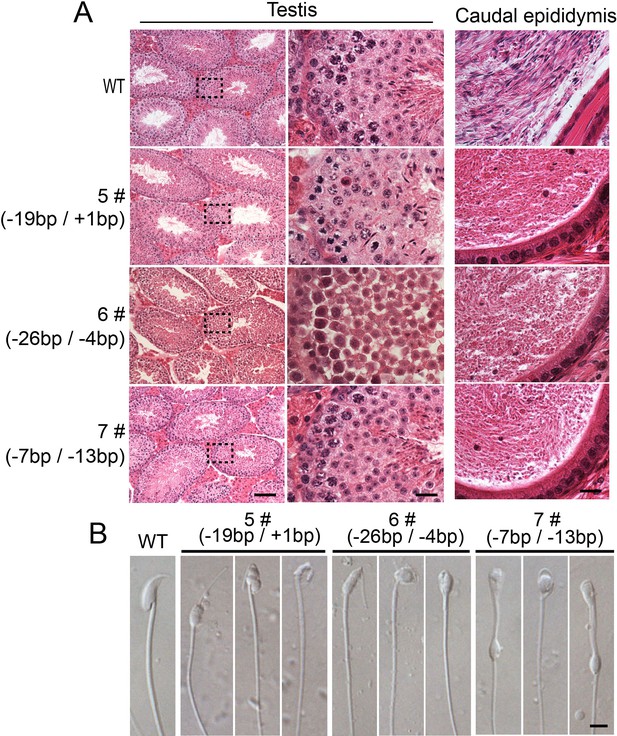
All of the biallelic Sun5 mutated male mice exhibit similar defects in spermiogenesis.
(A) All of the biallelic Sun5 mutated males showed normal testis structures but had abnormal HE staining in the caudal epididymis. Seminiferous tubules shown in the figures were at stage IV-VI. Scale bar: left panel, 50 μm; right two panels, 10 μm. (B) The spermatozoa in the biallelic Sun5-mutated males were all round-headed. Scale bar: 5 μm.
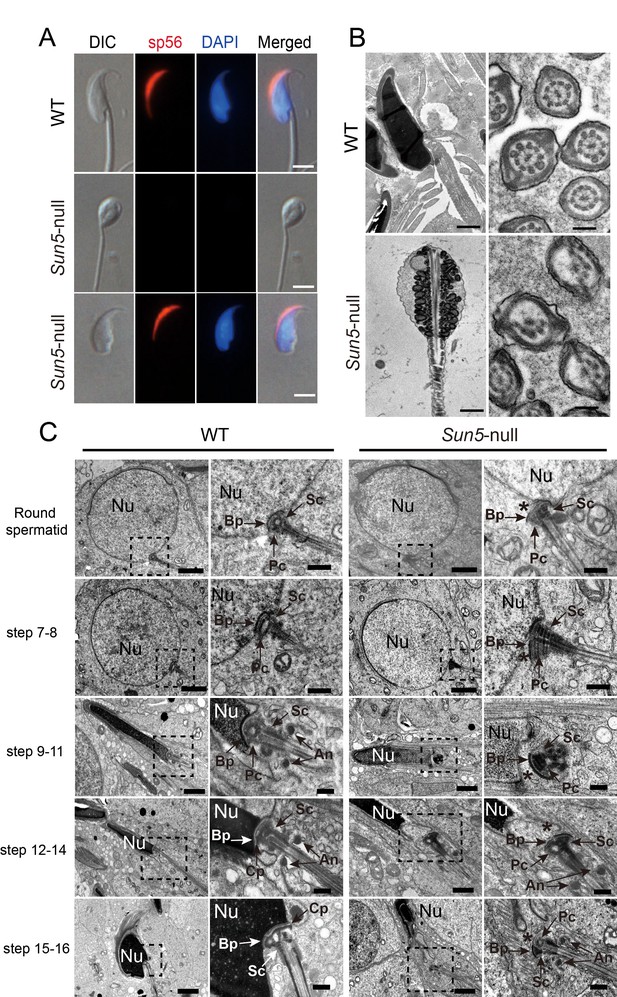
The absence of SUN5 has no effect on acrosome biogenesis but disrupts the development of the coupling apparatus between sperm head and tail.
(A) IF (immunofluorescence) staining of sp56 in WT and Sun5-null spermatozoon. The Sun5-null spermatozoa contains both round-headed spermatozoa and tailless heads (lower two panels). The proportion of these two types of spermatozoa were displayed in Figure 1J. Note that the round-headed Sun5-null spermatozoa do not contain nuclei and acrosomes, but the tailless Sun5-null sperm heads have nuclei and acrosomes. Scale bar: 5 μm. (B) Ultrastructure of WT and Sun5−/−caudal epididymides showing that the Sun5-null spermatozoon was filled with cytoplasm and misarranged mitochondria. Note that the axoneme of Sun5-null spermatozoon was also disrupted. Scale bar: left panel, 1 μm; right panel, 200 nm. (C) TEM analyses of the stepwise development of the coupling apparatus in WT and Sun5-null spermatozoa. In the round spermatid stage, the coupling apparatus can be assembled in both WT and Sun5-null spermatid, but the coupling apparatus could not be tightly attached to the nuclear envelope in Sun5-null spermatids. The asterisk indicates the gap between the nuclear (Nu) envelope and the basal plate (Bp). In the following developmental stages, the coupling apparatus was well-fixed on the nuclear envelope in WT spermatids, ensuring healthy spermatid differentiation. While in Sun5-null spermatids, the basal plate (Bp)-capitulum (Cp)-segmented column (Sc) together with the centriole (Pc) was detached from the nuclear envelope during spermatid elongation. An, annulus. Scale bar: the 1st and 3rd panel, 2 μm, 2nd and 4th panel, 0.5 μm.
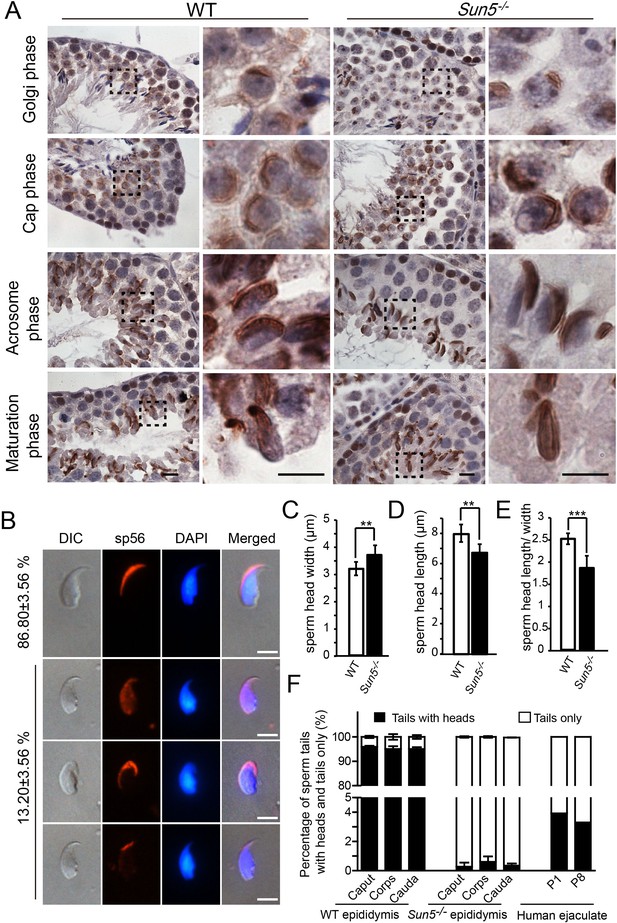
Acrosome biogenesis and epididymal spermatozoa in Sun5−/− testes.
(A) Immunohistochemistry staining of Afaf in WT and Sun5−/− testes, indicating that acrosome biogenesis was not affected in Sun5−/− testes. Scale bar: 10 μm. (B) sp56 IF staining in Sun5–null spermatozoa, showing that most Sun5–null spermatozoa had normal acrosome. Scale bar: 5 μm. (C) The width of sperm heads in WT and Sun5–null spermatozoa. (D) The length of sperm heads in WT and Sun5–null spermatozoa. (E) The ratio of sperm head length to width in WT and Sun5–null spermatozoa. Showing the differences between WT and Sun5–null sperm head morphology. (F) The percentage of spermatozoa with heads versus tails only in mouse caput, corpus, cauda epididymis and human ejaculate, and very few intact spermatozoa could be found in Sun5 mutant mouse and human.
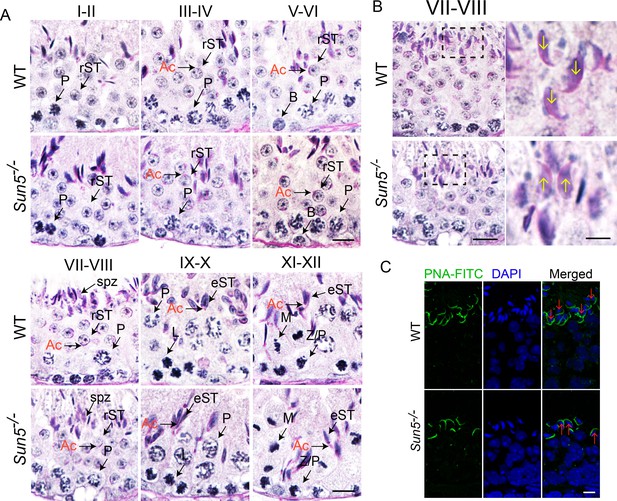
Spermiation defects in Sun5−/− mice.
(A) Periodic acid-Schiff (PAS) staining revealed histology of all twelve developmental stages in WT and Sun5−/− testes. The morphology of Acrosome (Ac) marks the specific developmental stages. Scale bar, 10 μm. (B) Sun5-null spermatids have lost their orientation toward the basement membrane during spermiation in stage VII-VIII seminiferous epithelia. The arrows indicate the orientation of the sperm heads. Scale bar: left panel, 10 μm; right panel, 5 μm. (C) Peanut agglutinin (PNA) staining of stage VII-VIII seminiferous epithelia showed the miss-arranged spermatids in Sun5−/− testes. The arrows indicate the orientation of the sperm heads. Scale bar, 5 μm. Ac: acrosome; B: B type spermatogonium; eST: elongating spermatid; L: leptotene spermatocyte; M: meiotic spermatocyte; P: pachytene spermatocyte; rST: round spermatid; spz: spermatozoon; Z: zygotene spermatocyte.
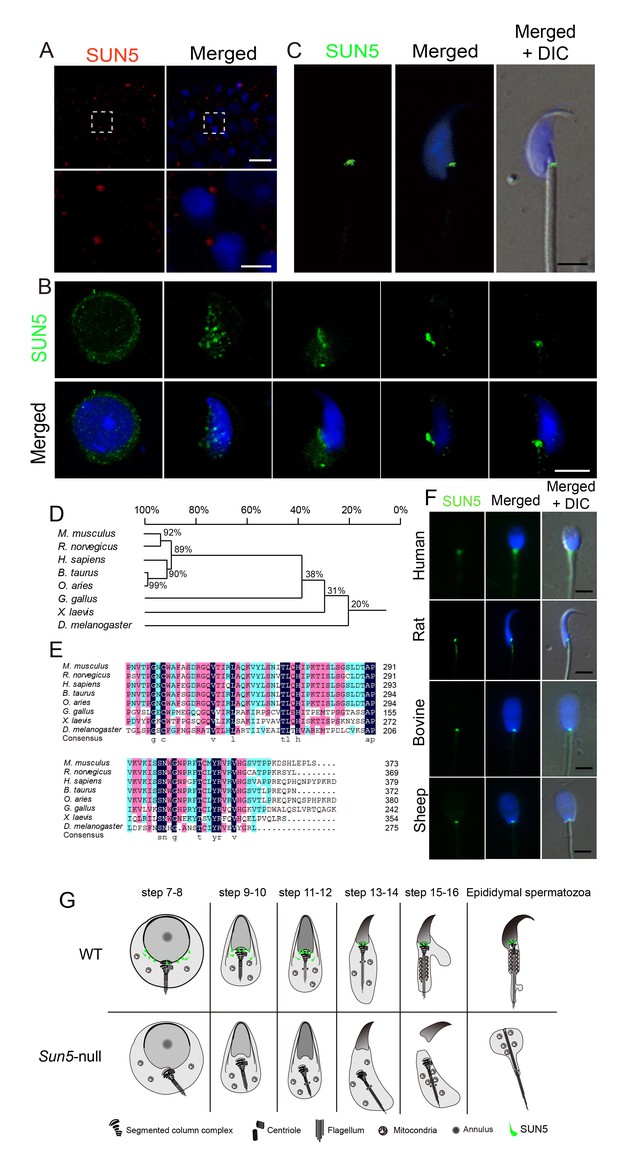
SUN5 localizes to the coupling apparatus between the sperm head and tail in mammals.
(A) IF of SUN5 in testes. Scale bar: upper panel, 10 μm; lower panel, 2.5 μm. (B) IF of SUN5 in spermatids at different developmental stages. Scale bar: 5 μm. (C) Single-sperm immunofluorescence of SUN5. Scale bar: 5 μm. (D) Phylogenetic tree of the SUN5 homolog proteins from different species. (E) Sequence alignment of the conserved SUN domain of SUN5 in different species. The dark blue labeled sequences showed 100% identity among species, pink labeled ones showed lower identity than the dark blue ones, then the green labeled ones showed lower identity than the pink ones. (F) SUN5 localizes to the sperm head-tail coupling apparatus in all tested mammals. Scale bar: 5 μm. (G) Schematic representation of the role of SUN5 in the development of the coupling apparatus in WT and Sun5-null spermatids based on TEM analyses and immuno-staining.
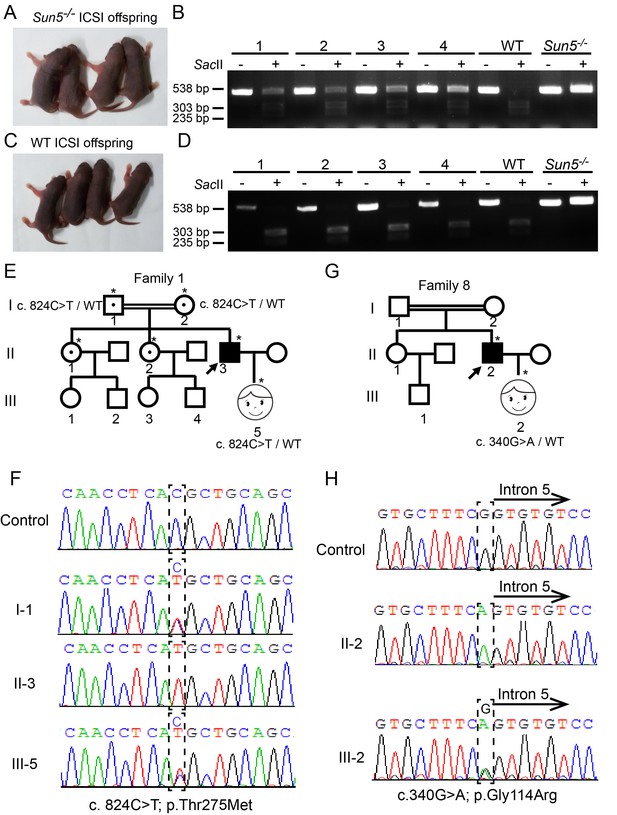
Infertility caused by SUN5 mutations could be overcome by ICSI.
(A) Representative images and (B) genotypes of the Sun5-null ICSI offspring. (C) Representative images and (D) genotypes of the WT ICSI offspring. (E) Pedigree of family 1 with inherited SUN5 mutations, and the healthy baby of the infertility patient after ICSI. The individuals with a single star were Sanger sequenced. (F) Sequences of the SUN5 mutation sites of the representative individuals from each generation of family 1. (G) Pedigree of family 8 with inherited SUN5 mutations, and the health baby of the infertility patient after ICSI. The individuals with a single star were Sanger sequenced. (H) Sequences of the SUN5 mutation sites of the representative individuals from each generation of family 8.
-
Figure 4—source data 1
The sperm motility and morphology analysis of the two patients underwent ICSI.
- https://doi.org/10.7554/eLife.28199.013
Development of WT and Sun5-null ICSI offspring.
(A) Body weight of WT and Sun5−/− ICSI offspring, statistical analysis was performed using at least three individuals. (B) Testis weight of adult WT and Sun5−/− ICSI offspring, statistical analysis was performed using at least three individuals. Data represent mean ±SEM.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.28199.014





