The deca-GX3 proteins Yae1-Lto1 function as adaptors recruiting the ABC protein Rli1 for iron-sulfur cluster insertion
Figures
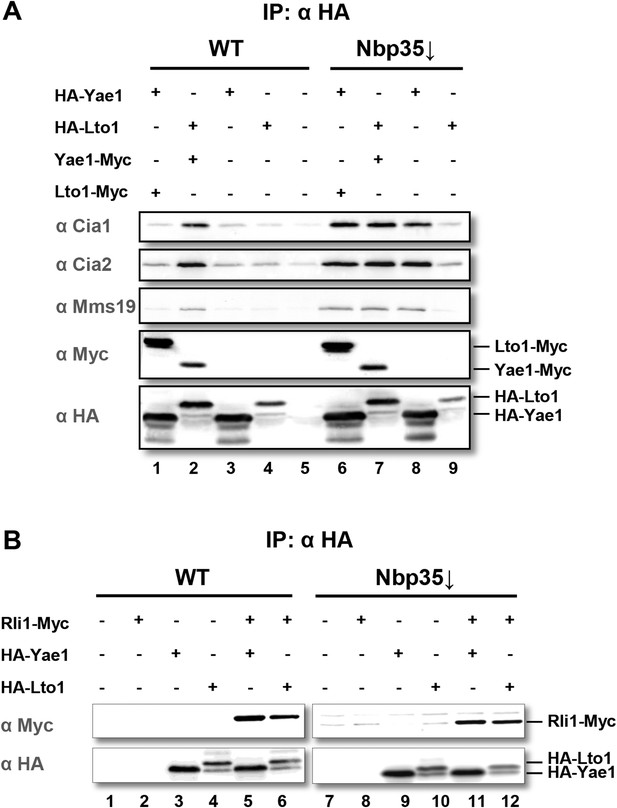
Yae1 and Lto1 interact with the cytosolic Fe-S protein assembly (CIA) targeting complex and the Fe-S protein Rli1.
(A) Wild-type (WT) and Gal-NBP35 yeast cells were co-transformed with 2µ vectors containing either no insert or genes encoding HA-Yae1, HA-Lto1, Yae1-Myc and Lto1-Myc as indicated. Cells were cultivated in minimal medium containing glucose leading to Nbp35 depletion (↓) in Gal-NBP35 cells. Cell lysates were prepared, and a HA-tag immunoprecipitation (IP) was performed. The immunoprecipitate was analyzed for the indicated proteins or tags by immunoblotting. (B) WT and Gal-NBP35 yeast cells were co-transformed with 2µ vectors containing either no insert or genes encoding HA-Yae1, HA-Lto1 and Rli1-Myc and treated as in part A. The lower band in HA-Lto1-containing lanes is a HA-Lto1 degradation product.
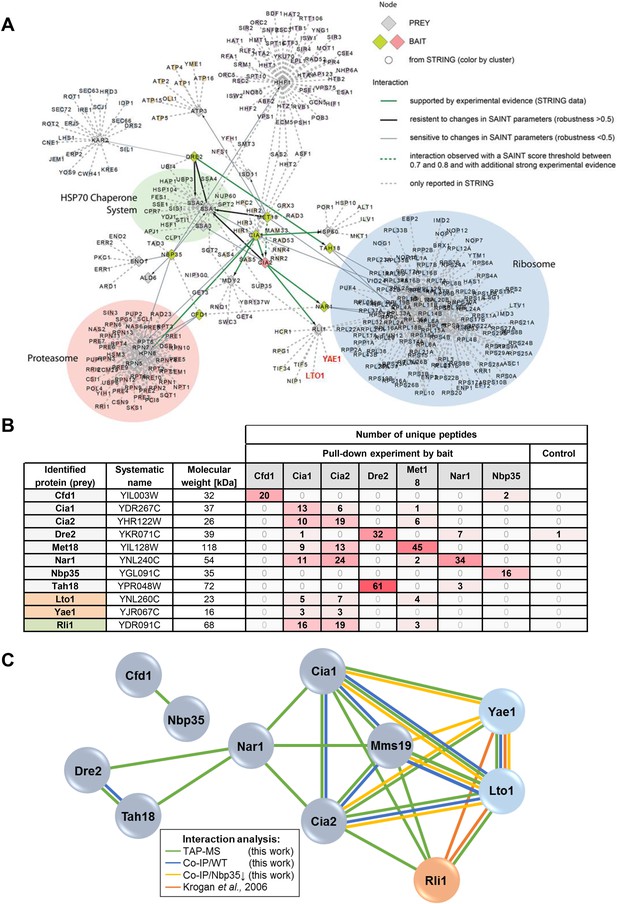
Yae1 and Lto1 interact with the CIA targeting complex and with the Fe-S protein Rli1.
(A) Interactions between core components of the CIA machinery (i.e., Tah18, Dre2, Cfd1, Nbp35, Nar1, Cia1, Cia2 and Mms19) identified by TAP-MS (tandem affinity purification (TAP) coupled to protein identification by mass spectrometry) with additional STRING evidence. Baits used for affinity purifications are represented as either red (CIA2) or olive diamonds, and preys are shown as grey diamonds. The interaction network around the core CIA components is extended by nodes imported from the STRING database with reported connections to baits and preys identified in our screen with a STRING confidence score >0.9. Edges between all nodes and their attributes correspond to bait-prey interactions of different confidence levels, either by probabilistic scoring (SAINT) of TAP-MS data and/or from experimental data (STRING). Thick green lines depict high confidence interactions observed by TAP-MS, which, in addition, are supported by experimental evidence in STRING. Thick black lines indicate robust connections, resistant to changes in parameters specified in the SAINT algorithm (SAINT score >0.8, and robustness >0.5). Thin grey lines represent connections of lower confidence level, which are sensitive to changes in parameters for the SAINT algorithm (>0.8 SAINT, robustness of <0.5). Dashed green lines show interactions observed with a SAINT score threshold between 0.7 and 0.8, and with additional experimental evidence. Dashed grey lines are interactions only reported in STRING with a STRING confidence score >0.9. We summarized nodes in a semi-automatic manner, and performed a GO enrichment analysis on the resulting clusters. Clusters were defined by STRING nodes that only had one neighbor which had to be either a prey or bait from our screen. Three additional clusters were defined manually based on common neighborhood. (B) Detection of selected CIA protein interaction partners by identification of unique peptide numbers. TAP was performed with CIA-TAP fusion proteins as baits and associated partners identified via mass spectrometry. The amount of unique peptides referring to selected interaction partners of each CIA protein was determined (n = 4). As a control, we used the strain SC0000, which does not express any TAP-fusion protein. (C) Interactions between the CIA proteins and some prominent potential partners were verified by dedicated co-IP experiments (Co-IP). Verification procedures are represented by different colors. Green: Detection via TAP-MS (based on identification of unique peptides, see also part B). Blue: Co-IP using C-terminally HA-tagged proteins as baits in WT background. Yellow: Co-IP using HA-tagged proteins as baits in a strain depleted for the early-acting CIA component Nbp35. Orange: Interactions reported by a previous systematic analysis (Krogan et al., 2006).
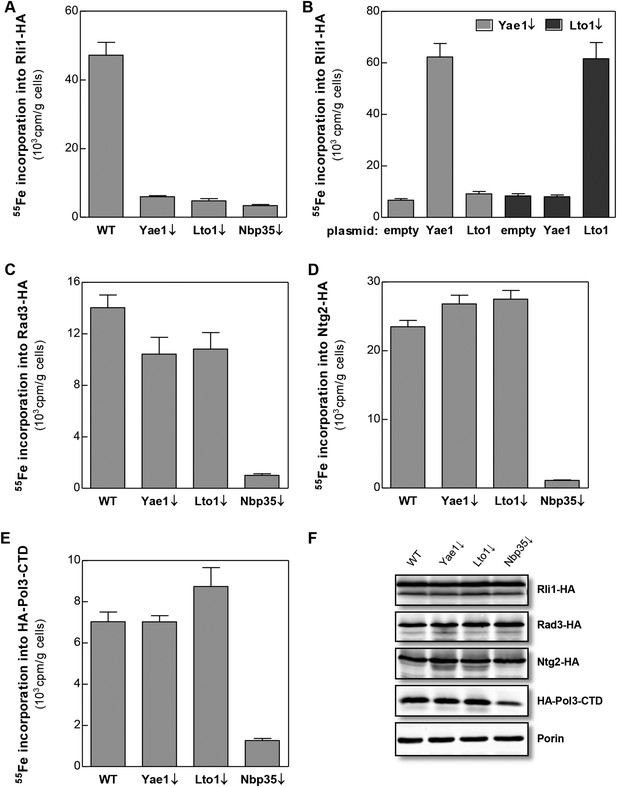
Yae1 and Lto1 specifically mediate Fe-S cluster association with Rli1.
WT, Gal-YAE1, Gal-LTO1 and Gal-NBP35 yeast cells were transformed with plasmids encoding Rli1-HA (A, B), Rad3-HA (C), Ntg2-HA (D) and the C-terminal domain of Pol3 N-terminally fused to HA (E). To increase the amount of Pol3-CTD-bound Fe-S clusters the accessory subunit Pol31 was co-expressed (Netz et al., 2012b). In part B Gal-YAE1 and Gal-LTO1 cells contained 2µ plasmid-borne YAE1 or LTO1 or the empty vector. Cells were cultivated for 24 hr in glucose-containing minimal medium for depletion of the respective proteins (↓). After an additional 16 hr in minimal medium lacking iron cells were radiolabeled for 2 hr with 55FeCl3, cell extracts were prepared, and the Fe-S target proteins were immunoprecipitated with anti-HA antibodies. The amount of 55Fe associated with target proteins was quantified by scintillation counting. Error bars indicate the SEM (n > 4). (F) A representative immunostain of the indicated proteins from parts A, C–E.
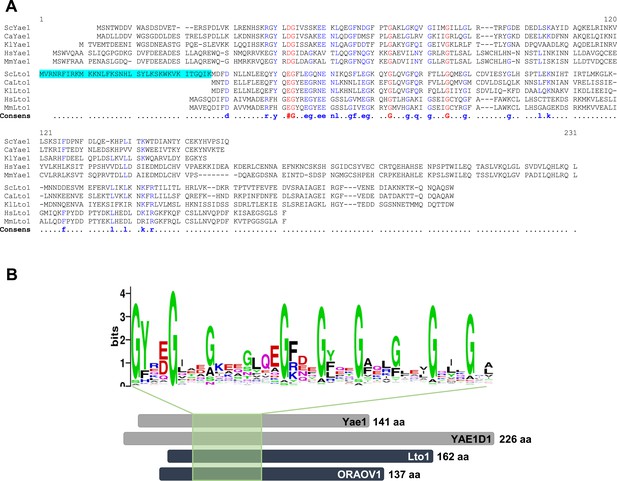
Yae1 and Lto1 contain a deca-GX3 sequence motif that is conserved in eukaryotes.
(A) The multi-sequence alignment of Yae1 and Lto1 from various eukaryotic species (Sc, Saccharomyces cerevisiae; Ca, Candida glabrata; Kl, Kluyveromyces lactis; Hs, Homo sapiens; Mm, Mus musculus) was calculated by Multalin (Corpet, 1988). The consensus sequence is shown below the alignment. The ScLto1 sequence highlighted in cyan corresponds to the erroneously annotated N-terminus of this protein (see also Figure 2—figure supplements 2, 3). (B) A multi-sequence alignment of the protein family pfam09811 was conducted and a weblogo generated to highlight the conserved deca-GX3 domain (weblogo.berkeley.edu). The bottom part shows a cartoon of Yae1 and Lto1 from S. cerevisiae and man (YAE1D1 and ORAOV1) to highlight the relative positions of the deca-GX3 motif. The number of amino acids (aa) of each protein is depicted at the C-terminus.
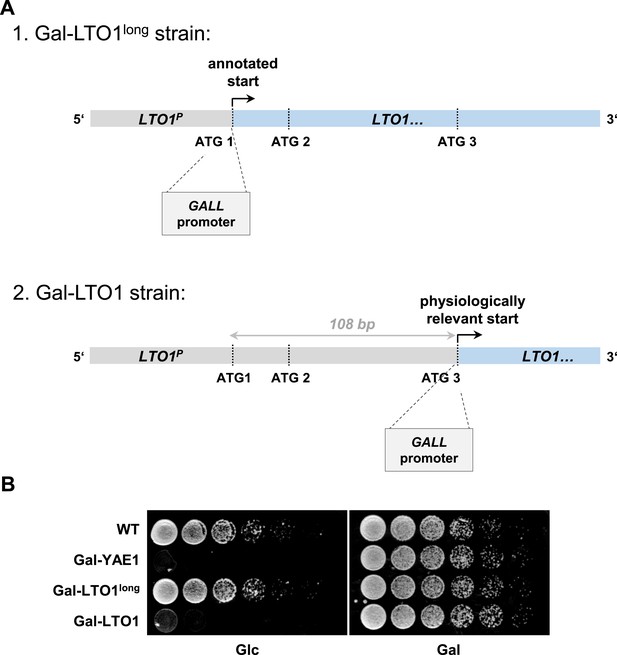
Generation of a regulatable strain for depletion of Lto1 questions the previously predicted physiological translation start site of LTO1.
During the generation of the galactose-regulatable strain of LTO1 we noted that the previously predicted translation start site may be incorrect. (A) Cartoon of the 5′ region of the LTO1 gene. For functional analyses of Lto1 and Yae1 the GALL cassette was inserted directly in front of the start codons of YAE1 and LTO1. In the latter case the GALL promoter was inserted at different sites. First, at the original translation start codon (ATG1) of Saccharomyces Genome Database (strain Gal-LTO1long), or second, at the corrected translation start site (ATG3) located 108 bp downstream of the annotated start as determined in this work (strain Gal-LTO1). LTO1P: promoter of LTO1. (B) Cells from part A were grown over night in glucose-containing rich medium. A serial dilution (1:5) was spotted onto rich medium agar plates supplemented with glucose (Glc) or galactose (Gal).
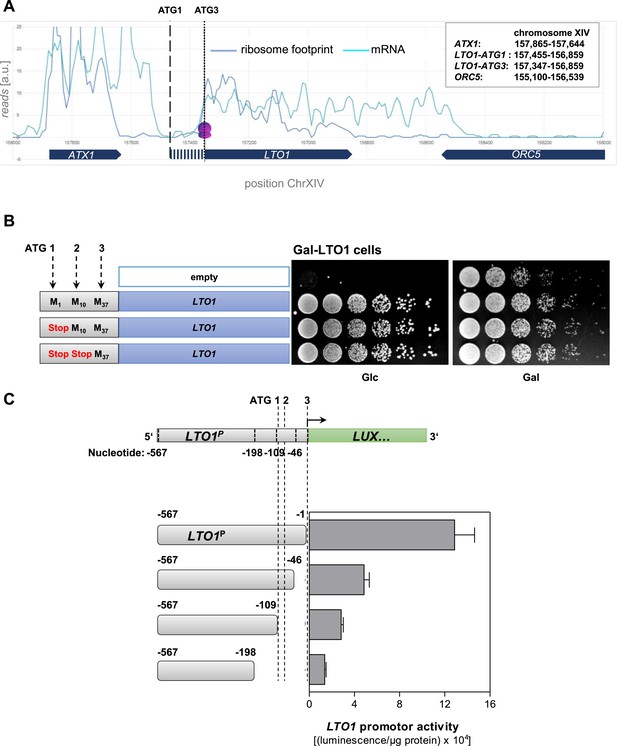
Determination of the correct physiological translation start site of LTO1.
(A) Ribosome footprinting analysis of LTO1. The physiological start site of LTO1 was determined by bioinformatic analysis using the published data from ribosome footprinting (Ingolia et al., 2009). Ribosome footprinting and mRNA data were obtained from GEO (Gene Expression Omnibus, #GSE13750) and imported into Microsoft Access. Reads corresponding to chromosome XIV and coordinates corresponding to the LTO1-coding region were selected and transferred to Excel. The sequence reads were binned in a 30 bp window. The vertical dashed line represents the translation start site as predicted by SGD (ATG1), and the vertical dotted line is the start site defined in this work (ATG3). ATX1 and ORC5 are neighboring reference genes. (B) The region in front of codon ATG3 of LTO1 is not essential for cell growth. Gal-LTO1 cells were transformed with an empty centromeric plasmid or plasmids containing various LTO1 genes under control of the natural promoter. As indicated on the left part the ATG codons 1 and 2 of LTO1 were mutated to stop codons and analyzed for their relevance to support cell growth. Cells were cultivated 16 hr in glucose-containing minimal medium to deplete endogenous Lto1 and spotted onto plates supplemented with glucose (Glc) or galactose (Gal) (serial 1:5 dilution, start OD600 = 0.5). (C) The region in front of codon ATG3 of LTO1 is critical for gene expression. LTO1 promoter (LTO1P) fragments were fused to the luciferase (LUX) reporter gene. WT cells harboring the luciferase-based reporter constructs were cultivated in glucose-containing minimal medium. Promoter activities were measured in exponentially growing cells and normalized to protein amounts. Error bars indicate the SEM (n > 4).
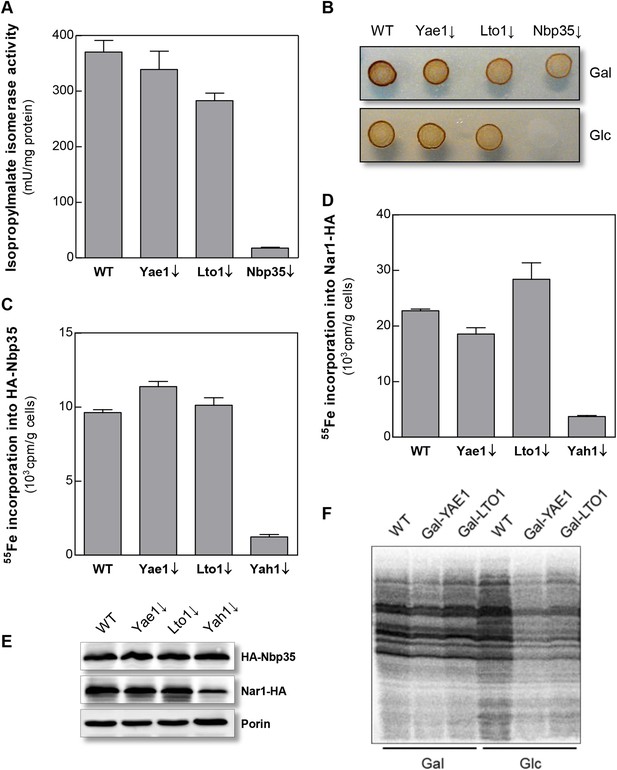
Yae1 and Lto1 do not perform a general role in Fe-S protein maturation.
(A) WT, Gal-YAE1, Gal-LTO1 and Gal-NBP35 cells were cultivated on glucose-containing minimal medium for 40 hr. Cell extracts were analyzed for the activities of isopropylmalate isomerase (Leu1). (B) Cells were analyzed for sulfite reductase activity in vivo after growth for 3 days on minimal medium agar plates supplemented with ammonium bismuth citrate and sodium sulfite. Sulfide produced by sulfite reductase yields the brown precipitate Bi2S3. (C, D) WT, Gal-YAE1, Gal-LTO1 and Gal-YAH1 cells were transformed with plasmids containing genes encoding HA-Nbp35 (C) and Nar1-HA (D). Cells were radiolabeled with 55FeCl3, and 55Fe-S protein formation was estimated as in Figure 2. (E) A representative immunostain of the indicated proteins from parts C, D. (F) WT, Gal-YAE1 and Gal-LTO1 cells were cultivated in rich medium containing galactose (Gal) or glucose (Glc) for 40 hr, and radiolabelled with 35S-methionine for 10 min at 30°C. Extracts were prepared, and the protein synthesis efficiency was analyzed by SDS-PAGE and autoradiography. Error bars indicate the SEM (n > 4).
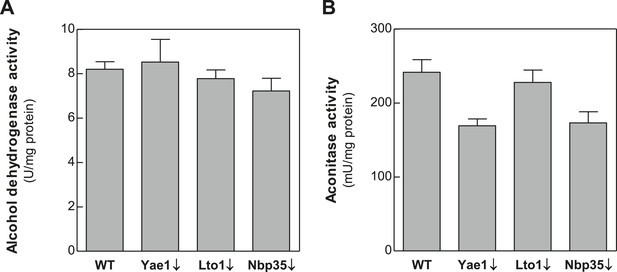
Depletion of Yae1, Lto1 and Nbp35 does not affect enzyme activities of the cytosolic alcohol dehydrogenase and the mitochondrial Fe-S enzyme aconitase.
WT, Gal-YAE1, Gal-LTO1 and Gal-NBP35 cells were cultivated in glucose-containing minimal medium to deplete the respective proteins (↓). Cell extracts were prepared using glass beads, and the activities of alcohol dehydrogenase (A) and the mitochondrial aconitase (B) were measured. Activities were normalized to protein amounts. Error bars indicate the SEM (n > 4).
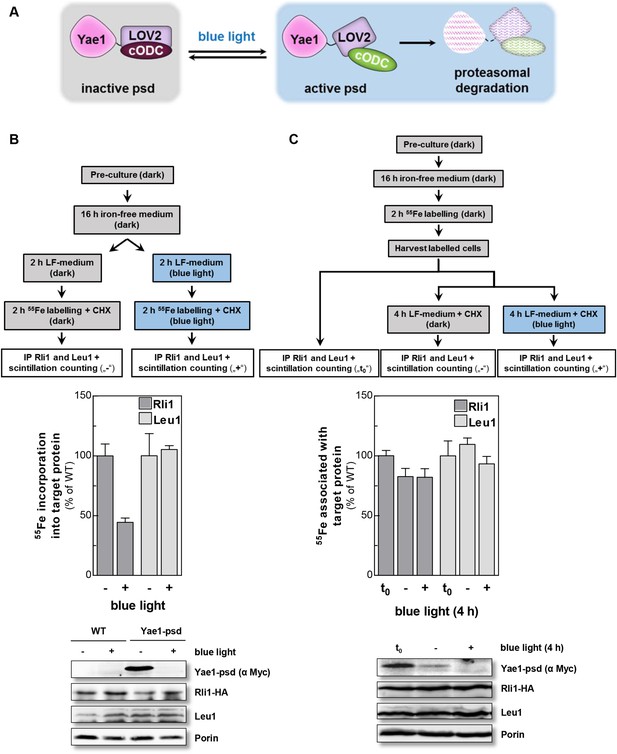
Yae1 is a Fe-S cluster maturation rather than stabilization factor for Rli1.
(A) Schematic representation of the light-induced rapid degradation of the Yae1-3Myc-AtLOV2-cODC1 fusion protein (psd; photosensitive degron). In the dark the cODC1 degron is inactive and thus the fusion protein is stable. Irradiation with blue light induces a structural rearrangement within the LOV2 domain leading to the activation of the degron and direct degradation of the fusion protein by the 26S proteasome (adapted from [Renicke et al., 2013]). (B) WT and Yae1-psd (photosensitive degron) cells were transformed with a 2µ plasmid encoding RLI1-HA. Cells were grown overnight in iron-free minimal medium in the dark. Half of the cells (0.5 g) were exposed to blue light in low fluorescence medium (LFM), and the other half was kept in the dark for 2 hr. Cycloheximide (200 µg/ml) was added, and cells were radiolabeled with 55Fe with or without blue light irradiation. The amount of 55Fe associated with Rli1 or Leu1 was quantified by IP and scintillation counting (cf. Figure 2). The radioactivity associated with Fe-S proteins in Yae1-psd mutant cells is presented relative to that of WT cells. Error bars indicate the SEM (n > 3). The bottom part shows a representative immunostain of the indicated proteins in cell extracts. (C) Three independent cultures of WT and Yae1-psd cells were grown overnight in iron-free minimal medium in the dark and radiolabeled with 55Fe for 2 hr. Culture 1 was assessed immediately for 55Fe-S cluster association to Rli1-HA or Leu1 by IP and scintillation counting (t0). Cultures 2 and 3 were washed with H2O and supplemented in LFM with cycloheximide. Cells were kept in the dark (−) or exposed to blue light (+) for additional 4 hr, and then analyzed for 55Fe-S cluster association. The bottom part shows a representative immunostain of the indicated proteins in extracts of Yae1-psd cells.
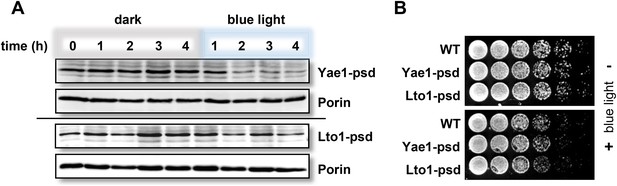
Fusion of Yae1 and Lto1 with a photosensitive degron allows their efficient degradation upon irradiation with blue light.
(A) Blue light-induced degradation of psd-fusion proteins. Yeast cells expressing Yae1-psd or Lto1-psd were grown in liquid LFM supplemented with glucose in the dark. After removal of an aliquot (t = 0 hr), cells were split and one half was exposed to blue light (LED lamp 465 nm, 30 µmol × m−2 × s−1), whereas the other half was kept in the dark. At indicated time points cells were lysed. The degradation of the fusion proteins was followed by immunostaining using α-Myc antibodies (porin served as a loading control). (B) WT, Yae1-psd and Lto1-psd cells were serially diluted (1:5, start OD600 = 0.5) and spotted onto YP agar plates supplemented with glucose. Plates were incubated in the dark or in blue light overnight.
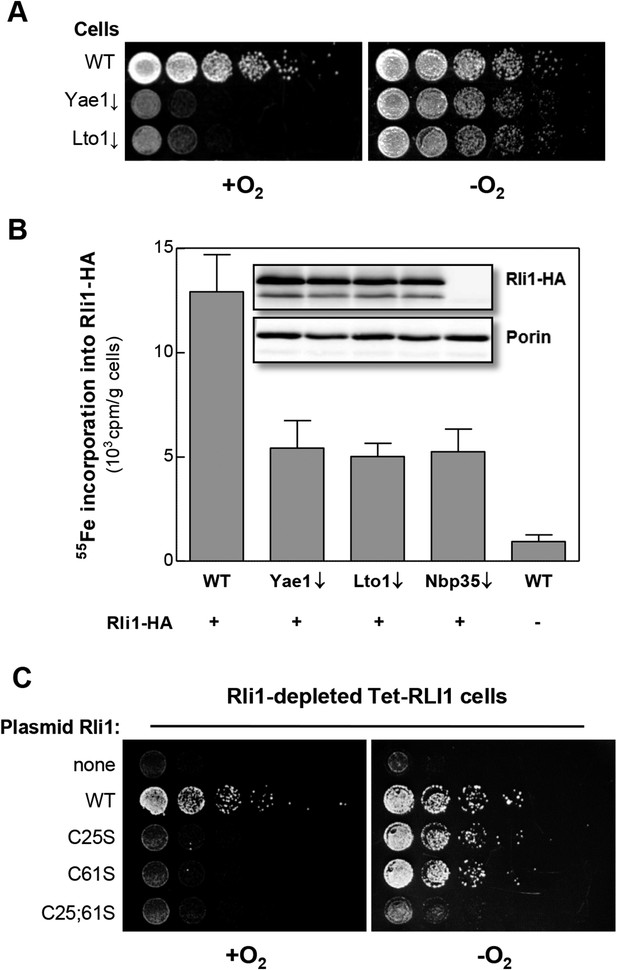
Yae1 and Lto1 are required for Fe-S cluster assembly on Rli1 also under anaerobic conditions.
(A) The indicated cells were cultivated overnight in rich medium containing glucose. Serial dilutions (1:5) were spotted onto glucose-containing rich medium agar plates. Growth was in the presence or absence of O2. (B) WT, Gal-YAE1, Gal-LTO1 and Gal-NBP35 cells were transformed with plasmids encoding RLI1-HA (+) or no gene (−). Cells were cultivated for 30 hr in glucose-containing minimal medium under anaerobic conditions and transferred to minimal medium lacking iron for additional 16 hr. After radiolabeling for 2 hr with 55Fe under anaerobic conditions, cell extracts were prepared anaerobically and analyzed for 55Fe incorporation into Rli1-HA (cf. Figure 2). Error bars indicate the SEM (n > 3). The inset shows a representative immunostain of Rli1-HA and porin. (C) Tet-RLI1 cells were transformed with an empty vector, a plasmid containing RLI1 under its native promoter (300 bp upstream of RLI1) or plasmids containing RLI1 coding for cysteine to serine mutants of Rli1 as indicated. Cells were cultivated in glucose-containing minimal medium for 16 hr. Serial dilutions (1:5) were spotted under anaerobic conditions onto glucose-containing minimal medium agar plates supplemented with 5 µg/ml doxycycline to deplete endogenous Rli1. Further growth was in the presence or absence of O2.
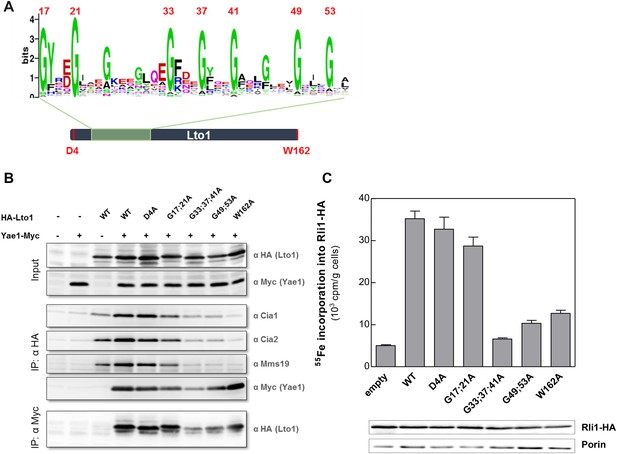
The conserved deca-GX3 motif and the C-terminal tryptophan are functionally crucial elements of Lto1.
(A) Cartoon of S. cerevisiae Lto1 to highlight the mutated residues (red) within the deca-GX3 motif and the N- and C-termini (cf. Figure 2—figure supplement 1B). (B) Gal-NBP35 yeast cells were co-transformed with 2µ vectors containing either no insert or genes encoding HA-Lto1, Yae1-Myc or HA-tagged Lto1 mutants as indicated. Cells were cultivated in minimal medium containing glucose. Cell lysates were prepared, and a trichloroacetic acid (TCA) precipitation was performed to assess the expression levels of the fusion proteins (Input). After HA- or Myc-tag IP the precipitate was analyzed for the indicated tags or proteins by immunoblotting. (C) Cells were co-transformed with plasmids containing RLI1-HA and single copy plasmids containing LTO1 or mutated versions as indicated. Cells were cultivated for 24 hr in glucose-containing minimal medium and the 55Fe radiolabeling-IP procedure was performed to estimate the amount of 55Fe associated with Rli1-HA. Error bars indicate the SEM (n > 4). The inset shows a representative immunostain of Rli1-HA and porin.
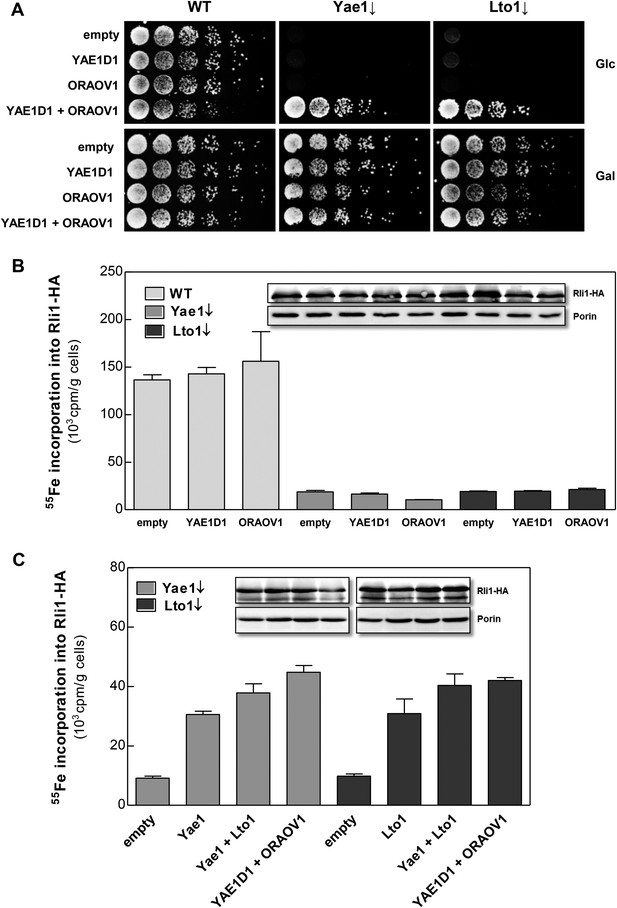
The human YAE1D1-ORAOV1 complex can functionally replace Yae1-Lto1.
(A) WT, Gal-YAE1 and Gal-LTO1 cells were transformed with 2µ vectors containing either no insert (empty) or genes encoding human YAE1D1 and/or ORAOV1 as indicated. Cells were cultivated overnight in minimal medium containing glucose to deplete Yae1 and Lto1 (↓). Serial dilutions (1:5) were spotted onto minimal medium agar plates supplemented with glucose (Glc) or galactose (Gal). (B) Cells expressing RLI1-HA plus YAE1D1 or ORAOV1 were radiolabeled with 55Fe, and the amount of 55Fe associated with Rli1-HA was quantified by scintillation counting. Error bars indicate the SEM (n > 4). The inset shows a representative immunostain of Rli1-HA and porin. (C) In a similar analysis as in part B the effects of the expression of YAE1, LTO1, YAE1D1 or ORAOV1 for Rli1-HA maturation in Yae1- and Lto1-depleted cells were estimated.
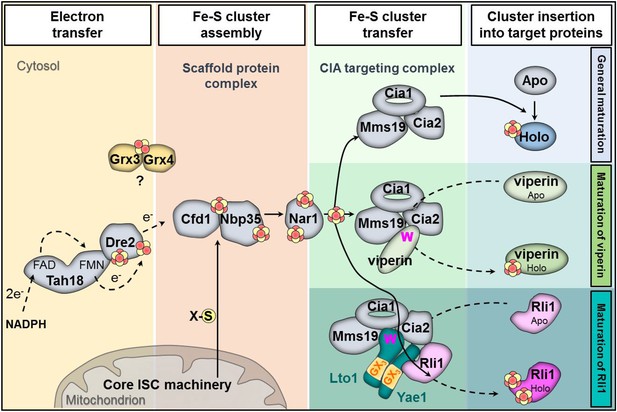
Working model for the specific function of the CIA proteins Yae1 and Lto1 in the maturation of the cytosolic Fe-S protein Rli1.
Maturation of cytosolic and nuclear Fe-S proteins is a multi-step process conducted by different CIA protein subcomplexes. First, a [4Fe-4S] cluster is assembled on the scaffold protein complex composed of Cfd1-Nbp35. This reaction depends on a yet unknown sulfur-containing compound (X-S) which is produced by the mitochondrial iron-sulfur cluster (ISC) assembly machinery and is exported by the mitochondrial ABC transporter Atm1 to the cytosol. Further, the electron transfer chain NADPH-Tah18-Dre2 is required. The Grx3-Grx4 complex mediates a yet undefined function, possibly in delivering iron, for cytosolic-nuclear Fe-S protein biogenesis. Second, the newly assembled [4Fe-4S] cluster is released from Cfd1-Nbp35 and inserted into target apoproteins via Nar1 and the CIA targeting complex (Cia1-Cia2-Mms19). The precise mechanism of the latter steps in still unclear. As reported in this study, maturation of the essential Fe-S protein Rli1 additionally depends on the function of the two specific adaptor proteins Yae1 and Lto1. The Yae1-Lto1 complex uses a unique binding cascade to recruit Rli1 to the CIA targeting complex for Fe-S cluster insertion. The CIA targeting complex interacts with the conserved C-terminal tryptophan residue (W) of Lto1; the conserved deca-GX3 motifs (yellow boxes) of Yae1-Lto1 are crucial for their complex formation; and Yae1 associates with Rli1. Such an adaptor function is dispensable in the maturation pathway of the virus-induced Fe-S protein viperin because it directly binds to the CIA targeting complex via its conserved C-terminal tryptophan residue.
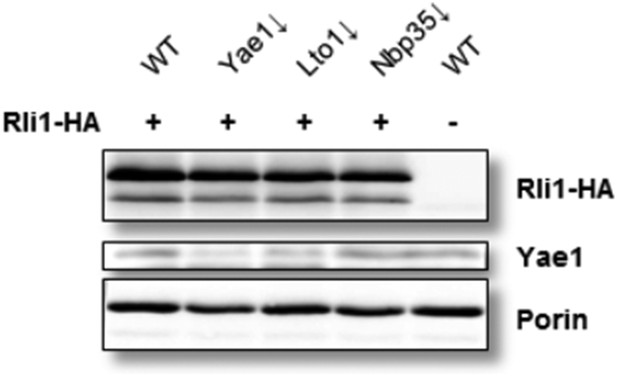
Yae1 depletion is efficient under anaerobic conditions. Wild-type (WT), Gal-YAE1, Gal-LTO1 and Gal-NBP35 cells were transformed with plasmids encoding RLI1-HA (+) or no gene (-). Cells were depleted for the indicated proteins (↓) by growth for 30 h in glucose-containing minimal medium under anaerobic conditions and transferred to minimal medium lacking iron for additional 16 h. After radiolabeling for 2 h with 55Fe under anaerobic conditions, cell extracts were prepared and levels of indicated proteins analyzed via immunoblotting. Due to the low abundance of Yae1, only weak signals were obtained with our available antiserum.
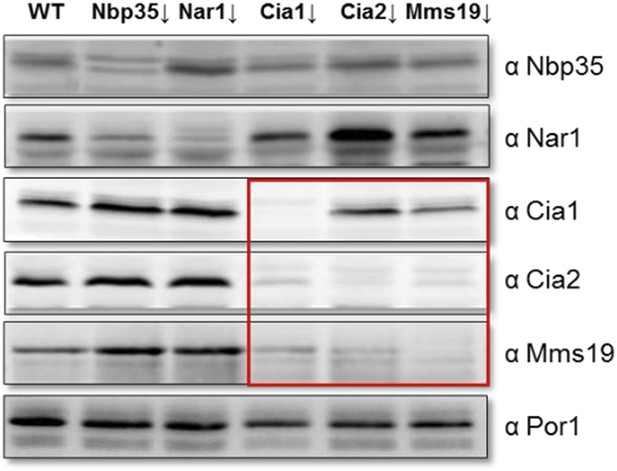
Depletion of individual CIA targeting complex components affects the levels of its other members. Gal-NBP35, Gal-NAR1, Gal-CIA1, Gal-CIA2, Gal-MMS19 and wild-type cells (WT) were grown in minimal medium supplemented with glucose for 40 h. Cell extracts were prepared, and proteins were analyzed by Western blotting using specific rabbit polyclonal antibodies. Porin was used as a loading control. The instability of the CIA targeting complex upon depletion of single members of this complex is indicated by the red box.
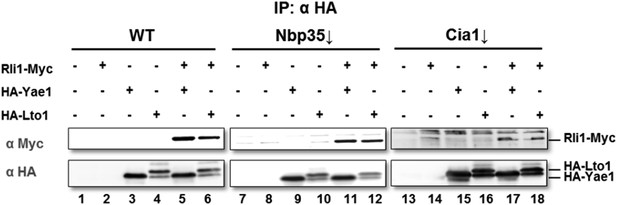
Wild-type (WT), Gal-NBP35 and Gal-CIA1 yeast cells were co-transformed with 2µ vectors containing either no insert or genes encoding HA-Yae1, HA-Lto1 and Rli1-Myc as indicated. Cells were cultivated in minimal medium containing glucose leading to Nbp35 and Cia1 depletion (↓) in Gal-NBP35 and Gal-CIA1 cells, respectively. Cell lysates were prepared, and a HA-tag immunoprecipitation (IP) performed. The immunoprecipitate was analyzed for the indicated proteins or tags by immunoblotting. The lower band in HA-Lto1-containing lanes is a HA-Lto1 degradation product.
Additional files
-
Supplementary file 1
S. cerevisiae strains, Oligonucleotides and Expression vectors.
- https://doi.org/10.7554/eLife.08231.017





