Switching of metabolic programs in response to light availability is an essential function of the cyanobacterial circadian output pathway
Figures
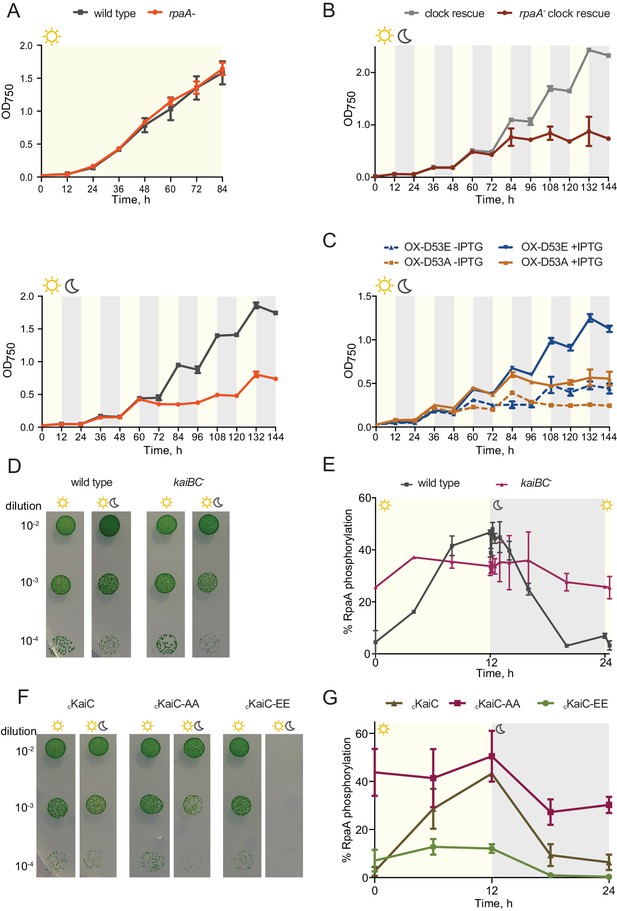
The rpaA- strain displays a defect in viability in light/dark conditions.
(A) Growth curves of wild type and the rpaA- strain in constant light (top) and in 12 hr light/12 hr dark conditions (below) in BG-11 medium. Points represent the mean of three independent experiments with error bars displaying the standard error of the mean. (B) Growth curves of the ‘clock rescue’ and the rpaA- ‘clock rescue’ strains in 12 hr light/12 hr dark conditions in BG-11 medium. Points represent the mean of three independent experiments with error bars displaying the standard error of the mean. (C) Growth curves of the rpaA- kaiBC- strains containing a Ptrc promoter driving expression of RpaA D53E (OX-D53E) or RpaA D53A (OX-D53A) in 12 hr light/12 hr dark conditions grown in the presence or absence of 20 μM IPTG. Points represent the mean of two biological replicates with error bars displaying the standard error of the mean. (D) Comparison of growth of wild type and the kaiBC- mutant in constant light and in light/dark conditions. The experiment was performed three independent times and a representative experiment is shown. (E) Quantification of RpaA phosphorylation levels measured by Phos-tag western blot in wild type and the kaiBC- strains in light/dark conditions. Each point represents the mean of two biological replicates with error bars displaying the standard error of the mean. (F) Comparison of growth of the cKaiC strain (subscript ‘c’ denoting constitutive, RpaA-independent expression) and strains expressing phosphomimetic variants of KaiC, cKaiC-EE or cKaiC-AA, in constant light and in light/dark conditions. The experiment was performed three independent times and a representative experiment is shown. (G) Quantification of phosphorylation levels of RpaA measured by Phos-tag western blot in the cKaiC strain or in the strains expressing phosphomimetic variants of KaiC, cKaiC-EE and cKaiC-AA, in light/dark conditions. Each point represents the mean of three biological replicates with error bars displaying the standard error of the mean.
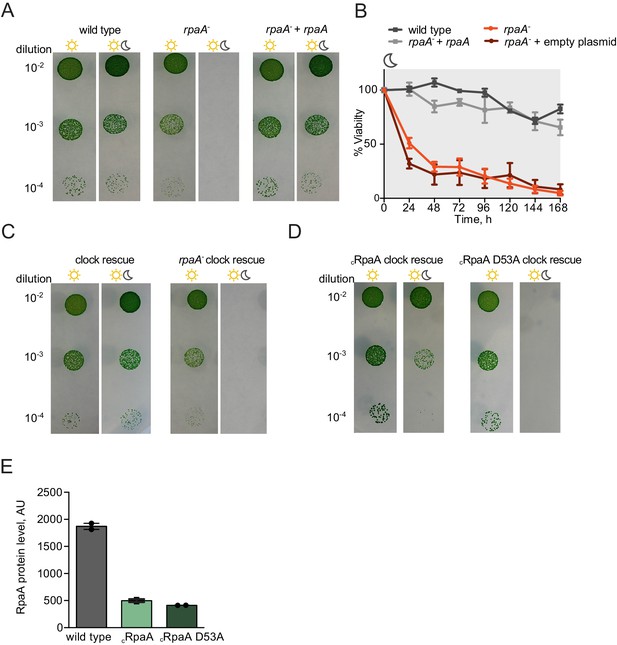
Characterization of growth and viability of the rpaA mutants.
(A) Complementation of the rpaA- strain with rpaA expressed from the neutral site NS1 rescues the viability defect in light/dark conditions. The experiment was performed three independent times and a representative experiment is shown. (B) Viability of the indicated strains after prolonged incubation in darkness. The survival of cells was assessed by a colony forming unit assay. Points represent the mean of two experiments with error bars displaying the standard error of the mean. (C) Comparison of growth of the ‘clock rescue’ strain and the rpaA- ‘clock rescue’ strain. The experiment was performed three independent times and a representative experiment is shown. (D) Comparison of growth of the cRpaA ‘clock rescue’ strain and the cRpaA D53A ‘clock rescue’ mutant, where cRpaA indicates constitutive expression of RpaA. In wild type strain RpaA activates its own expression. In the cRpaA strains the rpaA promoter was changed to a P0050 promoter allowing for constitutive expression of RpaA D53A. The experiment was performed three independent times and a representative experiment is shown. (E) Comparison of RpaA protein levels expressed from the native promoter and the P0050 clock-independent, constitutive promoter. Each point represents an individual measurement with error bars displaying the standard error of the mean.
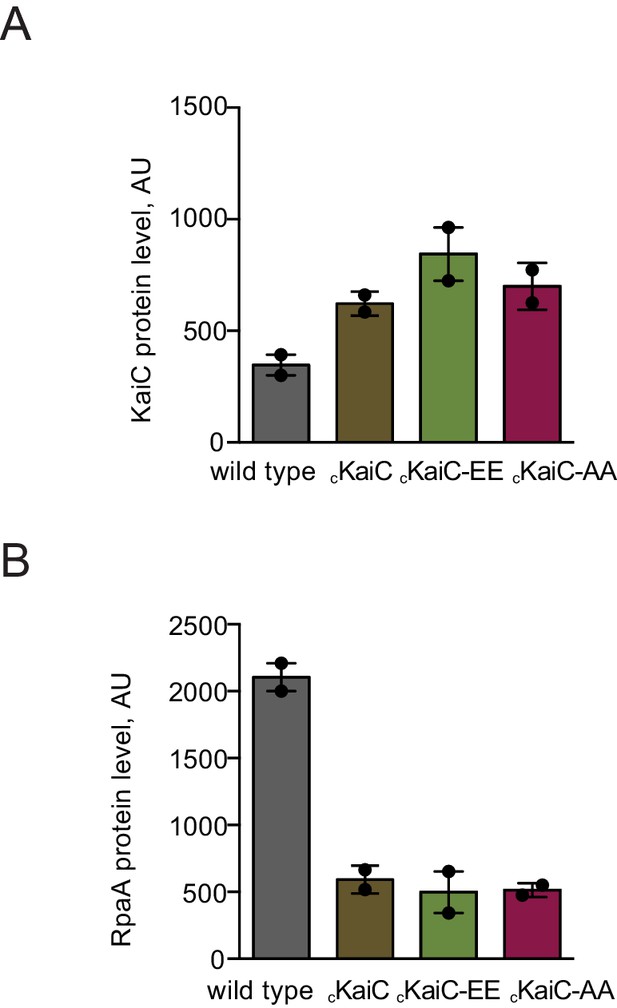
Characterization of protein levels of KaiC and RpaA in the cKaiC phosphomimetic mutants in comparison to wild type.
(A) Comparison of protein levels of KaiC variants expressed from the native and the P0050 promoters assessed by western blot. Each point represents an individual measurement with error bars displaying the standard error of the mean. (B) Comparison of protein levels of RpaA expressed from the native promoter and the P0050 promoter in the phosphomimetic strains assessed by western blot. Each point represents an individual measurement with error bars displaying the standard error of the mean.
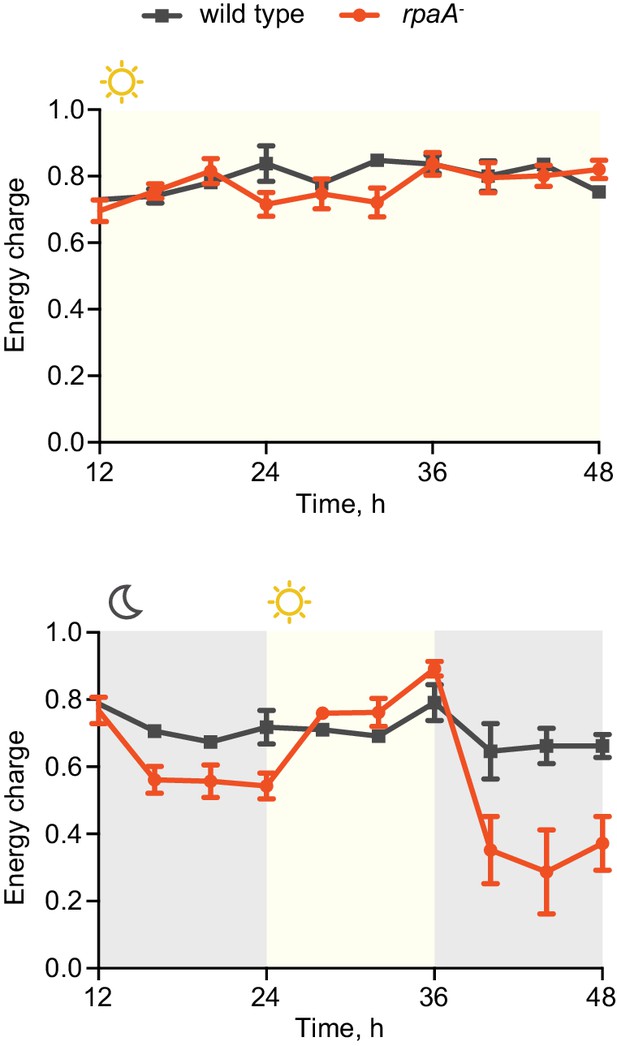
The rpaA- strain has a defect in maintenance of the cellular energy charge in oscillating light/dark conditions.
Changes in the energy charge, defined as ([ATP] +0.5*[ADP])/([ATP]+[ADP]+[AMP]), during growth of wild type and the rpaA- strain in constant light (top) or in 12 hr light/12 hr dark conditions (bottom) in BG-11 medium. Each point represents the mean of three independent experiments with error bars displaying the standard error of the mean.
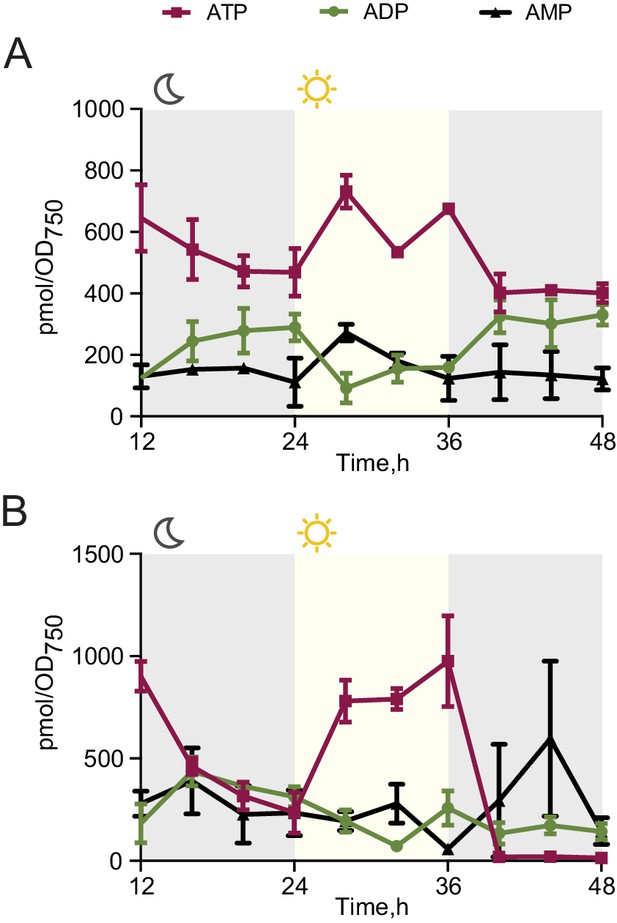
Changes in the ATP, ADP and AMP levels in cells grown in light/dark conditions.
Quantification of levels of ATP, ADP and AMP in wild type (A) and the rpaA- strain (B) grown in light/dark cycles. Points represent the mean of two experiments with error bars displaying the standard error of the mean.
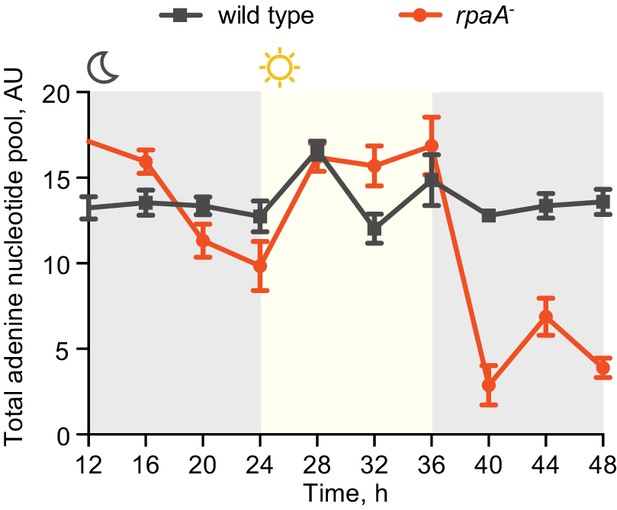
The rpaA- strain displays a defect in levels of adenine nucleotides in light/dark cycles.
Quantification of relative levels of all adenine nucleotides in wild type and the rpaA- strains grown in light/dark cycles. Points represent the mean of four experiments with error bars displaying the standard error of the mean.
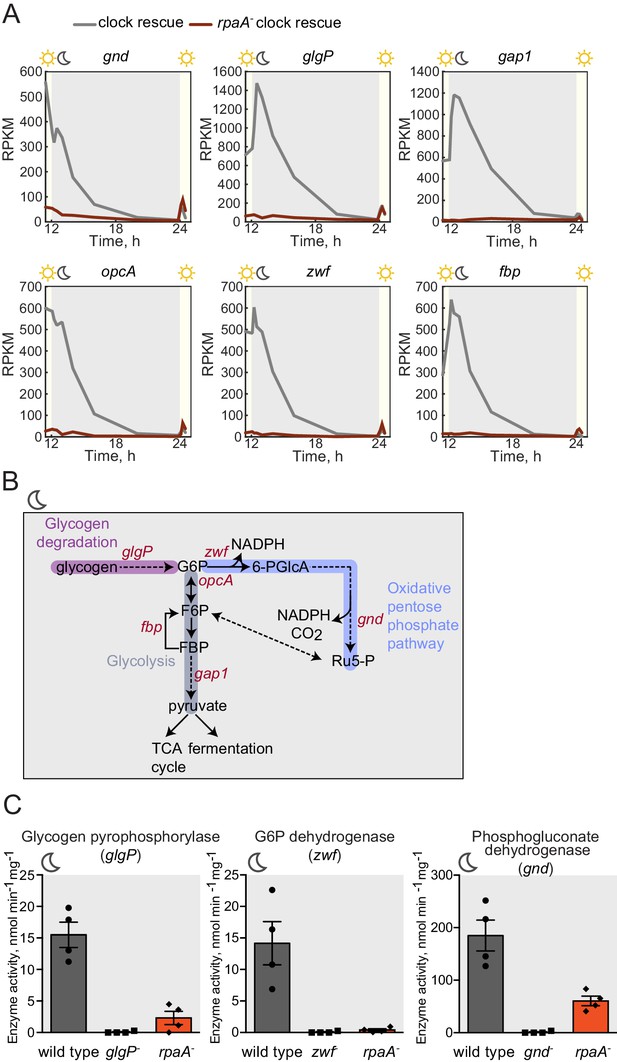
Sugar catabolism pathways are abrogated in the rpaA- strain.
(A) Temporal expression profiles of genes encoding enzymes involved in sugar metabolism whose expression is abrogated in the rpaA- ‘clock rescue’ strain. Relative RNA levels were measured in the light/dark conditions by RNA sequencing. (B) A diagram representing carbon metabolism in S. elongatus PCC7942 in the dark. At night glycogen constitutes the store for energy and carbon skeletons. Glycogen degradation, glycolysis and oxidative pentose phosphate pathways are the key metabolic pathways operating at night. Deletion of operons with genes colored red leads to a strong viability defect specifically in light/dark conditions (Figure 3—figure supplement 1C and Doolittle and Singer 1974; Scanlan et al., 1995). glgP, glycogen phosphorylase; gap1, glyceraldehyde-3-phosphate dehydrogenase; gnd, 6-phosphogluconate dehydrogenase; zwf, glucose-6-phosphate dehydrogenase; opcA, glucose-6-phosphate dehydrogenase assembly protein; fbp, fructose-1,6-bisphosphatase, G6P, glucose-6-phosphate; 6-PGlcA, 6-phosphogluconate; F6P, fructose-6-phosphate; FBP, fructose-1,6-bisphosphate, Ru5-P, ribulose-5-phosphate (C) Enzymatic activities of glycogen phosphorylase, glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase in wild type, the rpaA- and negative control strains measured 3 hr after exposure to expected darkness. Error bars represent standard error of the mean of four independent experiments.
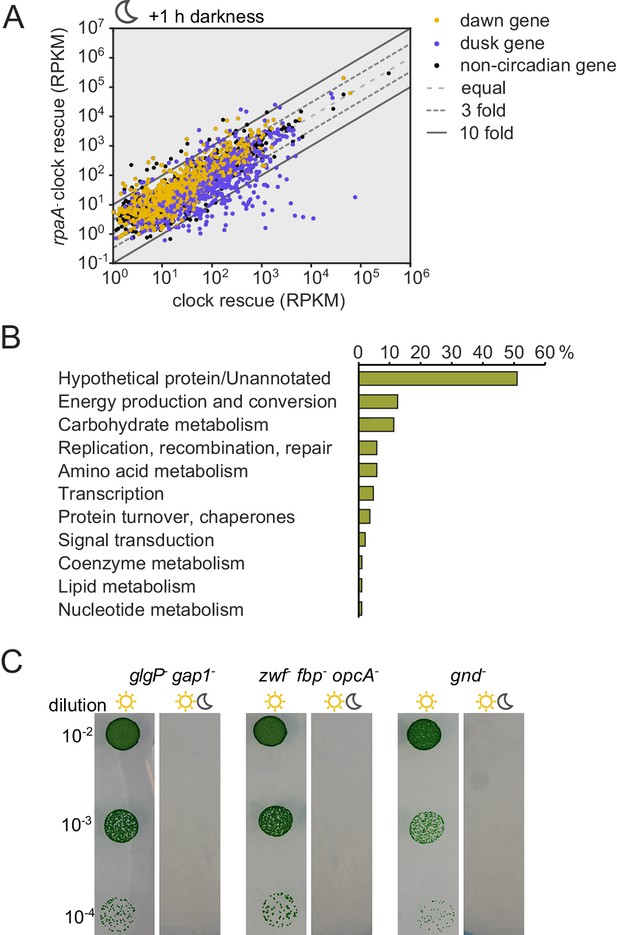
Expression of genes encoding enzymes involved in alternative sugar catabolism pathways is defective in the rpaA- strain in light/dark cycles.
(A) Comparison of the normalized gene expression in the ‘clock rescue’ and the rpaA- ‘clock rescue’ strains 1 hr after exposure to darkness. Circadian genes whose expression peaks at dawn in wild type strain are colored yellow, dusk-peaking genes are blue and non-circadian genes are black. Fold changes are displayed graphically as diagonal lines on the plot. (B) Analysis of the functional annotations of the protein coding genes whose expression is defective in the rpaA- ‘clock rescue’ strain. We identified 88 protein coding genes whose expression is at least 3-fold lower in the rpaA- ‘clock rescue’ strain than in the control strain, as described in the Materials and methods section. The detailed list of genes, whose expression is abrogated in the rpaA- ‘clock rescue’ strain, is presented in Supplementary file 1. (C) A representative photograph of a plate viability assay performed using mutants characterized by Doolittle and Singer (1974) and Scanlan et al. (1995). The deletion mutants of indicated genes involved in alternative sugar metabolism pathways were plated as a dilution series on BG-11 agar and grown in continuous light for 7 days or under light/dark conditions for 14 days. The experiment was performed four independent times.
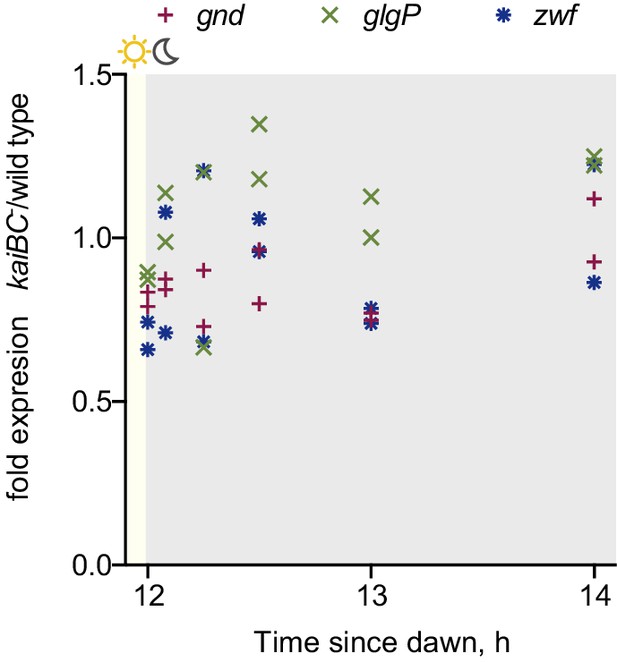
Expression of key metabolic dusk genes in the kaiBC- strain.
Expression of metabolic genes gnd, glgP and zwf measured by RT-qPCR in the kaiBC- strain relative to wild type at dusk and during the first two hours of darkness. Each point represents an independent measurement.
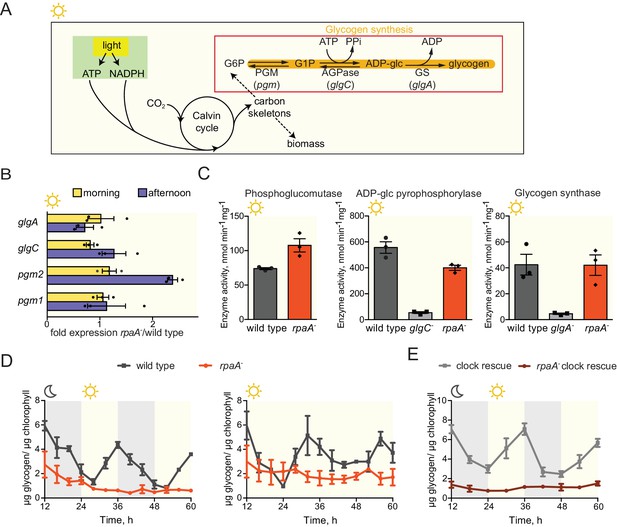
The rpaA- strain accumulates little glycogen.
(A) A diagram representing carbon metabolism in S. elongatus PCC7942 in light. During the day S. elongatus cells perform photosynthesis to produce carbon skeletons and energy for growth and for preparation of glycogen stores. The glycogen synthesis pathway is comprised of three enzymatic activities: phosphoglucomutase (PGM), ADP-glucose pyrophosphorylase (AGPase) and glycogen synthase (GS). G6P, glucose-6-phosphate; G1P, glucose-1-phosphate; pgm, phosphoglucomutase; glgC, ADP-glucose pyrophosphorylase; glgA, glycogen synthase. (B) Relative expression of genes encoding enzymes in the glycogen synthesis pathway measured by RT-qPCR in the morning and in the afternoon in wild type and the rpaA- strains. Error bars represent the standard error of the mean of three independent experiments. (C) Activities of enzymes in the glycogen synthesis pathway in wild type, the rpaA- and negative control strains measured during the day (at time = 10 hr). Error bars represent the standard error of the mean of three independent experiments. (D) Glycogen content in wild type and the rpaA- strains grown in light/dark and constant light conditions. Points represent the mean of two experiments with error bars displaying the standard error of the mean. (E) Glycogen content in the ‘clock rescue’ and the rpaA- ‘clock rescue’ strains grown in light/dark conditions. Points represent the mean of two experiments with error bars displaying the standard error of the mean.
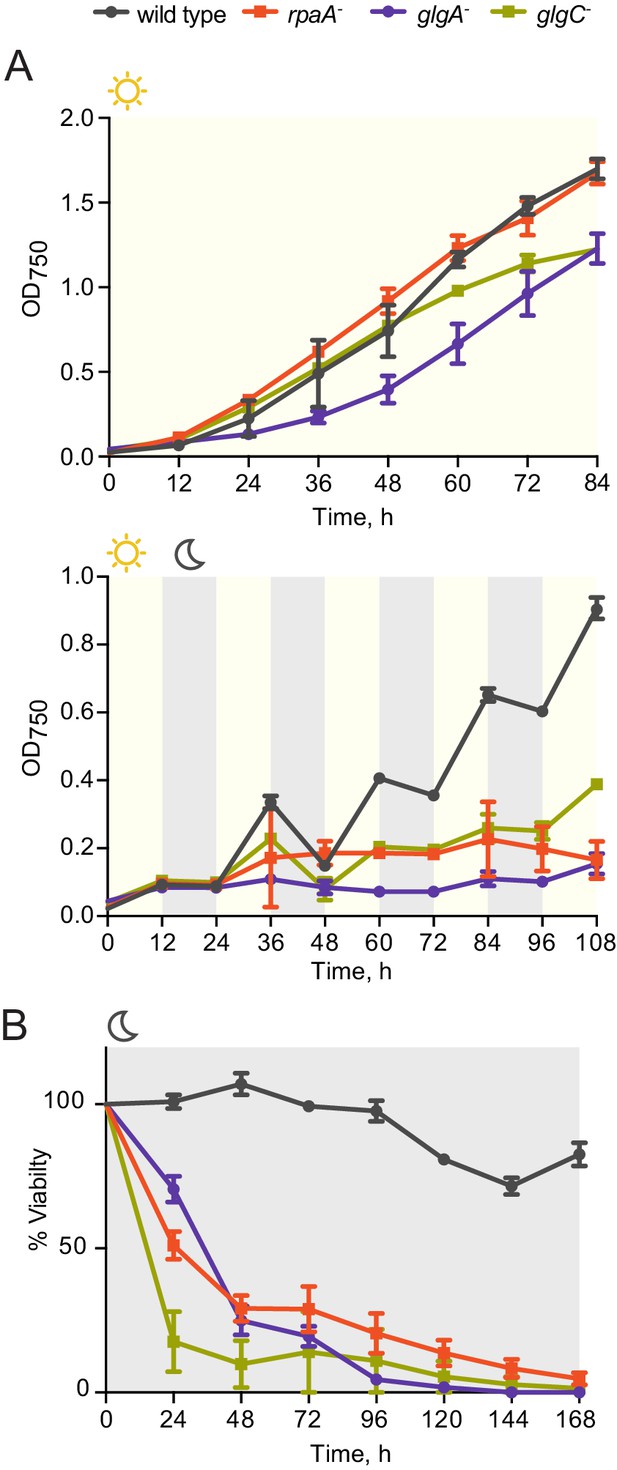
Comparison of growth and viability of the rpaA- strain and glycogen synthesis mutants.
(A) Comparison of growth curves of wild type, the rpaA- strain and the deletion mutants deficient in glycogen synthesis (glgA- and glgC-) in constant light (top) and in light/dark cycles (below). Points represent the mean of two experiments with error bars displaying the standard error of the mean. (B) Viability of wild type, the rpaA- strain and strains defective in glycogen synthesis after incubation in prolonged darkness. The survival of cells was assessed by a colony forming unit assay. Points represent the mean of two experiments with error bars displaying the standard error of the mean.
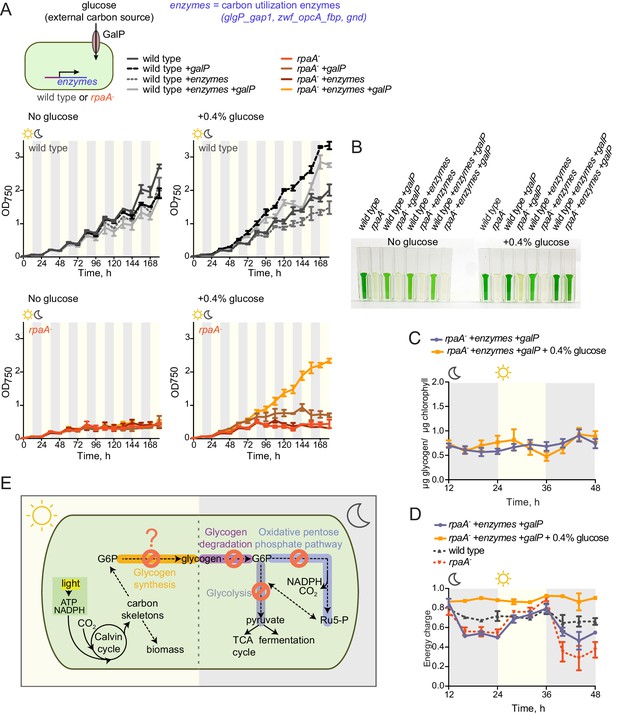
Restoration of sugar catabolism pathways and glucose feeding rescue the viability defect of the rpaA- strain.
(A) Growth curves of the indicated engineered strains in 12 hr light/12 hr dark conditions in BG-11 medium with 100 μM IPTG and with (right) or without (left) 0.4% glucose supplementation. Wild type (top) and the rpaA- (below) strains were engineered to express a sugar transporter GalP and/or sugar utilization enzymes using an IPTG-inducible promoter. Points represent the mean of two experiments with error bars displaying the standard error of the mean. (B) Representative cell cultures of the engineered strains photographed at the end of the growth experiment in light/dark conditions with or without 0.4% glucose supplementation. S. elongatus cells are blue-green in color. The darker the color of the culture, the higher is its optical density at 750 nm. (C) Glycogen content in in the rpaA- + enzymes + galP strain ±0.4% glucose in light/dark conditions. Points represent the mean of two experiments with error bars displaying the standard error of the mean. (D) Energy charge measurement in the rpaA- + enzymes + galP strain with or without 0.4% glucose supplementation in light/dark conditions. Points represent the mean of two experiments with error bars displaying the standard error of the mean. Data representing the energy charge in wild type and the rpaA- strain are reproduced from Figure 2 to facilitate comparison. (E) A model representing physiological processes in S. elongatus regulated by the activity of the circadian program that contribute to cell fitness. The circadian clock schedules periods of activity of RpaA to coordinate anticipatory carbon reserve formation during the day with carbon utilizing metabolic pathways activated at dusk to fuel cell integrity and viability in an environment in which light and dark periods alternate. Defects present in the rpaA deficient cells, indicated by red symbols on the diagram, lead to a low energy charge and reduced viability during periods of darkness.
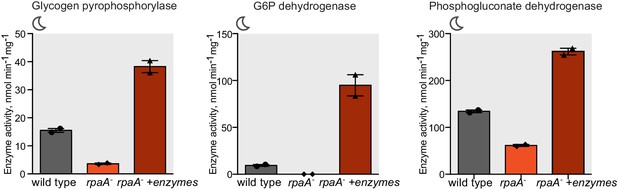
Characterization of strains used for the reconstitution experiments.
Enzyme activities of glycogen phosphorylase, glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase in wild type, the rpaA- and the rpaA- +enzymes strains. Strains were incubated in BG-11 with 100 μM IPTG for 12 hr in light and harvested for the assay 3 hr after exposure to darkness. Error bars display the standard error of the mean of two experiments.
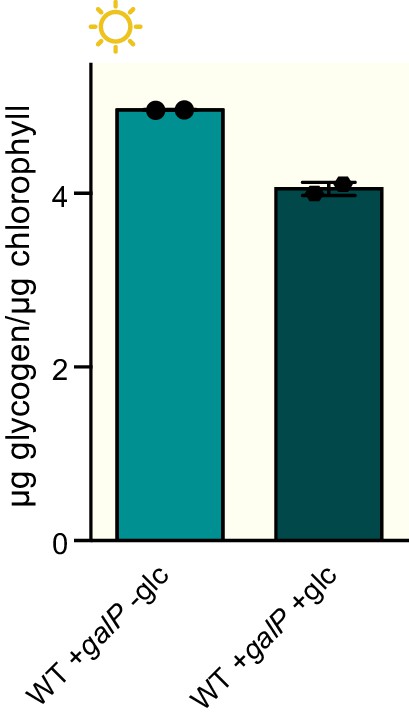
Glucose supplementation does not increase glycogen accumulation in wild type cells.
Glycogen content in wild type +galP strain grown in the presence of 100 μM IPTG with or without supplementation of the medium with 0.4% glucose. Error bars display the standard error of the mean of two independent experiments.
Additional files
-
Supplementary file 1
Genes whose expression is abrogated in the rpaA- ‘clock rescue’ strain.
- https://doi.org/10.7554/eLife.23210.017
-
Supplementary file 2
Strain list.
Cmr, chloramphenicol resistance; Gmr, gentamycin resistance; Kmr, kanamycin resistance; Ntr, nourseothricin resistance; Sp/St, spectinomycin/streptomycin resistance; NS 1, neutral site 1 (GenBank U30252)
- https://doi.org/10.7554/eLife.23210.018





