pp32 and APRIL are host cell-derived regulators of influenza virus RNA synthesis from cRNA
Figures
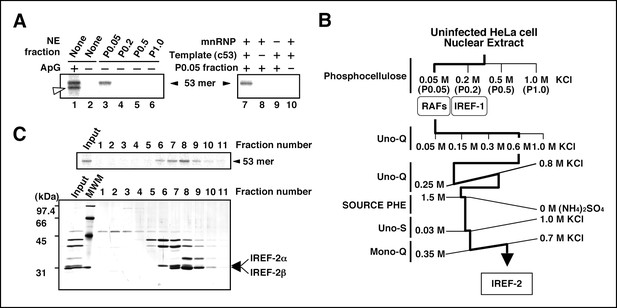
Purification of influenza virus replication factor-2 (IREF-2).
(A) IREF-2 activity in uninfected nuclear extracts (NEs). Biochemical complementation assays using a cell-free vRNA replication system for fractions separated by phosphocellulose column chromatography were performed. The fractions, P0.05 (lane 3), P0.2 (lane 4), P0.5 (lane 5), and P1.0 (lane 6), were individually assayed in the cell-free viral RNA synthesis reaction employing 5 ng PB1-equivalent micrococcal nuclease-treated vRNP (mnRNP) as an enzyme source and 10 ng of the complementary RNA (cRNA) model template (c53), as described in the 'Materials and methods'. Dinucleotide ApG, serving as primer for viral RNA synthesis, was added to a final concentration of 0.2 mM (lane 1). To confirm the components required for the reactions, cell-free viral RNA synthesis with mnRNP and c53 in the presence of the P0.05 fraction was carried out (lane 7; identical to the conditions of lane 3). Simultaneously, reactions omitting the cRNA model template (lane 8), mnRNP (lane 9), or P0.05 fraction (lane 10; identical to the conditions of lane 2) were also carried out. After incubation at 30°C for 2 hr, each reaction product was collected and subjected to 10% Urea-PAGE followed by autoradiography. (B) Purification scheme of IREF-2 from uninfected HeLa cell NEs. For details regarding the column chromatography, see 'Materials and methods'. (C) Profile of the fractions from Mono-Q column chromatography at the final purification step. Each Mono-Q fraction (fraction numbers 1–11) or input material for the Mono-Q column chromatography (i.e., unbound fraction of the Uno-S column chromatography) was individually added to this cell-free viral RNA synthesis reaction in the absence of any added primers (upper panel). Each Mono-Q fraction was subjected to 11.5% SDS-PAGE, and polypeptides were visualized by silver staining (lower panel). The closed arrowhead indicates 53 mer RNA products. The open arrowhead (50 mer) indicates the product possibly generated by internal priming of ApG. The arrows indicate two candidate peptides responsible for IREF-2 activity. The molecular weight (kDa) positions are denoted on the left side of the panel.
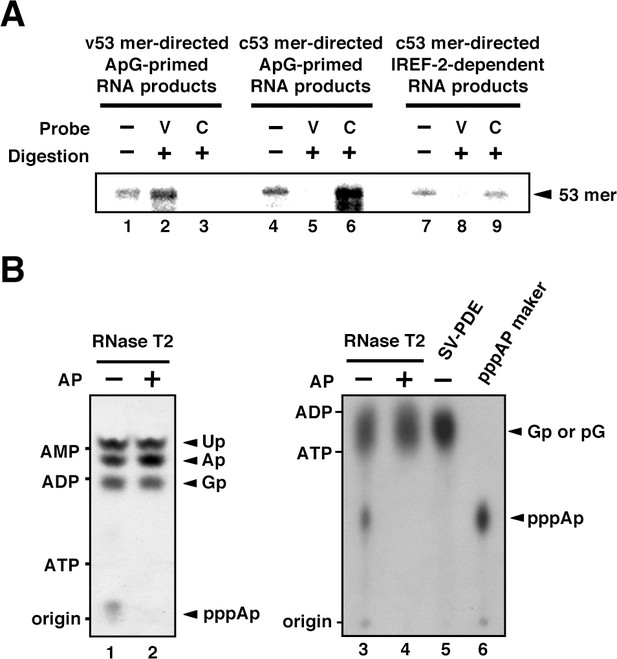
Products of influenza virus replication factor-2 (IREF-2)-dependent unprimed RNA synthesis.
(A) RNase T2 protection assay. Radioactively labeled vRNA products were synthesized in the cell-free viral RNA synthesis system with micrococcal nuclease-treated vRNP (mnRNP) and the v53 model template in the presence of ApG (lanes 1–3), the c53 model template in the presence of ApG (lanes 4–6), and c53 in the presence of the IREF-2 fraction and in the absence of ApG (lanes 7–9). Viral RNA products were hybridized with excess amounts of nonlabeled v53 (lanes 2, 5, and 8) or c53 (lanes 3, 6, and 9), which was followed by digestion with RNase T2. Hybridized and digested RNA samples were extracted, collected, and subjected to 10% Urea-PAGE followed by autoradiography visualization. The closed arrowhead indicates 53-nt-long RNAs. (B) Analysis of the 5'-terminal structure of IREF-2-dependent unprimed vRNA products. [α-32P] GTP-labeled IREF-2-dependent unprimed vRNA products were prepared in the cell-free viral RNA synthesis system. The radioactively labeled 53-nt-long vRNA products were isolated from 10% Urea-PAGE, which was followed by excision and elution from the gel. A portion of the isolated 53-nt-long products was treated with alkaline phosphatase (lanes 2 and 4). Both nontreated and alkaline phosphatase-treated [α-32P] GTP-labeled unprimed vRNA products were digested with RNase T2 (lanes 1–4) or snake venom phosphodiesterase (lane 5). The digested materials were spotted onto a polyethylenimine (PEI)-cellulose thin layer and developed with 1 N acetic acid-4 M LiCl (4:1, v/v) (left panel; lanes 1 and 2) or 1.6 M LiCl (right panel; lanes 3–6) and visualized by autoradiography. For mobility standards, nonradiolabeled AMP, ADP, and ATP were also subjected to thin-layer chromatography and are indicated on the left side of each panel. For a marker of pppAp, [γ-32P] ATP-labeled v53 synthesized using T7 RNA polymerase was also subjected to RNase T2 digestion and then to thin-layer chromatography (lane 6). The expected nucleotide positions are indicated on the right side of each panel by closed arrowheads.
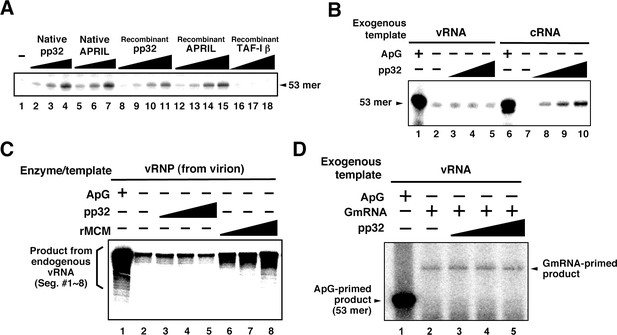
Characterization of influenza virus replication factor-2 (IREF-2) activities in the cell-free system.
(A) Dose response of IREF-2. Native or recombinant IREF-2 proteins were added to the cell-free viral RNA (vRNA) synthesis reaction using micrococcal nuclease-treated vRNP (mnRNP) and the c53 model template in the absence of primer. Native pp32 (lanes 2–4), native APRIL (lanes 5–7), recombinant pp32 (lanes 8–11), and recombinant APRIL (lanes 12–15) were used as follows: 5 ng (lanes 2, 5, 8, and 12), 15 ng (lane 3, 6, 9, and 13), 50 ng (lanes 4, 7, 10, and 14) and 150 ng equivalent (lanes 11 and 15) of IREF-2 proteins. Recombinant TAF-Iβ/SET protein prepared using an Escherichia. coli expression system (33, 110, and 330 ng) was also tested (lanes 16–18). After incubation at 30°C for 2 hr, the RNA products were collected and analyzed by 10% Urea-PAGE followed by autoradiography. (B) Template preference of IREF-2-dependent viral RNA synthesis. Viral RNA replication reactions were performed in the cell-free viral RNA synthesis system using mnRNP and either v53 (lanes 1–5) or c53 (lanes 6–10) as viral model templates for vRNA and complementary RNA (cRNA), respectively. ApG at a final concentration of 0.2 mM (lanes 1 and 6) or 30, 100, and 300 ng of recombinant pp32 (lanes 3–5 and 8–10) was added to the reaction. (C) Effect of IREF-2 on cRNA synthesis from vRNP. Cell-free viral RNA synthesis using 2 ng PB1-equivalent of vRNP as the enzyme source and an endogenous genomic vRNA template were carried out in the presence (lane 1) or absence (lanes 2–8) of 0.2 mM ApG. Recombinant pp32 (lanes 3–5, 30, 100, and 300 ng, respectively) was added to the reactions. As a positive control, 1.5, 5, and 15 ng of recombinant IREF-1/MCM were also used (lanes 6–8). The RNA products were collected and analyzed by 4% Urea-PAGE followed by autoradiography. One-third (33%) of the total products derived from the ApG-primed cRNA synthesis were subjected to Urea-PAGE (lane 1). (D) Effect of IREF-2 on cap-snatching viral transcription. Cell-free viral RNA synthesis reactions were performed using mnRNP as the enzyme source and the exogenous model vRNA template (v53) in the presence of 0.2 mM ApG (lane 1) or globin mRNA as the 5’-capped RNA donor (lanes 2–5). Recombinant pp32 (lanes 3–5, 30, 100, and 300 ng, respectively) was added to the reaction.
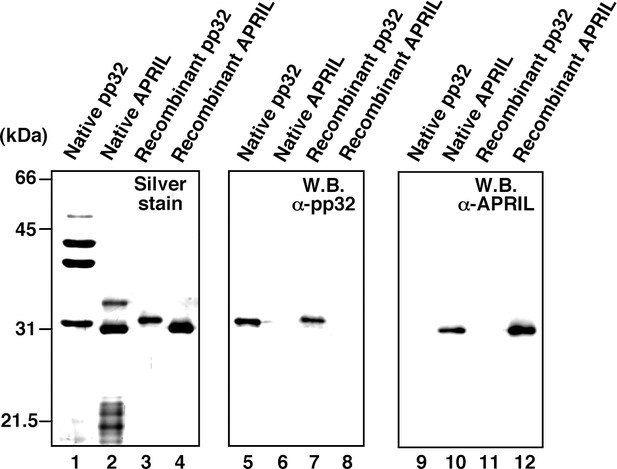
Protein profiles of native and recombinant influenza virus replication factor-2 (IREF-2s).
Native or recombinant IREF-2 proteins (50 ng) were subjected to 11.5% SDS-PAGE followed by silver staining (lanes 1–4) and western blot analysis with anti-pp32 antibody (lanes 5–8) or anti-APRIL antibody (lanes 9–12). The Mono-Q fraction 6 (shown in Figure 1C) was used as native pp32 (lanes 1, 5, and 9). Mono-Q fractions 9 and 10 (also shown in Figure 1C) were further purified with Mono-Q and used as native APRIL (lanes 2, 6, and 10). Recombinant pp32 (lanes 3, 7, and 11) and APRIL (lanes 4, 8, and 12) were prepared using the Escherichia. coli expression system, as described in 'Materials and methods'.
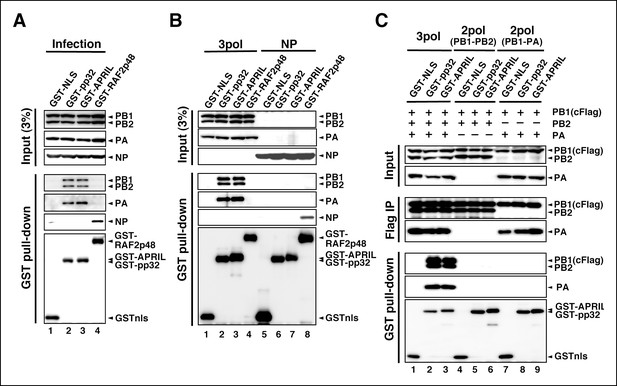
Interaction of influenza virus replication factor-2 (IREF-2) proteins with free forms of viral polymerase trimeric complexes.
(A) Interaction between IREF-2 and viral proteins in infected cells. HEK293T cells were transfected with 10 μg of plasmids expressing GSTnls (lane 1), GST-pp32 (lane 2), GST-APRIL (lane 3), and GST-RAF2p48 (lane 4). At 48 hr post transfection, influenza virus was infected at an multiplicity of infection (MOI) of 3. At 6 hr post infection, the transfected and infected cells were collected, lysed, and subjected to GST pull-down assays, as described in 'Materials and methods'. The pulled-down materials and 3% equivalent of the input samples were subjected to SDS-PAGE followed by western blot analysis with anti-PB1, -PB2, -PA, -NP, and -GST (only the pull-down sample) antibodies. (B) Interaction between IREF-2 and viral proteins in the transfected cells. HEK293T cells were transfected with 5 μg of plasmids expressing GSTnls (lanes 1 and 5), GST-pp32 (lanes 2 and 6), GST-APRIL (lanes 3 and 7), and GST-RAF2p48 (lanes 4 and 8) and also co-transfected with the plasmids for the viral polymerase subunits (5 μg of pCAGGS-PB1, 12.5 μg of pCAGGS-PB2, and 2.5 μg of pCAGGS-PA [lanes 1–4] or 10 μg of pCAGGS-NP for NP expression [lanes 5–8]). Forty-eight hours post transfection, the cotransfected cells were collected, lysed, and subjected to GST pull-down assays. The input samples (3%) and pulled-down materials were subjected to SDS-PAGE followed by western blot analysis. (C) Interaction between IREF-2 and trimeric or binary complexes of vRdRP. HEK293T cells were cotransfected with 5 μg of plasmids expressing GSTnls (lanes 1, 4, and 7), GST-pp32 (lanes 2, 5, and 6), GST-APRIL (lanes 3, 6, and 9), 5 μg of pCAGGS-PB1cFlag (lanes 1–9), 12.5 μg of pCAGGS-PB2 (lanes 1–6), and 2.5 μg of pCAGGS-PA (lanes 1–3 and 7–9). Forty-eight hours post transfection, the lysates from the cotransfected cells were subjected to GST pull-down assays or immunoprecipitation assays with anti-Flag antibody. The precipitated materials were subjected to SDS-PAGE followed by western blot analysis.
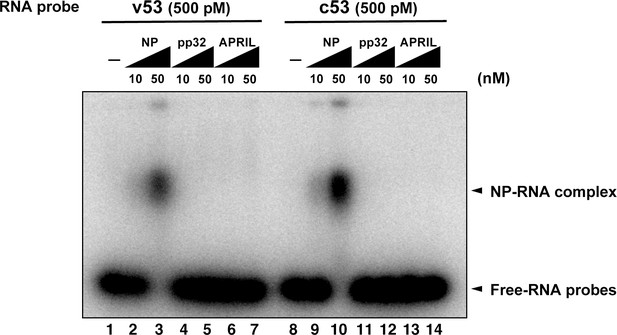
Electrophoresis mobility shift assay for influenza virus replication factor-2 (IREF-2) and viral RNA.
Radioactively labeled 53-nt-long model vRNA and complementary RNA(cRNA) probes (v53 and c53; 246.9 cpm/fmol) were synthesized by T7 RNA polymerase using [α-32P] GTP and isolated by gel excision. Each 500 pM (final concentration) of the labeled viral RNA probes, v53 (lanes 1–7) and c53 (lanes 8–14) was incubated with 10 nM or 50 nM of recombinant NP prepared using the Escherichia. coli expression system (lanes 2, 3, 9, and 10), recombinant pp32 (lanes 4, 5, 11, and 12), and recombinant APRIL (lanes 6, 7, 13, and 14) in 50 mM HEPES-NaOH (pH 7.9), 50 mM KCl, 0.5 U/μl of RNase inhibitor, and 15% (v/v) glycerol at 30°C for 30 min. After incubation, each binding mixture was loaded onto 0.6% agarose gel (buffered with TBE) and separated by electrophoresis (50 V for 3 hr). The gel was dried and visualized by autoradiography.
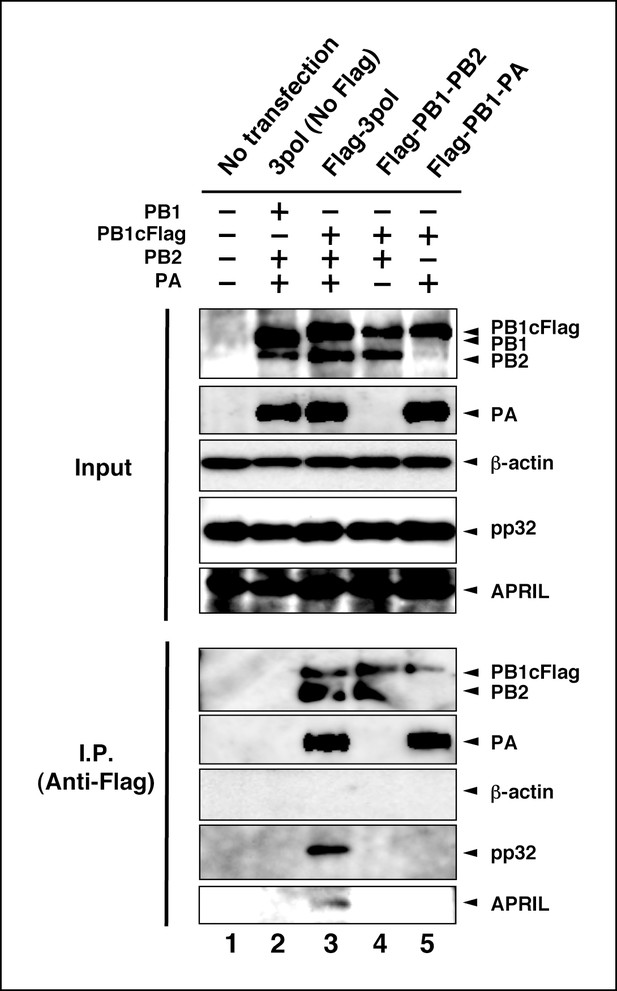
Interaction between viral RNA-dependent RNA polymerase (vRdRP) complexes and endogenous influenza virus replication factor-2 (IREF-2).
HEK293T cells were transfected with plasmids expressing PB1 (lane 2) or PB1cFlag (lanes 3–5), PB2 (lanes 2–4), and PA (lanes 2, 3, and 5), as indicated. At 48 hr post transfection, the cells were harvested, lysed, and subjected to immunoprecipitation assays using anti-Flag antibody, as described in 'Materials and methods'. Input and precipitated materials were analyzed by SDS-PAGE, followed by western blot analysis using the antibodies indicated on the right side of each panel.
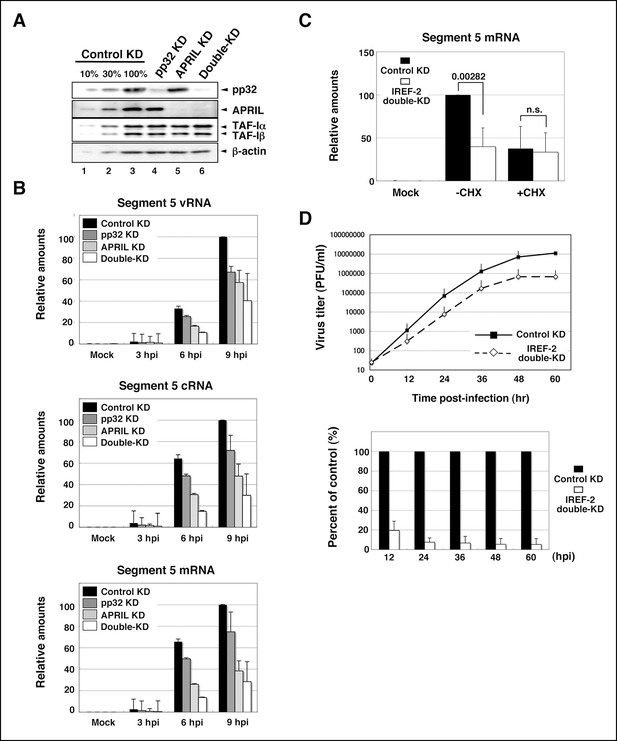
Effect of influenza virus replication factor-2 (IREF-2) knockdown in infected cells.
(A) Endogenous IREF-2 protein levels in knockdown (KD) HeLa cells. The whole-cell lysates of control KD cells (lane 3), pp32 KD cells (lane 4), APRIL KD cells (lane 5), and both pp32 and APRIL KD cells (termed double-KD cells; lane 6) were subjected to SDS-PAGE followed by western blot analysis with antibodies, as indicated on the right side of the panel. To measure the KD levels, 10% and 30% of the lysates of the control KD cells were also subjected to SDS-PAGE followed by western blot analysis with antibodies, respectively (lanes 1 and 2). (B) Viral RNA levels in infected KD HeLa cells. Total RNA was extracted from mock-infected or infected KD cells at an multiplicity of infection (MOI) of 1 at 3, 6, and 9 hr post infection. vRNA, complementary RNA(cRNA), and viral mRNA derived from segment 5 were quantitatively determined by RT-mediated qPCR (RT-qPCR), as described in 'Materials and methods'. (C) Primary transcription level in infected KD HeLa cells. Total RNA was extracted from mock-infected or infected KD cells at an MOI of 10 for 3 hr in the presence or absence of cycloheximide (CHX). Viral mRNA derived from segment 5 was quantitatively determined by RT-qPCR. (D) Effect of IREF-2 on progeny virus production. Control KD and IREF-2 double-KD A549 cells were infected with FluV (WSN/33 strain) at an MOI of 0.001. The number of infectious progeny viruses produced from control KD and IREF-2 double-KD A549 cells at each time point were plotted (upper panel), and the ratios of progeny viruses produced from IREF-2 double-KD A549 cells compared with those from control KD cells were also represented as a percentage of the control (lower panel). Each quantitative result is presented as the average with the standard deviation from at least three independent experiments. Significance was determined using Student’s t test. n.s.: not significant.
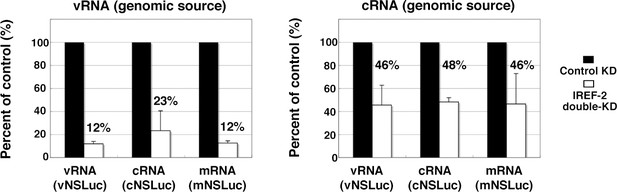
Effect of influenza virus replication factor-2 (IREF-2) on viral reporter RNA syntheses from a reconstituted model replicon.
HEK293T of control and IREF-2 knockdown (KD) cells (approximately, 3 × 105 cells in a 22-mm-diameter dish) were transfected with expression plasmids pCAGGS-PB1, -PB2, -PA, and -NP (15 ng each). In addition, 1.5 ng of either phPolI-vNS-Luc (for expression of vNS-Luc; left panel) or phPolI-cNS-Luc (for expression of cNS-Luc; right panel) was co-transfected as a viral genomic source. At 24 hr post co-transfection, viral reporter levels in both the control KD and the IREF-2 KD cells were quantitatively determined by RT-qPCR as described previously (Kawaguchi et al., 2011) and in 'Materials and methods'. Quantification and standard deviations of the viral reporter RNA levels from IREF-2 KD cells expressed as a percentage of the value from the control KD cells. Each experiment was repeated three times.


Additional files
-
Supplementary file 1
Molecular masses of trypsin-digested peptides from IREF-2 proteins and amino acid sequences of IREF-2 proteins corresponding to the tryptic cleavage molecular mass database.
- https://doi.org/10.7554/eLife.08939.012





