Major satellite repeat RNA stabilize heterochromatin retention of Suv39h enzymes by RNA-nucleosome association and RNA:DNA hybrid formation
Figures
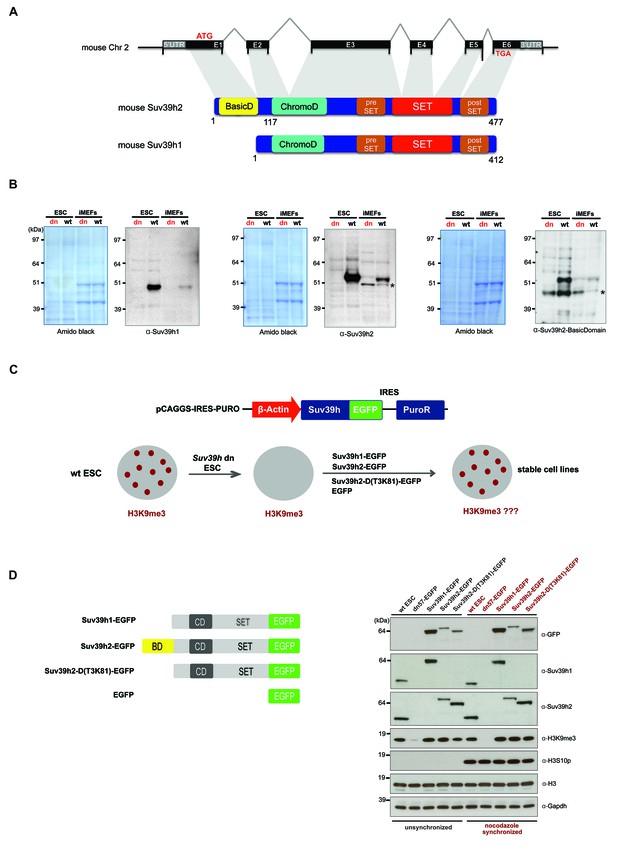
Characterization of the Suv39h2 protein and generation of rescued Suv39h dn mouse ES cells.
(A) Schematic representation of the mouse Suv39h2 gene locus and domain structure of the Suv39h1 and Suv39h2 enzymes showing the N-terminal basic domain of Suv39h2 in yellow. (B) Western blot of chromatin extracts from wild type and Suv39h dn mouse ES cells (ESC) and fibroblasts (iMEF) to detect endogenous Suv39h1 (48 kDa) and Suv39h2 (53 kDa). An antibody specific for the basic domain of Suv39h2 (Figure 1—figure supplement 1) also detects endogenous Suv39h2 at 53 kDa in wild type but not in Suv39h dn chromatin extracts. The asterisks indicate nonspecific bands. (C) Generation of rescued Suv39h dn mouse ES cell lines that express the indicated Suv39h-EGFP constructs under the control of a β-actin promoter. (D) Western blot of whole cell extracts from unsynchronized and nocodazole-synchronized mouse ES cell lines to examine expression of the various EGFP-tagged Suv39h products with an α-GFP antibody or with α-Suv39h1 and α-Suv39h2 antibodies to compare their expression levels with regard to the endogenous Suv39h1 and Suv39h2 proteins. H3K9me3 and H3S10phos levels were also analyzed. Histone H3 and Gapdh served as loading controls.
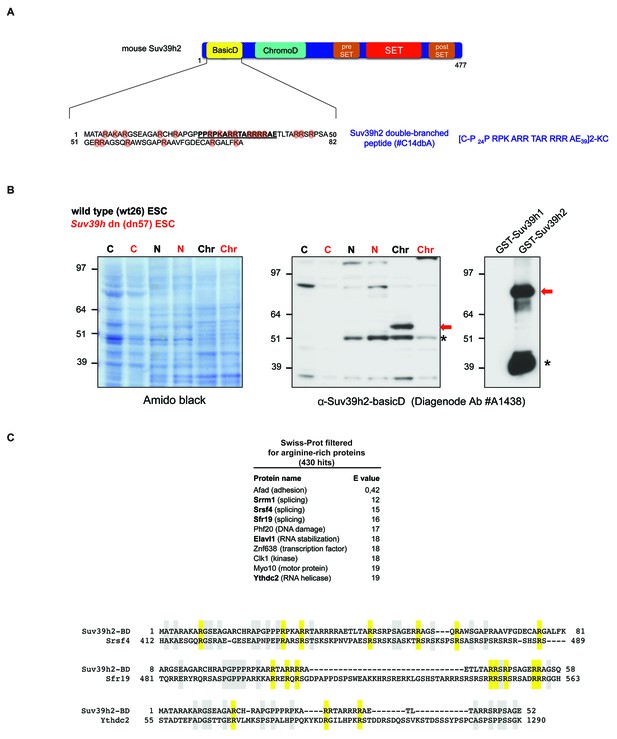
Generation of polyclonal antibodies against the basic domain of mouse Suv39h2 and amino acid sequence alignment of the basic domain of Suv39h2 with arginine-rich RNA binding factors.
(A) Schematic representation of the domain structure of the Suv39h2 enzyme highlighting the N-terminal basic domain (encoded by exon 1) in yellow. The amino acid sequence of this basic domain (aa 1–81) contains 19 arginines (R) and 3 lysines (K) denoted by orange circles. The underlined amino acids correspond to the sequence of the double-branched peptide used for the generation of rabbit polyclonal α-Suv39h2-basicD antibodies. (B) Western blot analysis of cytoplasmic (C), nucleoplasmic (N) and chromatin extracts (Chr) from mouse ES cells (ESC) using the α-Suv39h2-basicD antibody (Diagenode #A1438) indicates the presence of endogenous Suv39h2 (~53 kDa) in chromatin extracts of wild-type (wt26) but not in Suv39h dn (dn57) mutant ESC (middle panel). Immunoblotting of GST-Suv39h1 and GST-Suv39h2 recombinant proteins with the α-Suv39h2-basicD antibody (Diagenode #A1438) shows the specific recognition of full-length GST-Suv39h2 (~80 kDa) but not of GST-Suv39h1 (right panel). The asterisks indicate nonspecific bands. (C) The sequence of the basic domain of mouse Suv39h2, spanning amino acids 1–81, was queried using UniProt BLAST against a preselected mouse Swiss-Prot database that was filtered with the Entrez query ‘arginine rich’. The top ten hits from this query are listed and proteins with a described RNA binding activity are indicated in bold. Also shown are amino acid sequence alignments of the basic domain of Suv39h2 with two splicing factors (Srsf4 and Sfr19) and one RNA helicase (Ythdc2). Conserved arginine residues are highlighted in yellow and other conserved residues in light blue.
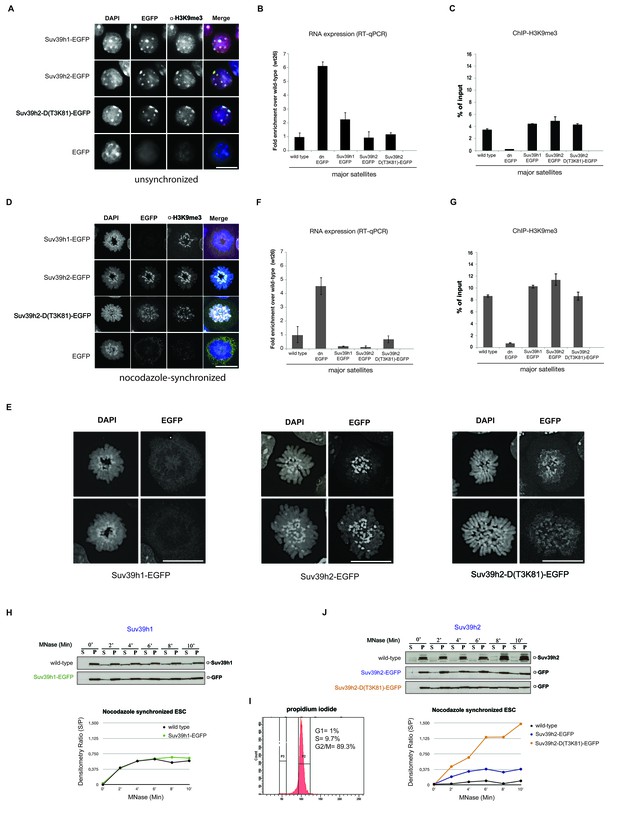
Distinct functions of Suv39h1 and Suv39h2 at mitotic chromatin.
(A) Immunofluorescence analysis for the various Suv39h-EGFP products and for H3K9me3 in interphase nuclei of the rescued mouse ES cell lines. The heterochromatic foci were counterstained with DAPI. Scale bar, 10 μm. (B) Reverse transcription quantitative PCR (RT-qPCR) with total RNA isolated from unsynchronized wild type, Suv39h dn or Suv39h-EGFP rescued mouse ES cell lines to detect expression from the major satellite repeats with MSR-specific primers. The amplified signals were normalized to Gapdh and are plotted in the histogram. The data represent the mean ± SD of at least two independent experiments. (C) Directed ChIP for H3K9me3 at the major satellite repeats in unsynchronized wild type, Suv39h dn or Suv39h-EGFP rescued mouse ES cell lines. The data represent the mean ± SD of at least two independent experiments. (D) Confocal spinning disc immunofluorescence analysis for the various Suv39h-EGFP products and for H3K9me3 at mitotic chromosomes that were presented in nocodazole-synchronized mouse ES cells. For each image, between 30–50 nuclei displaying mitotic chromosomes were analyzed. Scale bar, 10 μm. (E) Enlarged images of representative confocal IF analyses of mitotic chromosomes as described in (D). Only DAPI and EGFP signals are shown. Scale bar, 10 μm. (F) RT-qPCR as described in (B), but with total RNA preparations from nocodazole-synchronized mouse ES cells. (G) Directed ChIP for H3K9me3 as described in (C), but with chromatin material from nocodazole-synchronized mouse ES cells. (H) Chromatin release assay for endogenous Suv39h1 (wild type) and Suv39h1-EGFP in rescued mouse ES cells. Proteins were detected by Western blot in the soluble (S) or pellet (P) fractions after progressive (0, 2, 4, 6, 8 and 10 min.) MNase digestion of chromatin from nocodazole-synchronized mouse ES cells. Intensity of protein bands in the S or P fraction was quantified by ImageJ software. Chromatin release is measured by the S/P ratio, which is plotted in the indicated graph. (I) FACS profile (propidium iodide labeling) of nocodazole-synchronized wild-type mouse ES cells. (J) Chromatin release assay for endogenous Suv39h2 (wild type), Suv39h2-EGFP and the Suv39h-D(T3K81)-EGFP mutant as described in (H).
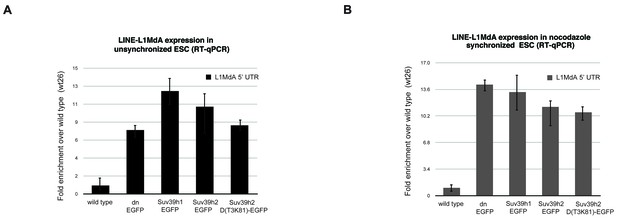
Reintroduced Suv39h-EGFP products cannot restore silencing of LINE L1MdA repeats in Suv39h dn mouse ES cells.
Total RNA isolated from wild type, Suv39h double null and Suv39h double null mouse ES cells rescued with EGFP-tagged Suv39h1, Suv39h2 or Suv39h2-D(T3K81) was used for reverse transcription quantitative PCR (RT-qPCR). The transcriptional output of LINE L1MdA-5’ UTR repeats was analyzed in unsynchronized (A) and nocodazole-arrested cells (B). The amplified signal was normalized to Gapdh and the results were plotted relative to the wild type signal. Error bars represent the standard deviation calculated from two biological replicates.
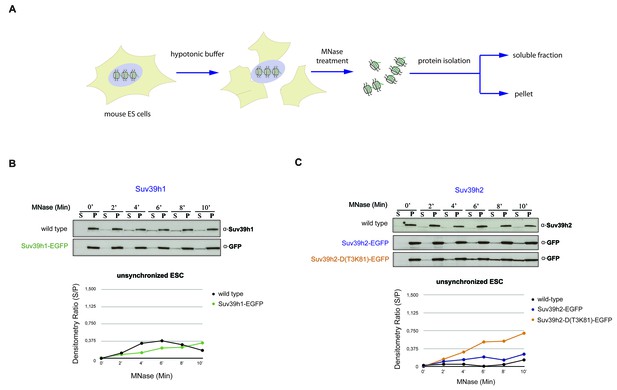
Chromatin release assay in unsynchronized mouse ES cells.
(A) Workflow of the chromatin release assay in unsynchronized mouse ES cells. Whole cell lysates from mouse ES cells were digested with 10 U of MNase for different time points (0,2,4,6,8 and 10 min) and proteins from the soluble and insoluble chromatin fractions were detected by western blotting. (B) Chromatin release for endogenous Suv39h1 in wt ES cells and for Suv39h1-EGFP in rescued ES cells. Protein bands in the soluble fraction (S) or in the pellet (P) were quantified by ImageJ software. Chromatin release is measured by the S/P ratio, which is plotted in the indicated graph. (C) Chromatin release assay for endogenous Suv39h2 in wt ES cells and for Suv39h2-EGFP and the Suv39h2-D(T3K81)-EGFP mutant in rescued ES cells as described in (B).
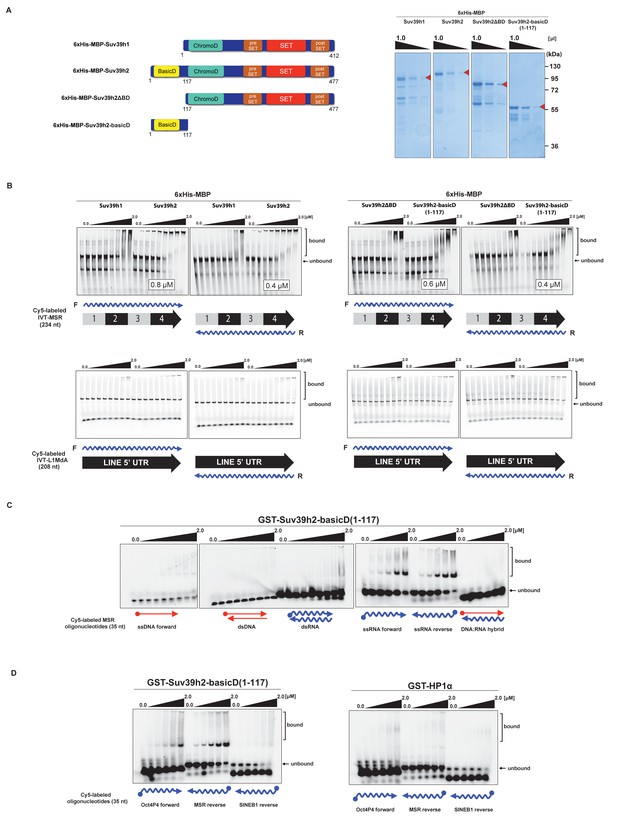
The basic domain of Suv39h2 preferably interacts with ssMSR RNA in vitro.
(A) Schematic representation of full-length Suv39h1, full-length Suv39h2, the Suv39h2ΔBD mutant and the Suv39h2-basicD(1-117) proteins. These were expressed as recombinant 6xHisMBP-fusion proteins and their purity is visualized by Coomassie staining. (B) Electrophoretic mobility shift assays (EMSA) with the indicated recombinant 6xHisMBP-Suv39h products and 3'-Cy5-labeled in vitro transcribed full-length (234 nt) ss-forward or ss-reverse MSR transcripts (upper panel) or with 3'-Cy5-labeled in vitro transcribed full-length (208 nt) ss-forward or ss-reverse LINE 5'UTR transcripts (lower panel). KD values that are within the tested protein concentration range of 16 nM to 2 μM were calculated with GraphPad Prism6 software and are indicated in the white boxes. The same amount (50 nM) of IVT MSR or LINE 5'UTR transcripts was used although there was reduced 3'-Cy5 labeling efficiency with the LINE 5'UTR ssRNA. (C) EMSA with recombinant GST-Suv39h2-basicD(1-117) and 5’-Cy5-labeled DNA or RNA oligonucleotides (35 nt each) from subunit 2 of the MSR that are probed as single-stranded, double-stranded or as RNA:DNA hybrid binding substrates. (D) Same EMSA as in (C), but with 5’-Cy5-labeled ssRNA oligonucleotides (35 nt) from an Oct4P4 lnc RNA (Scarola et al., 2015), the MSR reverse RNA and a SINE B1 reverse RNA. For comparison, this EMSA was also done with recombinant GST-HP1α.
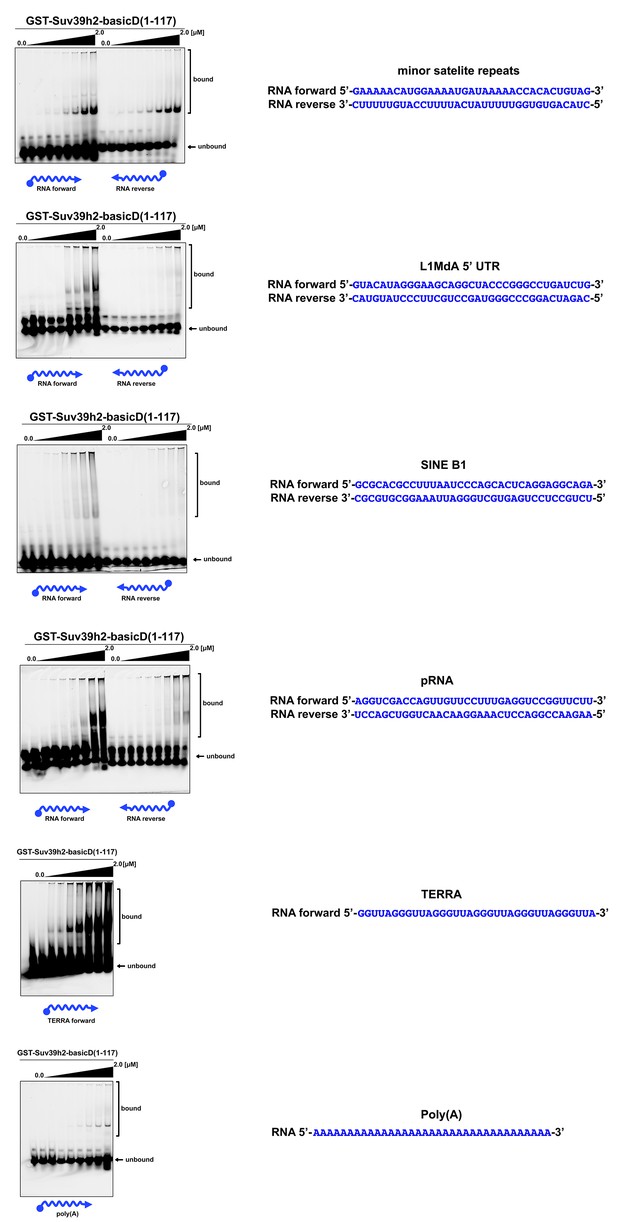
In vitro RNA binding of GST-Suv39h2-basicD(1-117) towards several distinct RNA oligonucleotides.
In vitro RNA binding assays with recombinant GST-Suv39h2-basicD(1-117) and a variety of 35 nt 5’-Cy5-labeled RNA oligonucleotides. The sequence of these RNA oligonucleotides corresponds to regions of repetitive DNA, such as minor satellite repeats, LINE L1MdA 5'UTR and SINE B1, as well as pRNA (Schmitz et al., 2010) and TERRA (Porro et al., 2014). A poly(A) RNA oligonucleotide was also included to test for binding to unstructured RNA. ssRNA oligonucleotides (50 nM) were incubated with increasing amounts (16 nM − 2 μM) of recombinant GST-Suv39h2-basicD(1-117) and separated by PAGE. The Cy5 signal was scanned on a Typhoon FLA 9500 fluorescence scanner at 800V and quantified using ImageJ software. The sequence and direction (forward or reverse) of the RNA oligonucleotides are indicated to the right of the respective EMSA-PAGE.
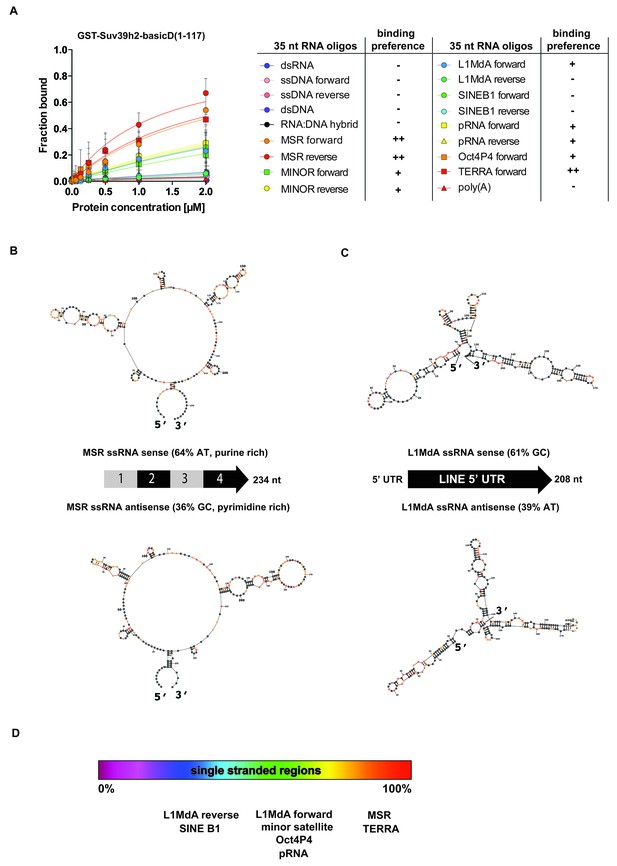
SHAPE-directed secondary structure models of major satellite repeat and LINE L1MdA 5'UTR RNA.
(A) Quantification of in vitro RNA binding for GST-Suv39h2-basicD(1-117) with 12 distinct ssRNA oligonucleotides as shown in Figure 3C,D and in Figure 3—figure supplement 1. In vitro RNA binding was tested with a protein concentration between 16 nM to 2 μm and is classified as robust binding (KD ≤ 2 μM, ++), intermediate binding (KD > 2 μM, +) or no binding (no interaction within this protein concentration range, −). (B) Secondary structure models for full-length (234 nt) ss-forward (purine-rich) or ss-reverse (pyrimidine-rich) MSR RNA based on in vitro probing by SHAPE analysis. (C) Secondary structure models for full-length (208 nt) ss-forward or ss-reverse LINE L1MdA 5'UTR RNA based on in vitro probing by SHAPE analysis. For both (A) and (B), nucleotide positions of the MSR or LINE L1MdA 5'UTR RNA are indicated and colors reflect normalized SHAPE reactivities of unreactive (0–0.4), moderately reactive (0.4–0.85) and highly reactive (>0.85) nucleotide positions. (D) Classification of the EMSA-tested RNA oligonucleotides (Figure 3C,D and Figure 3—figure supplement 1) based on their in silico structural prediction of single-stranded regions.
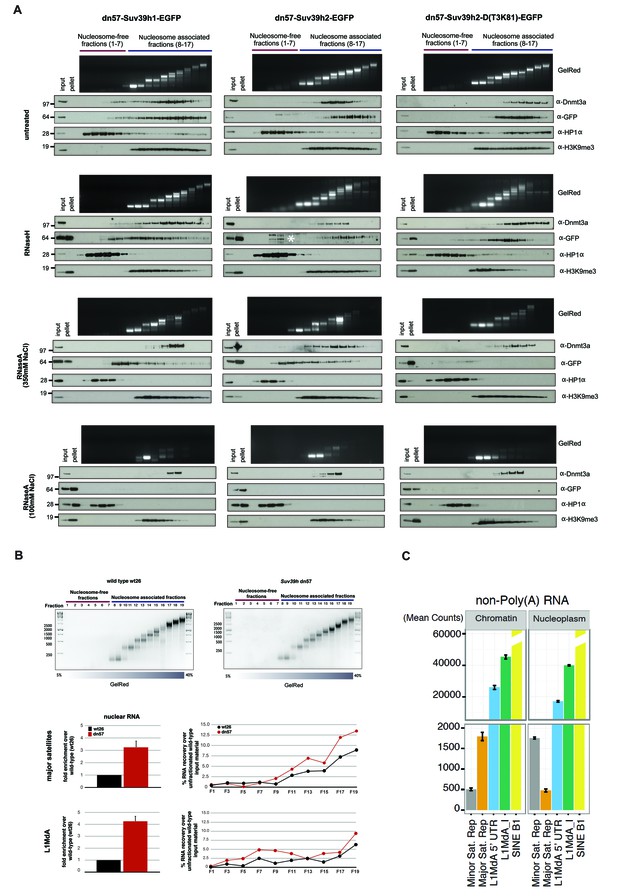
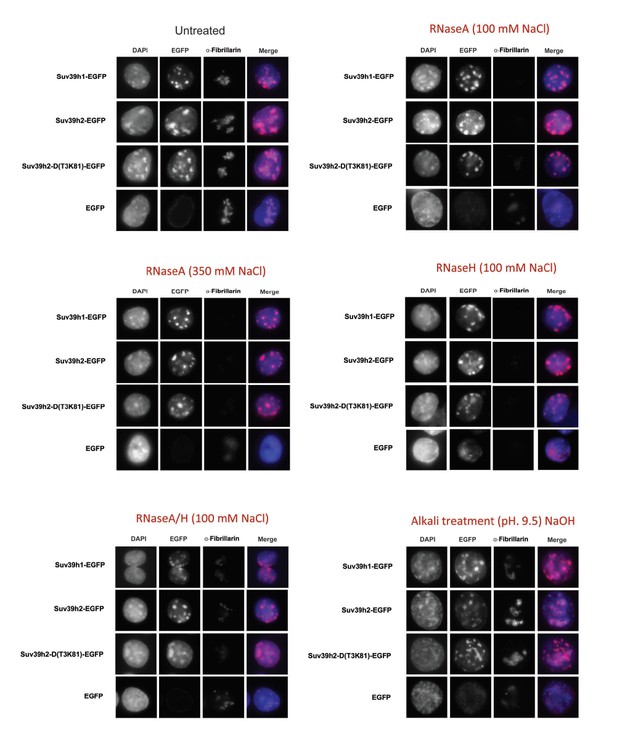
RNA-dependent association of Suv39h enzymes to native nucleosomes.
(A) Sucrose gradient fractionation of MNase-digested chromatin from the Suv39h1-EGFP, Suv39h2-EGFP and Suv39h2-D(T3K81)-EGFP mouse ES cell lines. The separation between nucleosome-free and nucleosome-containing fractions was monitored by Gelred staining of DNA. The sedimentation profile of the various Suv39h-EGFP products and of endogenous Dnmt3a, HP1α and of H3K9me3 was analyzed by western blotting with α-GFP, α-Dnmt3a, α-HP1α and α-H3K9me3 antibodies (first panel). To address whether RNA is associated with the nucleosomal fractions, MNase-digested soluble chromatin was incubated with RNaseH (second panel) or with RNaseA at 350 mM salt (third panel) or with RNaseA at 100 mM salt (fourth panel) before being loaded on the sucrose gradient. The asterisk indicates unspecific bands. All of these experiments were performed with two biological replicates and the RNaseA (100 mM salt) and RNaseH treatments were done three independent times. (B) Gelred DNA staining of sucrose gradient fractionation of MNase-digested chromatin from wild type and Suv39h dn ES cells and RT-qPCR to detect MSR and LINE L1MdA 5'UTR transcripts in RNA preparations from every second fraction of the sucrose gradient. No signal was detected in the control reactions lacking RT. The histogram on the left shows expression of MSR and LINE L1 MdA transcripts in nuclear RNA of wild-type and Suv39h dn ES cells. (C) Hiseq RNA sequencing of chromatin-associated and nucleoplasmic cDNA libraries (non-poly(A) selected) that were generated from wild-type ES cells to quantify the relative abundance of minor satellite repeat, major satellite repeat, LINE L1MdA and SINE B1 transcripts. Plotted are the mean counts of three biological replicates.
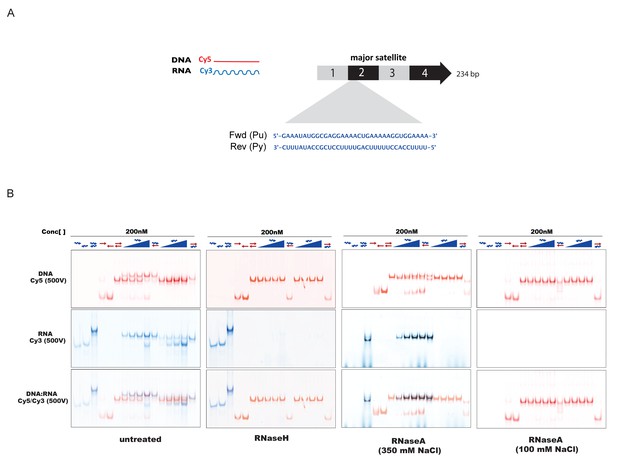
In-vitro characterization of substrate specificity of RNaseH and RNaseA.
(A) 35 nt 5’-Cy3-labeled RNA and 5’-Cy5-labeled DNA oligonucleotides spanning the second sub-repeat of the MSR consensus sequence were used to characterize the in vitro substrate specificity of RNaseH (Epicenter) or RNaseA (ThermoFisher Scientific). (B) ssRNA and ssDNA oligonucleotides were heat-denatured and gradually cooled to allow the formation of duplex structures (dsDNA, dsRNA and RNA:DNA hybrids). Formation of RNA:DNA hybrids was achieved by titrating increasing amounts of ssRNA (100 nM – 400 nM) onto a dsDNA template (200 nM). Equimolar amounts of single and double-stranded oligonucleotides were subjected to native PAGE and their migration was visualized by scanning the Cy3 and Cy5 fluorescent signals on a Typhoon FLA 9500 fluorescence scanner at 500V (first panel, untreated). Single-stranded and double-stranded oligonucleotides were incubated with 10 U of RNaseH before being resolved by PAGE. Scanning of the Cy3 fluorescent signal shows depletion of ssRNA oligonucleotides only when these are forming a heteroduplex with ssDNA (second panel, RNaseH). Incubation of single and double-stranded oligonucleotides with 10 μg of RNaseA at high salt concentrations (350 mM NaCl) reveals digestion of ssRNA, while dsRNA and RNA:DNA hybrids remain mostly intact (third panel, RNaseA 350 mM NaCl). Complete digestion of ssRNA, dsRNA and RNA forming RNA:DNA hybrids was observed when RNaseA treatment was performed under low salt conditions (100 mM NaCl) (fourth panel, RNaseA 100 mM NaCl).
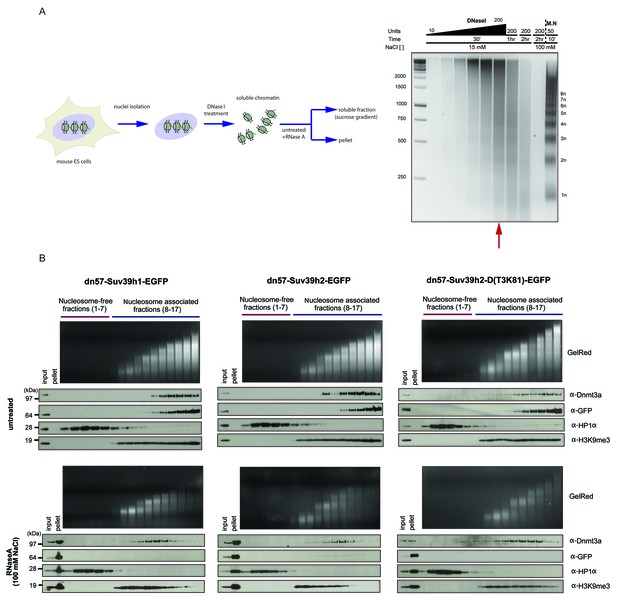
RNaseA-mediated dissociation of Suv39h-EGFP products from DNase1-generated soluble chromatin fractions.
(A) Workflow of the experimental strategy used to generate DNase1-digested soluble chromatin. Purified ES cell nuclei were incubated with increasing amounts of DNase1 (10, 20, 40, 70, 100 and 200 units) and for different time points (30 min, 1 hr and 2 hr). DNA was then extracted from the soluble fractions and resolved on an agarose gel. The size distribution of the DNase1-digested chromatin was compared to the nucleosomal ladder obtained after MNase treatment (M.N.). The red arrow indicates the conditions (200 units of DNase1 for 30 min at 15 mM NaCl) that were used to generate DNAse1-solubilized chromatin from the various Suv39h-EGFP rescued ES cell lines (B) DNase1-digested and soluble chromatin from Suv39h1-EGFP, Suv39h2-EGFP and Suv39h2-D(T3K81)-EGFP rescued ES cells was either left untreated (upper panels) or incubated with RNaseA at 100 mM NaCl (lower panels) before fractionation on a linear sucrose gradient and Western blot analysis of the sedimentation profile of the various Suv39h-EGFP proteins and endogenous Dnmt3a and HP1α as described for Figure 5A.
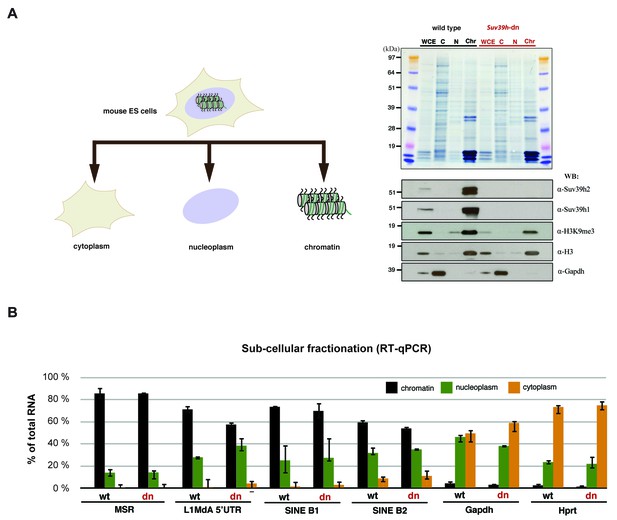
Relative abundance of major satellite and other repeat transcripts in cytoplasmic, nucleoplasmic and chromatin fractions.
(A) Wild type (wt26) and Suv39h double null (dn57) ES cells were separated into cytoplasmic, nucleoplasmic and chromatin fractions and the fractionation profile of Suv39h1, Suv39h2, H3K9me3, histone H3 and Gapdh was analyzed by Western blotting. (B) RNA was isolated from the cytoplasmic, nucleoplasmic and chromatin fractions and quantified by RT-qPCR with primers that are specific for major satellite repeats, LINE L1MdA 5'UTR, SINE B1 and SINEB2 repeat elements and for the house-keeping genes Gapdh and Hprt. The relative abundance of each of these transcripts in the three sub-cellular fractions is plotted and is normalized to the total (i.e. the sum of cyt + nuc + chr) RNA signal for each distinct RT-qPCR. Error bars represent the standard deviation from two biological replicates.
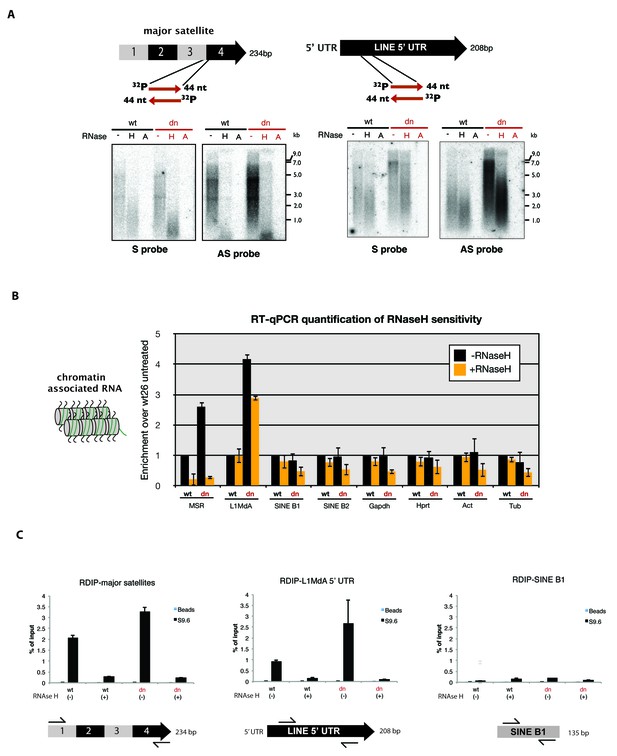
RNaseH sensitivity of MSR transcripts and detection of RNA:DNA hybrids.
(A) Northern blot analysis of chromatin-associated RNA from wild type and Suv39h dn mouse ES cells. Equal amounts (5 μg) of trizol-purified chromatin RNA was either left untreated or incubated with RNaseH or RNaseA (100 mM salt) before being separated in a denaturing agarose gel and transferred to a nylon membrane. Hybridization was performed using strand-specific DNA oligonucleotide probes to detect major satellite (left panel) or LINE L1MdA 5' UTR (right panel) repeat RNA. (B) RT-qPCR for repeat RNA (MSR, LINE L1MdA, SINE B1, SINE B2) and several housekeeping transcripts (Gapdh, Hprt, actin, tubulin) in chromatin-associated RNA isolated from wild type and Suv39h dn mouse ES cells and either untreated or digested with RNaseH prior to reverse transcription. The data are normalized to the untreated wild-type control and the experiments were performed with two biological replicates. (C) RDIP analysis of chromatin-associated RNA in wild type and Suv39h dn mouse ES cells. Trizol-purified chromatin RNA was either untreated or digested with RNaseH and then immunoprecipitated with the S9.6 antibody, followed by directed amplification with primers that are specific for major satellite repeats, the LINE L1MdA 5'UTR or SINE B1 elements. The data represent the mean ± SD of at least two independent experiments.
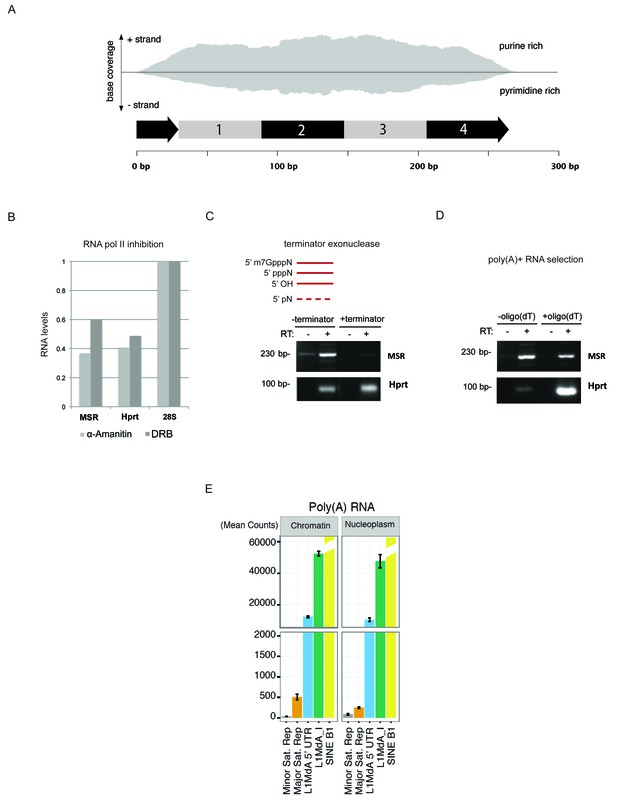
Characterization of major satellite RNA in mouse ES cells.
(A) HiSeq RNA sequencing reads from nuclear ribo-depleted RNA preparations of wild type mouse ESC (wt26) were randomly mapped to the major satellite repeat consensus sequence in a strand-specific manner (Bulut-Karslioglu et al., 2014). Distribution of reads in forward (purine rich) and reverse orientation (pyrimidine rich) reveals transcriptional activity from both strands. (B) Wild type (wt26) ES cells were treated with 100 μg/ml α-amanitin (Sigma) or 0.1 mM 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) (Sigma) to inhibit RNA polymerase II transcription. Total RNA isolated from treated and untreated ESC was reverse transcribed with random hexamers and then amplified with primers that are specific for MSR, Hprt or 28S rRNA as negative control. The RT-qPCR amplification signal after RNA polymerase II inhibition was normalized to the untreated samples. (C) Total RNA isolated from mouse ESC was left untreated or incubated with a terminator-5’-phosphate dependent exonuclease (Epicenter) that selectively digests RNA with a free 5’-monophosphate but cannot digest RNA containing a 5’-triphosphate (pppN), 5’ cap (m7GpppN) or 5’ hydroxyl group. RNA was then reverse transcribed into cDNA and the cDNA was amplified using specific primers for MSR or Hprt. (D) Total RNA isolated from mouse ESC was incubated with non-coated beads or with oligo(dT) coated beads to enrich for poly(A)+ RNA. Total RNA and poly(A)+ selected RNA were reverse transcribed into cDNA using random hexamers in the presence or absence of reverse transcriptase (RT). Enrichment for Hprt in the poly(A)+ fraction was observed whereas major satellite RNA was not enriched, indicating that only a fraction of major satellite RNA posses a poly-(A) tail. (E) HiSeq RNA sequencing of chromatin and nucleoplasmic cDNA libraries (poly(A)-selected RNA) from wild type (wt26) ES cells. Distribution of RNA sequencing reads in the different sub-cellular fractions was quantified for various repetitive elements and plotted as the mean count of reads for three independent sequencing runs (± SEM) and normalized for sequencing depth.
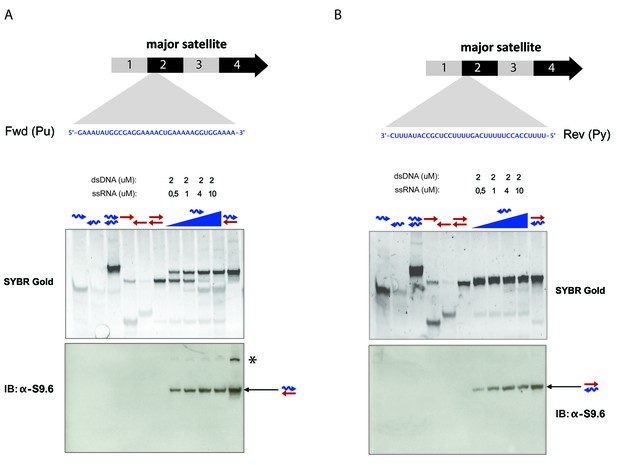
Specificity of the S9.6 monoclonal antibody towards RNA:DNA hybrids.
(A) RNA and DNA oligonucleotides (35 nt) spanning the second sub-repeat of the major satellite consensus sequence were used in the formation of single and double-stranded molecules. Formation of RNA:DNA hybrids was further achieved by titrating increasing amounts of ssRNA in forward (purine-rich) orientation onto a dsDNA template. Equimolar amounts of single and double stranded oligonucleotides were subjected to native PAGE and detected by SYBR gold staining. Following transfer to a nylon membrane, immunoblotting with the S9.6 antibody was performed. The signal from the S9.6 antibody detecting RNA:DNA hybrids is indicated with a black arrow and the asterisk shows a weak reactivity of the S9.6 antibody with other non-canonical nucleic acid structures that could reflect triple helices (B) Formation of RNA:DNA hybrids with the reverse (pyrimidine-rich) RNA strand of the sub-repeat 2 of the MSR and detection by SYBR gold staining of nucleic acids and reactivity with the S9.6 antibody.
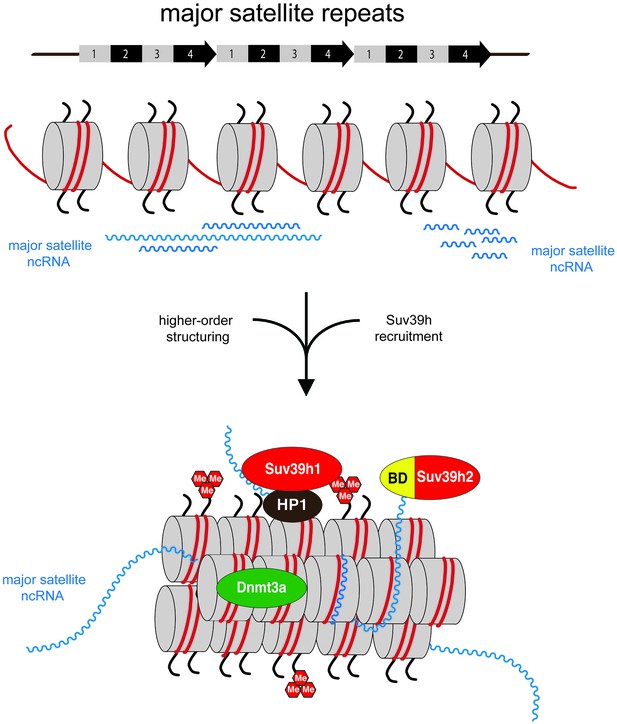
Model for an RNA-nucleosome scaffold as the underlying structure of mouse heterochromatin.
Model depicting a higher-order RNA-nucleosome scaffold that is established by chromatin association of major satellite repeat (MSR) RNA. In this model, initial transcriptional activity of the MSR repeats is needed to build heterochromatin. The intrinsic property of MSR repeat sequences to form RNA:DNA hybrids will facilitate their chromatin retention and most likely occurs in inter-nucleosomal regions. Additional portions of ssMSR RNA organize the assembly of a higher-order RNA-nucleosome structure and are also important for the recruitment and stabilization of the Suv39h enzymes to heterochromatin. MSR RNA decorated heterochromatin will provide multiple affinities for the Suv39h KMT, such as ssRNA binding by the basic domain (BD) of Suv39h2 (this study), H3K9me3 (Wang et al., 2012) and RNA binding by the chromo domains of both mouse Suv39h1 (Shirai et al., 2017) or human SUV39H1 (Johnson et al., 2017) enzymes and HP1 interaction (Yamamoto and Sonoda, 2003; Maison et al., 2011). Additional protein-protein contacts with other chromatin-associated components (Maison et al., 2016), histone H1 (Lu et al., 2013) and transcription factors (Bulut-Karslioglu et al., 2012) are not shown.
Additional files
-
Supplementary file 1
The Table lists DNA and RNA oligonucleotide sequences that were used as primers or nucleic acid substrates in a variety of assays described in this study.
Additional sequences of RNA oligonucleotides (minor satellite repeats, LINE L1 MdA, SINE B1, pRNA and TERRA) used for EMSA are indicated in Figure 3—figure supplement 1.
- https://doi.org/10.7554/eLife.25293.018





