A deep proteomics perspective on CRM1-mediated nuclear export and nucleocytoplasmic partitioning
Figures
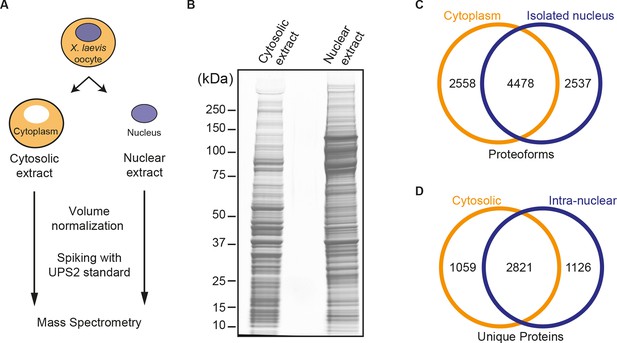
Spatial proteomics of Xenopus laevis oocytes.
(A) Workflow for mass spectrometric analysis of cytosolic and nuclear proteins. For details, see Materials and methods and main text. (B) Analysis of obtained cytosolic and nuclear fractions by SDS-PAGE and Coomassie-staining. The loads correspond to 750 nanolitres of either yolk-free cytoplasm or nuclear contents. (C) Venn diagram of proteoforms (including all allelic variants of a given gene product) that have been identified in the manually isolated cytoplasms and nuclei. (D) Venn diagram is similar to (C), but proteoforms corresponding to a given gene (foremost allelic variants) have been merged down to ‘unique proteins’. Also, proteins were subtracted that just co-purified with nuclei, but do not represent intranuclear proteins; this applied to constituents of the nuclear envelope (ER) and the nucleus-associated mitochondrial cloud.
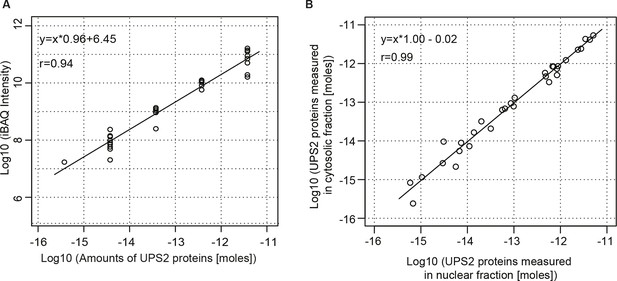
Estimation for the accuracy of mass spectrometric protein quantitation.
(A) Log-log regression between amounts of added UPS2 standard proteins and measured iBAQ intensities. (B) Correlation of measured amounts of UPS2 standard proteins added to either the nuclear or cytoplasmic fraction.
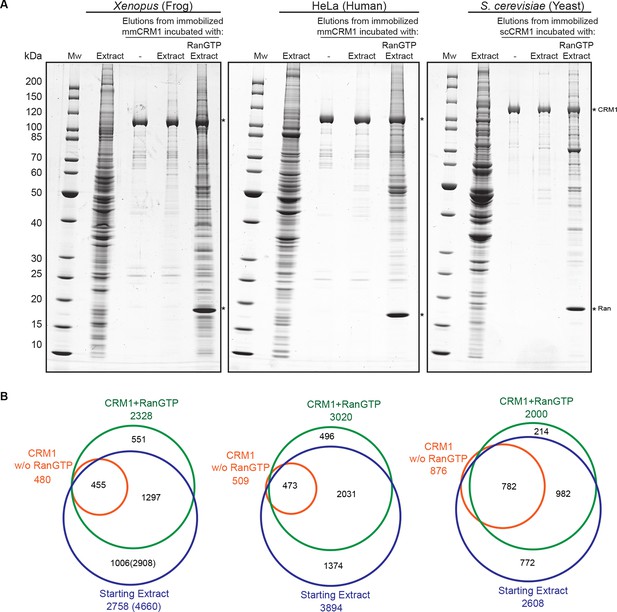
Identification of potential CRM1 cargoes from 3 species.
(A) Mouse (mm) or yeast (sc) CRM1 were immobilised through a biotinylated Avi-tag to streptavidin agarose, and incubated with indicated extracts (1 ml), without or with the addition of 5 μM RanQ69L5-180GTP. The beads were thoroughly washed and subsequently eluted at 45°C with SDS sample buffer (which leaves the biotin-streptavidin interaction largely intact). Analysis of indicated samples was by SDS-PAGE and Coomassie-staining. 1/200 of the starting extracts and 1/5 of eluates were loaded. (B) Starting extracts, CRM1 w/o Ran, and CRM1+RanGTP samples were analysed by mass spectrometry. Venn diagrams represent identified unique proteins. Numbers in parenthesis include also proteins that were not identified in a total Xenopus extract or the ‘CRM1+RanGTP’ sample, but in the isolated nuclear and cytoplasmic fractions; these proteins extend the list of ‘CRM1-non-binders’.
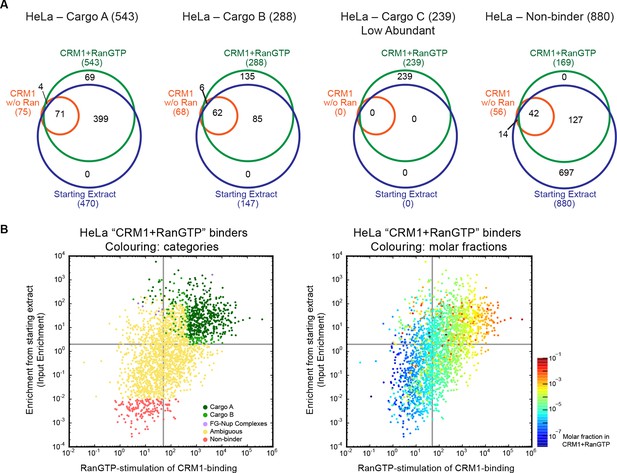
Categories of CRM1-binders from HeLa cells.
For each identified CRM1-binder, we calculated or estimated three parameters from measured iBAQ intensities: its abundance (molar fraction) within the ‘CRM1+RanGTP’-bound sample, the RanGTP-stimulation of its CRM1-binding, and how strongly it became enriched by the ‘CRM1+RanGTP’-affinity chromatography. These numbers where then used to group binders into distinct categories, ranging from ‘A’ (the most probable cargoes) to ‘non-binders’. (A) Venn diagrams representing the indicated cargo classes with respect to their identification in the starting extract, ‘CRM1 w/o Ran’- and/or ‘CRM1+RanGTP’-bound samples. (B) Scatter plot representing ‘CRM1+RanGTP’-binders from HeLa cells, using the parameters ‘RanGTP-stimulation’ and ‘input-enrichment’ as coordinates. Colouring is according to classification. Most ‘non-binders’ had not been identified in the ‘CRM1+RanGTP’ sample; they are therefore also not plotted. Measurement of the parameters ‘RanGTP-stimulation’ and ‘input-enrichment’ required the identification a given candidate in input, ‘CRM1 w/o Ran’, as well as in the ‘CRM1+RanGTP’- sample. If undetected in either ‘input’ or ‘CRM1 w/o Ran’, then the missing parameter was estimated as a lower bound (based on the detection sensitivity of our MS setup). Candidates detected only in ‘CRM1+RanGTP’ were not plotted (because for them, both parameters would have to be estimated). (C) Scatter plot is as in (B), but colour code is used to indicate the abundance in the ‘CRM1+RanGTP’-bound sample.
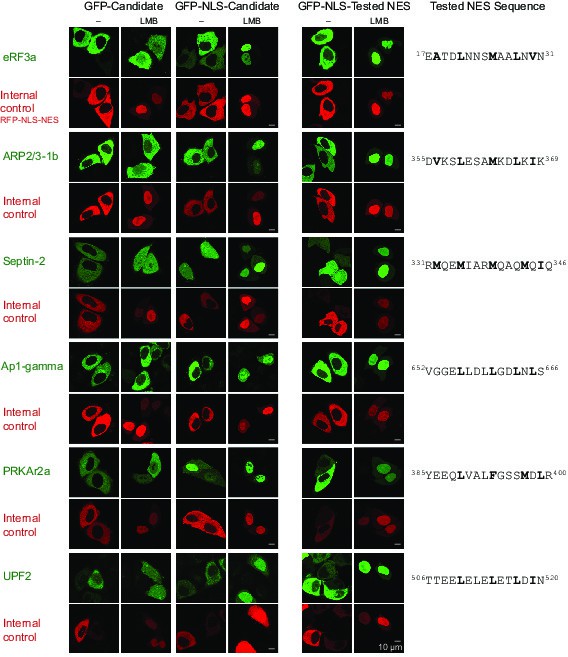
Validation of Xenopus CRM1-cargo candidates and identification of NESs.
HeLa cells were transfected to express GFP- or GFP-NLS-fused candidate proteins, then incubated with or without the CRM1 inhibitor leptomycin B (LMB), fixed, and analysed by confocal laser scanning microscopy (CLSM). The co-transfected RFP-NLS-NES was detected in a separate channel as a control for the LMB-effect. Tested candidates: eukaryotic peptide chain release factor eRF3a (Q91855), subunit 1b of the ARP2/3 complex (Q6GNU1), Septin-2 (B7ZR20), Ap1-gamma subunit of the clathrin-associated adapter complex (Q6GPE1), the cAMP-dependent kinase type II-alpha regulatory subunit pRKAr2a (F7CZT8), and the regulator of nonsense transcripts UPF2 (Q498G1). UniProt entry names are given in parentheses. Figure also shows sequences of identified NESs, and their validations as transfected GFP-NLS-fusions with an LMB-sensitive cytoplasmic localisation.
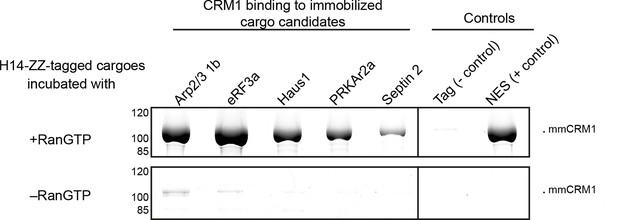
Identification of cargo candidates as direct CRM1-binders.
The H14-ZZ-Sumo tagged candidate proteins ARP2/3 1b (Q6GNU1), eRF3a (Q91855), Haus1 (Q3B8L5), pRKAr2a (F7CZT8), Septin-2 (B7ZR20) were expressed in E. coli, purified, immobilised on anti-zz beads, and incubated with CRM1 in the absence or presence of RanGTP. Immobilised candidate proteins were released, after washing, by Sumo-protease cleavage and co-eluting materials were analysed by SDS-PAGE (Note that Septin-2 elution was less efficient than the others). An unfused H14-zz-Sumo module served as a negative and a fusion with a PKI-NES as a positive control for CRM1-binding.
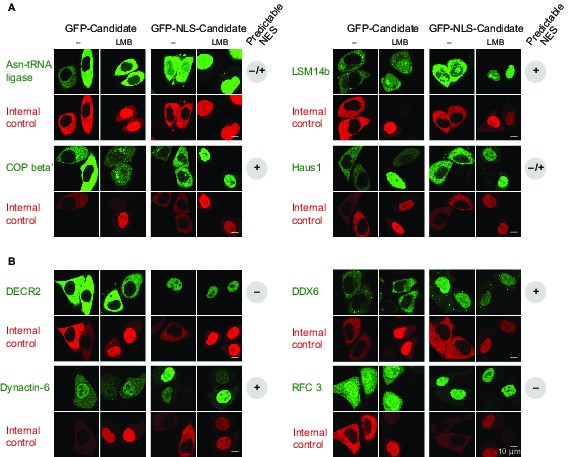
Validations of additional CRM1 cargo candidates from Xenopus.
Analysis was as in Figure 4. (A) Tested candidates that behave like true CRM1 cargoes: Asn-tRNA ligase (Q6DD18), LSM14b (L14BB), COP beta’ (Q7ZTR0), and Haus1 (Q3B8L5). (B) Tested candidates that are not CRM1 cargoes: the peroxisomal 2,4-dienoyl-CoA reductase DECR2 (Q6GR01), the RNA helicase DDX6, dynactin 6 (Q6IRC3) and the replication factor complex subunit RFC 3 (Q4QQP4). DDX6 had been in cargo category A, but probably requires Lsm14 (see panel A and main text) for CRM1 interaction. We assume an analogous scenario for DECR2. Dynactin 6 is in category ‘ambiguous’ and was therefore not considered a CRM1 cargo in the first place.
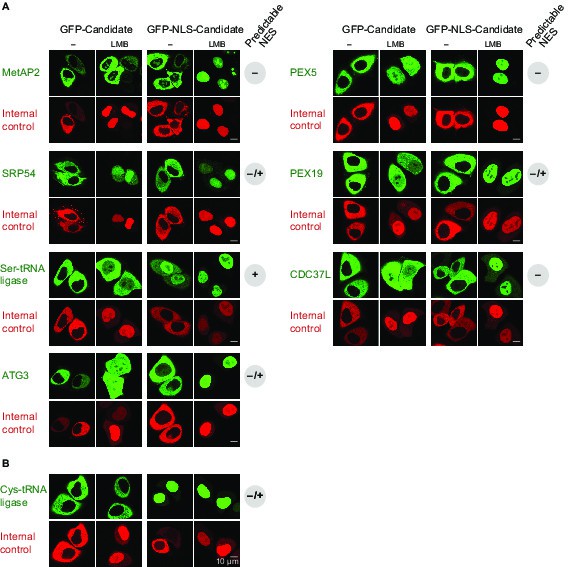
Validation of human CRM1 cargo candidates.
Analysis was as in Figure 4. UniProt identifiers correspond either to abbreviated protein names or are given in parentheses. (A) Positively tested CRM1 cargoes: the co-translational methionine aminopeptidase MetAP2 (MAP2), PEX5, SRP54, PEX19, the Ser-tRNA ligase (SYSC), CDC37L (CD37L), and ATG3. (B) The Cys-tRNA ligase (SYCC) was classified as a ‘CRM1-non-binder’ and accordingly shows a CRM1-independent nuclear exclusion.
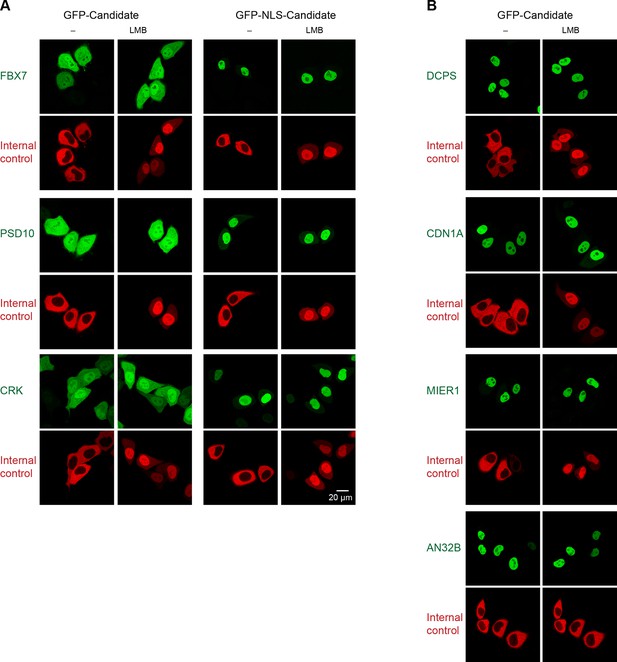
Validation of human CRM1 non-binders.
(A) Re-tested CRM1 non-binders, which showed a nucleocytoplasmic equilibration: the F-box only protein 7 (FBX7), the 26S proteasome non-ATPase regulatory subunit 10 (PSD10), and the Adapter molecule CRK (CRK). Note that leptomycin B shifted none of them to a more nuclear localization. Analysis was as in Figure 4. (B) The following proteins were tested: the m7GpppX diphosphatase (DCPS), the Cyclin-dependent kinase inhibitor 1 (CDN1A), the mesoderm induction early response protein 1(MIER1), and the Acidic leucine-rich nuclear phosphoprotein 32 family member B (AN32B). Note that all these proteins showed an exclusively nuclear localization already without leptomycin B treatment, which is consistent with the assumption of negligible steady state export.
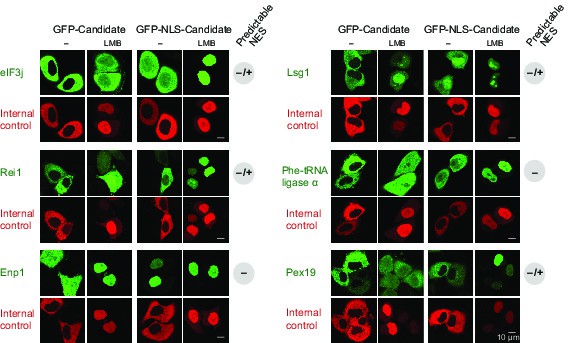
Validation of CRM1 cargo-candidates from S. cerevisiae.
Analysis was as in Figure 4 and included: the translation initiation factor eIF3j, the ribosome biogenesis factors LSG1, REI1, and ENP1, the α-subunit of the Phe-tRNA ligase (SYFA), as well the peroxisome biogenesis factor PEX19. This positive validation of yeast cargoes in HeLa cells also emphasises the extreme conservation of NES-recognition by CRM1.
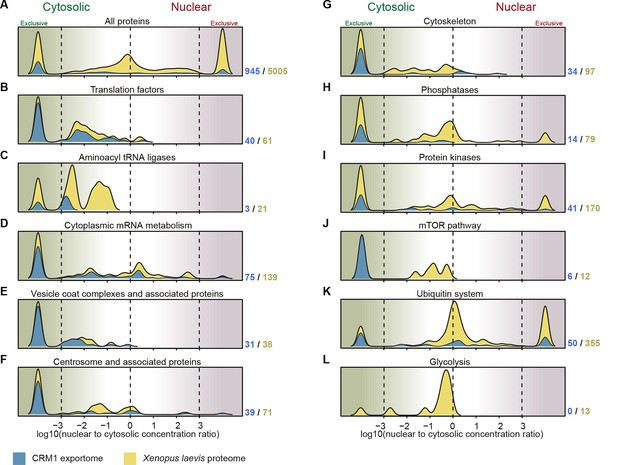
Correlation between functional groups, nucleocytoplasmic partitioning and CRM1-interaction.
(A) Panel shows a ‘density plot’ to illustrate how many unique proteins (see Figure 1D) show a given N:C partition coefficient in Xenopus oocytes. The yellow area covers all proteins, the blue area only proteins that belong to CRM1 cargo categories ‘A’-‘C’. (B–L) Density plots are analogous to (A), but each panel represents just one functional group and the curves were re-scaled to account for the smaller number of proteins in a given group. Functional groups were initially defined by KEGG BRITE hierarchies and then manually refined (see Supplementary file 2 for included proteins).
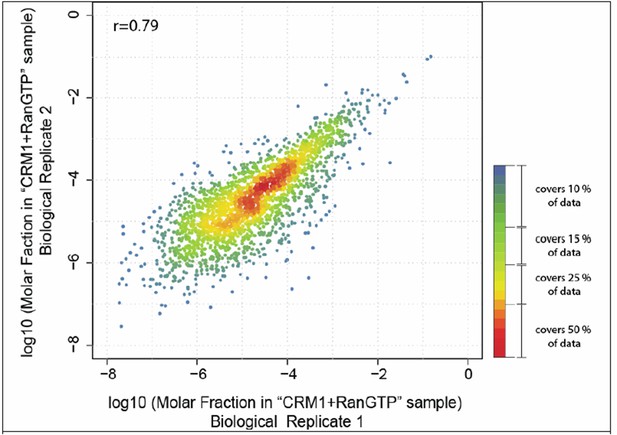
Scatter plot shows correlation of molar fractions of given protein species within the ‘CRM1+RanGTP’-bound fractions of the two biological replicates.
Note that many of the data points overlap at the diagonal. The local density of data points in the plot was therefore color-coded like a heat map, with red indicating the highest density and blue the lowest density. The red-orange region represents already 50% of the data points.
Additional files
-
Supplementary file 1
Nucleocytoplasmic protein partitioning in Xenopus oocytes.
- https://doi.org/10.7554/eLife.11466.014
-
Supplementary file 2
CRM1 binder analysis of Xenopus proteins.
- https://doi.org/10.7554/eLife.11466.015
-
Supplementary file 3
CRM1 binder analysis of HeLa proteins.
- https://doi.org/10.7554/eLife.11466.016
-
Supplementary file 4
CRM1 binder analysis of yeast proteins.
- https://doi.org/10.7554/eLife.11466.017
-
Supplementary file 5
Compares newly identified CRM1 cargoes from human HeLa cells with previous identifications listed in BioGRID, the NESdb database, and by Thakar et al. (2013).
- https://doi.org/10.7554/eLife.11466.018
-
Supplementary file 6
A fasta file with sequences of proteins that have been tested for a leptomycin B-sensitive localisation in HeLa cells (Figures 3, 5–8, and Figure 7—figure supplement 1)
- https://doi.org/10.7554/eLife.11466.019





