Neurofibromin controls macropinocytosis and phagocytosis in Dictyostelium
Figures
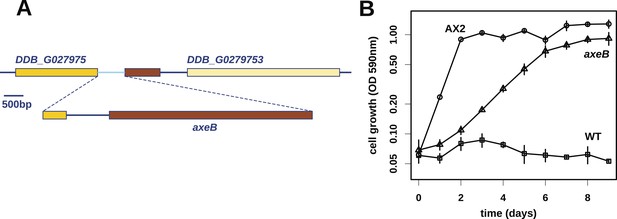
Discovery of the D. discoideum axeB locus.
(A) The region of chromosome 3 spanning the genes DDB_G0279751 and DDB_G0279753 in AX4 genome (top line) contains a conversion mutation in which almost 9 kilobases of sequence (lower line) were lost and replaced by sequence (pale blue) resembling a short region of chromosome 1. The deleted segment contains most of the D. discoideum orthologue of NF1, axeB (brown). (B) NF1 knockout cells can grow in the standard axenic medium, HL5. Amoebae of strains Ax2, DdB (WT), and HM1591 (axeB, an engineered NF1 knockout strain in the DdB background; in this and subsequent figures, ‘axeB’ refers to this strain), were incubated in tissue culture plates in HL5 medium, and growth measured at indicated timepoints using a crystal-violet binding assay. See also Figure 1—figure supplements 1, 2. The AX4 reference genome is at dictyBase (http://dictybase.org).
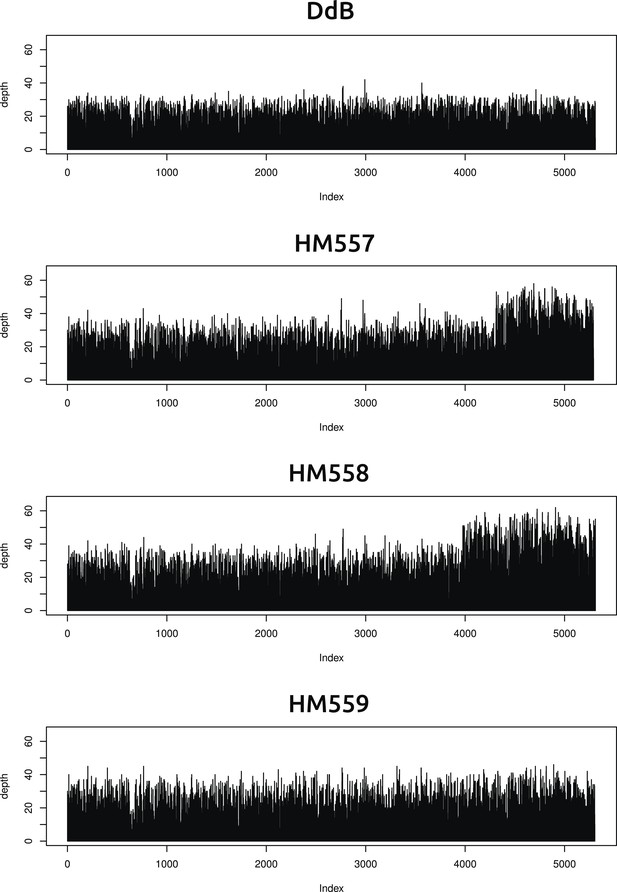
Two new axenic mutant strains possess overlapping duplications on the same chromosome.
The samtools ‘depth’ command was used to calculate the depth of coverage at each position along the chromosomes. A rolling median was obtained (window size 999) to remove outliers, then each chromosome examined by sampling every 1000th position and plotting them sequentially using R (www.r-project.org); the ‘index’ in the plots refers to these 1000 nucleotide divisions. Contiguous segments with approximately double the average depth reflect likely duplication events. Only two such segments could be identified, on overlapping regions at one end of chromosome four in strains HM557 and HM558: shown here are the plots for this chromosome in all four strains resequenced. These duplications are large, spanning hundreds of kilobases and many genes, and it is possible that they contribute to these strains' growth phenotypes; this hypothesis remains to be tested.
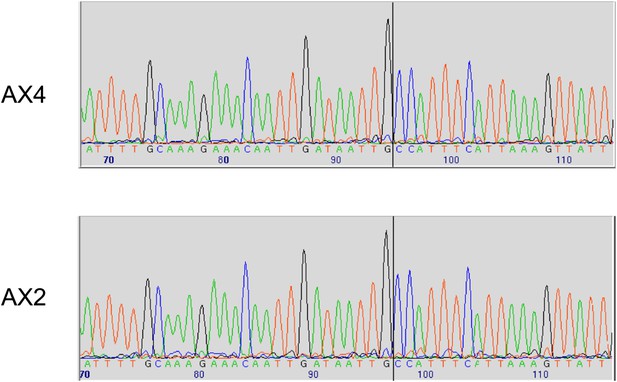
Two established axenic mutants possess identical complex mutations affecting the axeB gene.
Genomic DNA isolated from the Kuspa laboratory stock AX4 and the Kay laboratory stock Ax2 was amplified using primers spanning the deletion-insertion mutation identified in the AX4 reference sequence, and sequence using a primer within the upstream gene. A black vertical line shows the 5′ boundary of the mutation; the boundaries and inserted sequence are identical. The mutation's effects on the parental DdB sequence are annotated in the sequence file deposited in the ENA database as HF565448. Resequencing of these strains' genomes, to be described elsewhere, confirmed this result.
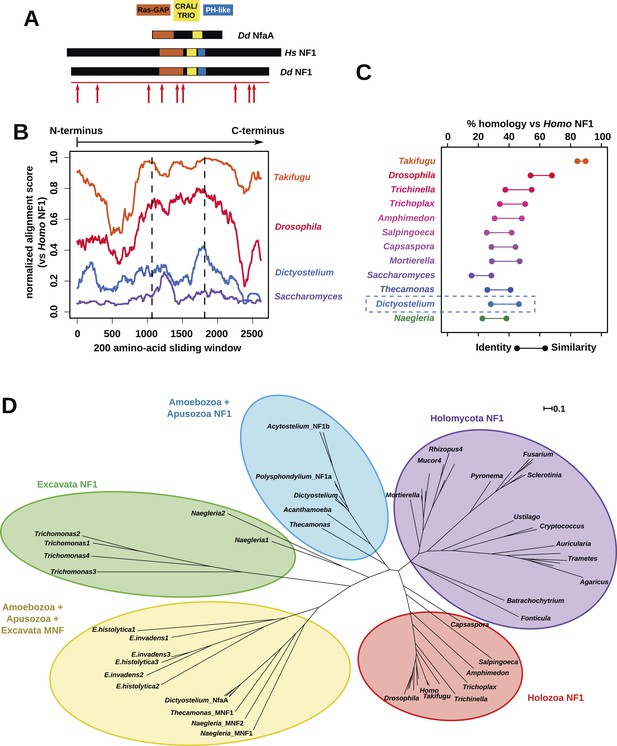
NF1 is broadly conserved in a range of amoeboid species as well as animals and fungi.
(A) NF1 and related proteins have a characteristic domain organisation. The RasGAP domain and adjacent CRAL/TRIO and PH-like domains can be used to identify NF1-like proteins, although the PH-like domain is divergent. Approximate locations of mutations identified in axenic mutants are indicated with arrows; these are described precisely in Table 1. (B) The D. discoideum (Dd) NF1 sequence shows homology to the Homo sapiens protein along its entire length: the sequence of the Hs protein was split into segments with a sliding window of 200 amino acids, and these globally aligned to the Dd, Takifugu rubripes, and Drosophila melanogaster NF1 orthologues, and the Saccharomyces cerevisiae Ira1p sequence. Dashed lines mark the outermost windows containing parts of the central domains. (C) NF1 protein sequences from Takifugu rubripes, Drosophila melanogaster, Trichinella spiralis, Trichoplax adhaerens, Salpingoeca rosetta, Capsaspora owczarzaki, Mortierella verticillata, Saccharomyces cerevisiae (Ira1p), Dd, and Naegleria gruberi (EFC40840.1) were globally aligned with the Homo sapiens NF1 sequence. The bars display the percentage similarity and identity of the protein to the human sequence. (D) Phylogram of NF1 and MNF homologues; the Dictyostelium AxeB protein is an NF1 homologue, while homologues of NfaA form the MNF class of RasGAP, defined here. The presence of NF1 and MNF in Naegleria and Thecamonas as well as amoebozoans indicates that MNF was ancestral and then lost in a common ancestor of the Holozoa and Holomycota after the divergence of apusozoans. The scale shows substitutions/site. See Figure 2—figure supplement 1 for a version with all species labelled, and also Figure 2—figure supplements 2 and Figure 2—source data 1 for illustration of the wider pattern of conservation of RasGAPs.
-
Figure 2—source data 1
Examples of RasGAPs and NF1 orthologues in different lineages.
- https://doi.org/10.7554/eLife.04940.009
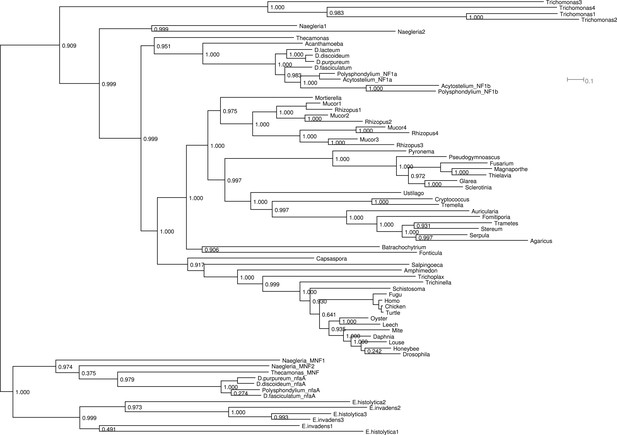
Phylogram of NF1 and MNF homologues.
This represents the same tree as Figure 2D, displayed rectilinearly instead of radially. Selected NF1 homologues from Metazoa and Fungi are included; outside of these taxa all identified homologues are included. The code used, aligned sequences and tree files have been deposited in FigShare (with DOIs 10.6084/m9.figshare.1057805–808). For NfaA-related proteins we suggest the name MNF for ‘miniature Neurofibromin’ to avoid confusion with the unrelated Naegleria Nfa1 protein (Shin et al., 2001).
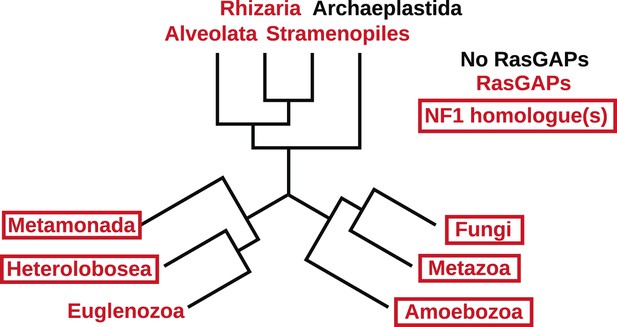
The presence of NF1 homologues and other RasGAPs in the three main eukaryotic supergroups.
While the corticates evidently ancestrally possessed RasGAPs (and Ras signalling), no NF1 homologues are detectable in the genomes of any presently available in the public databases. A previously discussed putative homologue in Stramenopiles has a START domain next to its RasGAP domain, not the unrelated CRAL/TRIO domain found in NF1 (van Dam et al., 2011). The other two supergroups, podiates and excavates, both possess NF1 (and MNF) homologues; if the root of the eukaryotic tree lies between the podiates and either Naegleria or Trichomonas this implies that NF1 was present in the LECA. Examples of RasGAPs and NF1 orthologues in the lineages shown here are given in Figure 2—source data 1.
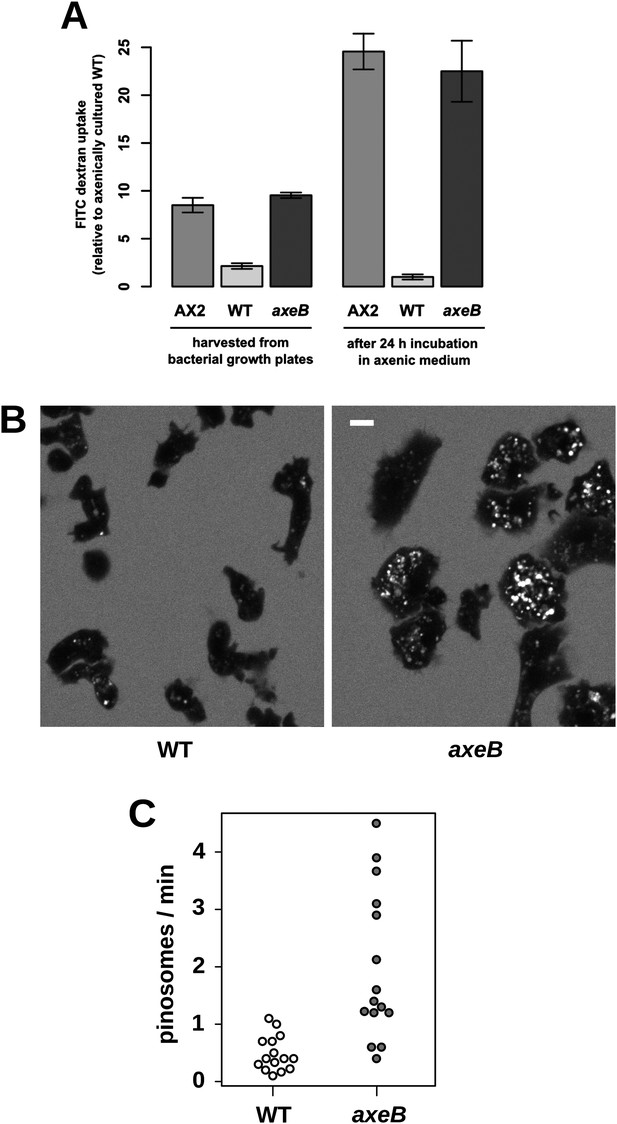
NF1 mutants grow axenically in HL5 medium and have increased fluid uptake.
(A) NF1 knock-out mutants accumulate fluid more quickly than wildtypes. Fluid uptake was measured by shaking cells, either fresh from bacterial growth plates or after 24 hr incubation in axenic medium, with fluorescent dextran in buffer for 1 hr. (B) NF1 mutants accumulate fluorescent dextran in large endosomes, and exhibit a flattened phenotype compared to wildtypes. Cells were harvested from bacterial growth plates and incubated in Loflo medium plus TRITC-dextran for 30 min then imaged by confocal microscopy; cells' cytoplasm appears dark since no dextran penetrates it while endosomes are bright as their contents become concentrated. NF1 mutants tend to assume a flattened morphology; since only a single confocal section is shown this will tend to exaggerate the apparent number of endosomes per cell and so these images should not be relied on for comparison of cumulative fluid uptake. (C) NF1 knock-out mutants form macropinosomes more frequently than wildtypes, as assessed by confocal imaging. 15 cells of each strain were tracked in total in three independent experiments. Scale = 5 μm. Data points are the means of three independent experiments plus and minus the standard error. See also Figure 3—figure supplements 1–3.
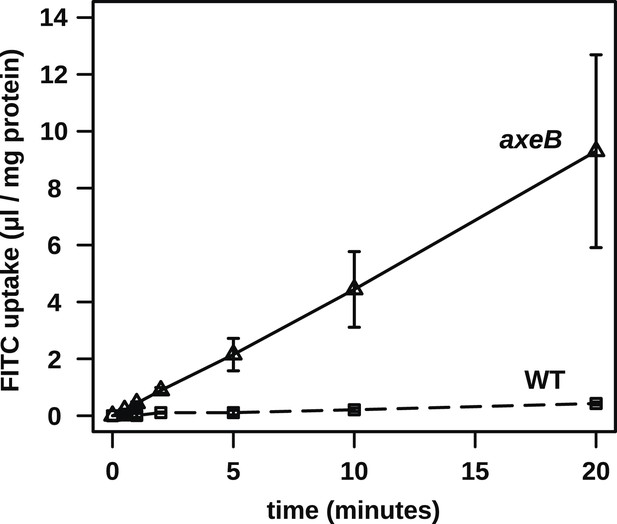
No evidence for fast recycling of ingested fluid.
Although wildtype cells appear to have a lower rate of macropinocytosis as assessed by confocal microscopy, it was formally possible that their lower rate of fluid accumulation could be explained by a faster rate of fluid release, for instance by recycling of fluid from endosomes before fusion with lysosomes. To test this, we incubated cells in axenic medium overnight before performing a timecourse of FITC dextran uptake. Fast recycling would be revealed by a high initial rate of uptake followed by a more moderate rate as a portion of ingested dextran is expelled. We found no evidence for such recycling, although a small amount might occur. Means plus and minus standard errors are given for three independent experiments.
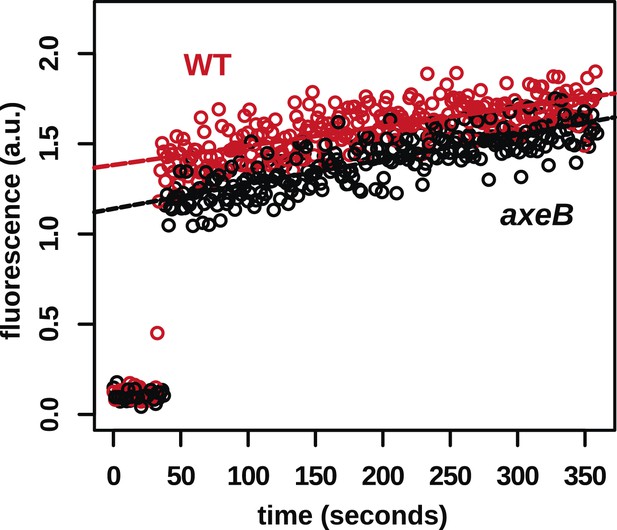
In contrast to fluid uptake, membrane uptake is not increased in NF1 mutants.
DdB (WT) and the NF1 null mutant HM1591 (axeB) were harvested from bacterial growth plates, washed, and resuspended in KK2 buffer and shaken at room temperature for 15 min before being added a stirred fluorimeter cuvette containing FM1-43 dye. When cells are added, the fluorescence of the sample increases rapidly as dye enters the plasma membrane; as membrane is internalised, compensating fresh membrane is exposed to the surface enabling more dye to bind and fluoresce. While fluid uptake is several-fold higher in mutants, we found no evidence of increased membrane uptake rates, indicating that most membrane is taken up as small vesicles or narrow tubules with large surface-area to volume ratios.
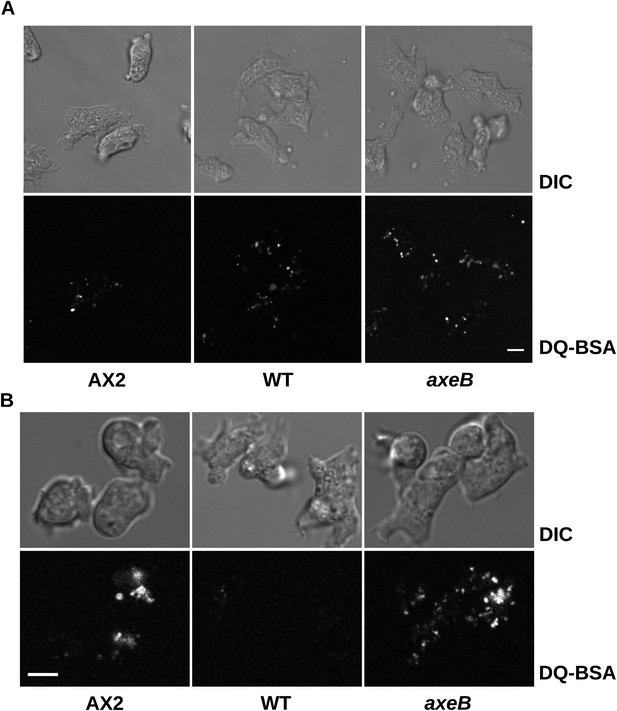
Intracellular degradation of proteins occurs normally in NF1 mutants.
To image degradation of internalized protein, cells of strains Ax2, DdB, and HM1591 were incubated in Loflo medium plus 50 µg/ml DQ Green BSA, either (A) for 60 min for cells taken directly from bacterial growth, or (B) for 15 min for cells incubated for 24 hr in loflo medium before DQ Green BSA was added; cells were imaged for green fluorescence of degraded peptides using the same laser power and gain in each case. Scale = 5 μm.
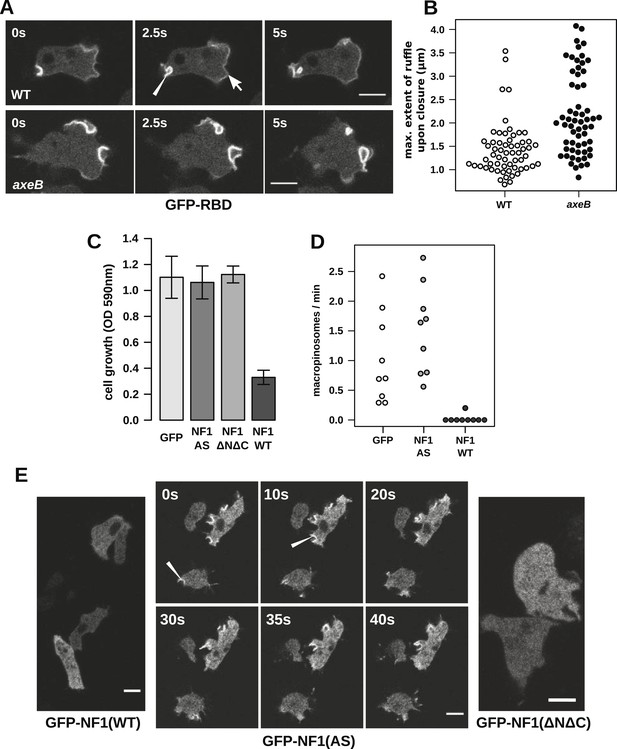
NF1 localises to membrane ruffles, its loss potentiates Ras signalling at macropinosomes, and its over-expression represses macropincytosis.
(A) Ras activity, as reported by GFP-tagged Raf1 Ras-binding domain (GFP-RBD), is exhibited at sites of macropinocytosis (pointer) in wildtype DdB cells as well as at the leading edge (arrow) as the cells move; the distribution of the reporter is qualitatively similar in NF1 knock-out amoebae, but ruffling is more extensive than in wildtypes. (B) The Ras-marked membrane ruffles tend to be larger in knock-out mutants prior to closure into pinosomes. Mutant or wildtype GFP-RBD reporter strains were harvested from bacterial growth plates and Ras-marked ruffles were measured across their longest visible axis just after they closed; data are from 60 events for each strain in total from three independent experiments. (C) Introduction of N-terminally GFP-tagged Dictyostelium NF1 proteins into axeB mutants reduces axenic growth in the case of the wildtype sequence (NF1-WT) but not when two consecutive arginine residues in the protein's ‘arginine finger’ are mutated to alanine and serine (NF1-AS′), nor when only the central region of the protein encompassing the RasGAP, CRAL-TRIO, and PH-like domains (NF1ΔNΔC) is expressed, when compared to a GFP control. Data are means plus and minus standard error for three independent experiments using the crystal violet assay to assess growth after 7 days incubation in tissue culture plates. (D) The active NF1-RR construct almost completely abolishes macropinosome formation when expressed in NF1 mutants, while the inactive NF1-AS form does not inhibit macropinocytosis. Bacterially grown cells were monitored by confocal microscopy as in Figure 3C; rates for nine cells of each line from three independent experiments are shown. (E) The NF1-AS mutant protein is recruited to membrane ruffles and sites of macropinocytosis (examples indicated by pointers), whereas the wildtype version (NF1-RR) has an even cytoplasmic distribution, as does the truncated NF1ΔNΔC protein. The scale bars represent 5 μm. See also Figure 4—figure supplements 1–5.
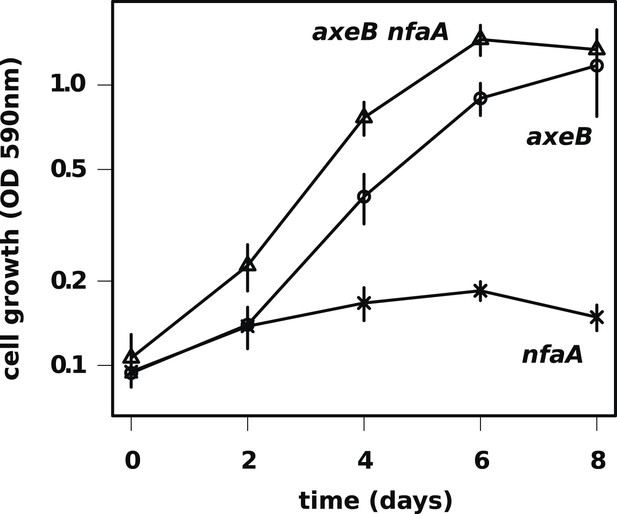
The axenic growth phenotype is specific to loss of the NF1 RasGAP protein.
All axenic mutants we have examined so far possess mutations in axeB, the gene encoding NF1. To test whether other RasGAPs might also have related functions (but, for instance, a lower rate of spontaneous mutation), we also deleted the related RasGAP gene nfaA in both the wildtype and axeB null background. The nfaA single mutant (HM1709) does not grow axenically, as assessed by the crystal violet binding assay, while the axeB nfaA double mutant (HM1710) has slightly potentiated the axeB phenotype, suggesting that in the absence of NF1, the NfaA protein can substitute for it to some extent. Data are the means plus and minus the standard error for three independent experiments.
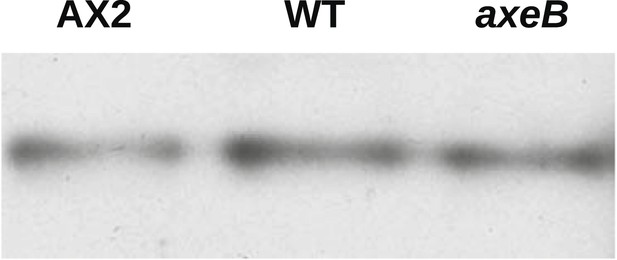
NF1 mutants do not have an increase in overall Ras activity as assayed using RBD pulldowns.
Using GST-Raf1-RBD beads to pull down GTP-bound Ras proteins and an anti-Ras antibody to compare samples by immunoblotting, no increase in Ras activity could be found in vegetative axenic mutants Ax2 and HM1591 (axeB) compared to wildtype DdB cells (WT); a single representative experiment is shown.
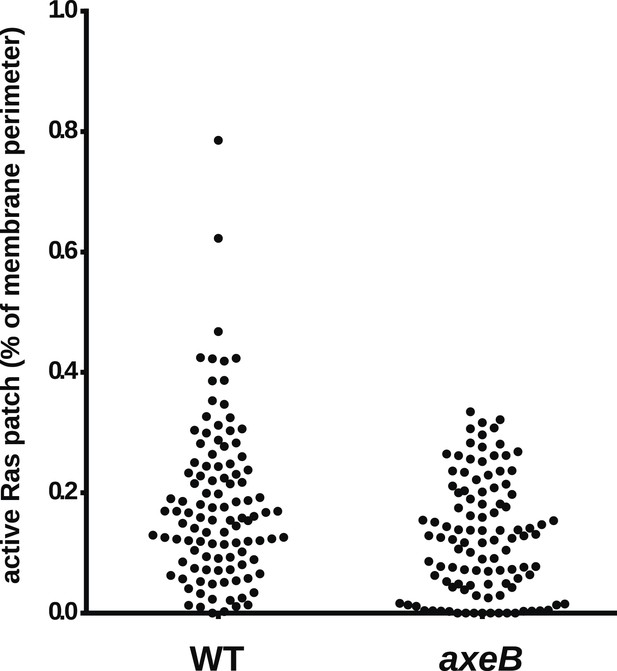
NF1 mutants do not have an increase in overall Ras activity as assessed by confocal microscopy.
Using GFP-Raf1-RBD reporter constructs, no increase in plasma-membrane associated active Ras was observed in the axeB null: this was quantified from tilescans of cells from three independent experiments. In all cases, to ensure that the cells were in comparable state, they were used within 30 min of harvesting from bacterial growth.
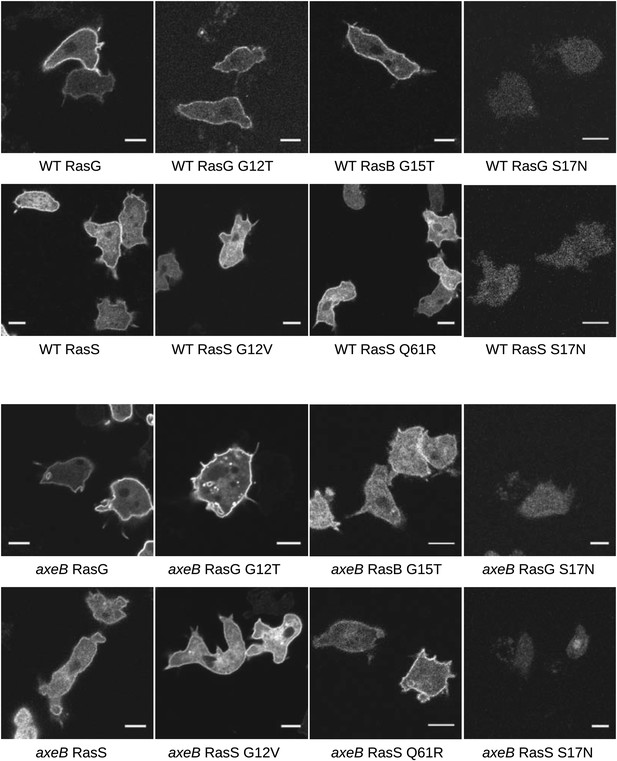
Localisation of GFP-Ras fusion proteins.
DdB cells (WT) or HM1591 (axeB) cells expressing GFP or GFP-tagged RasG (‘G’), RasGG12T (‘G12’), RasS (‘S’), RasSG12V (‘S12’), RasSQ61R (‘S61’), or RasBG15T (‘B15’) were imaged by confocal microscopy to confirm proper localisation. All show some degree of enrichment on the plasma membrane except the dominant negative S17N mutants, which are very weakly fluorescent and do not show membrane localisation, presumably because they are deleterious and only cells with restricted expression grow through the selection procedure. Scale = 5 μm.
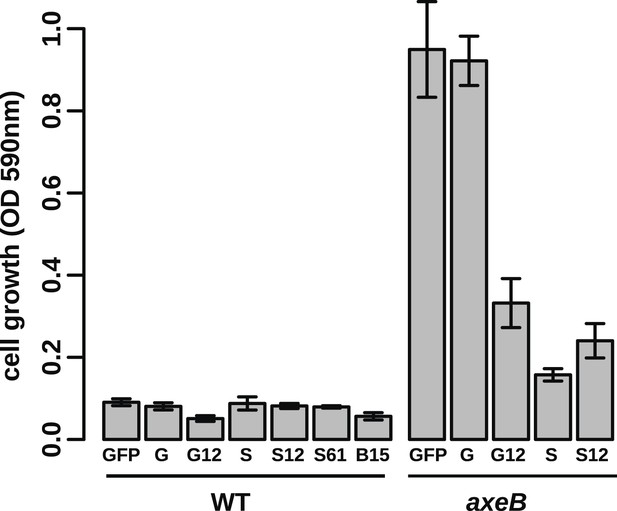
Growth phenotypes of Ras expression lines.
GFP-tagged D. discoideum Ras proteins were expressed in either or both DdB and HM1591 to test their effects on growth in HL5 medium in 24-well tissue culture plates; growth was assessed by the crystal violet assay after 7 days' incubation (B = RasB, G = RasG, S = RasS): no active Ras constructs are able to stimulate axenic growth of wildtype DdB cells; expression of constitutively active RasG (G12) or RasS (S12), or expression of wildtype RasS (S) is actually deleterious towards axenic growth. Data are means plus and minus standard error for three independent experiments.
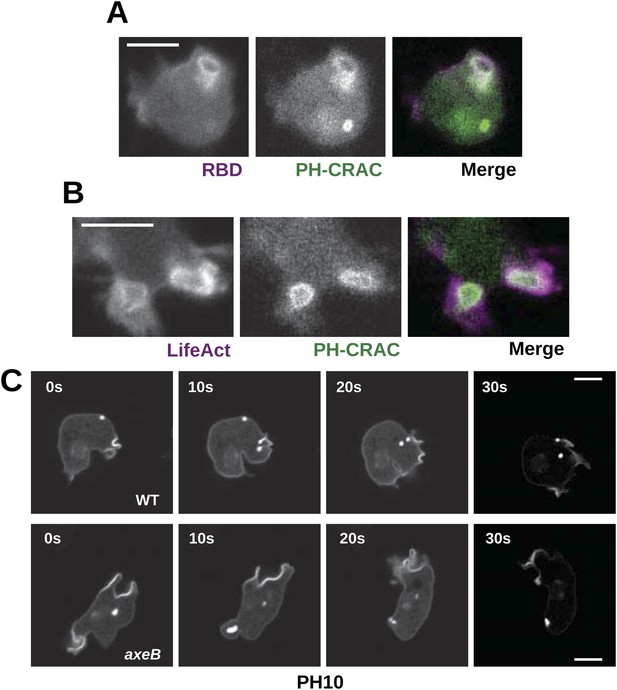
Downstream signalling: connections between Ras and PI3K activity during macropinocytosis.
(A) Ras activity (mCherry-Raf1-RBD reporter, magenta) is accompanied by phosphoinositide 3-kinase activity (PH-CRAC-GFP reporter, green) on macropinosomes in axeB mutants; note the green endosome where PI3K products remain but Ras signalling has terminated. (B) Actin polymerisation (labelled with mRFP-LifeAct, magenta) occurs around the structures marked by the PH-domain reporter (green). (C) PH domains (GFP-PH10) are also recruited to macropinosomes in vegetative wildtype DdB cells; the kinetics of recruitment and retention are similar in axeB cells. The scale bars represent 5 μm. See also Figure 5—figure supplement 1.
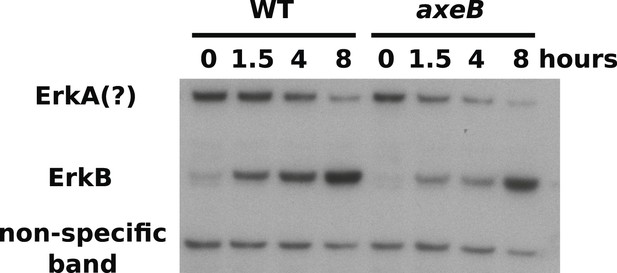
ERK phosphorylation is not increased in NF1 mutants.
WT (DdB) and axeB knockout (HM1591) cells were harvested from bacterial growth plates then washed and incubated in HL5 medium and shaken for the indicated times before ERK activity was assessed using an antibody raised against a phosphorylated TEY motif. A band of the expected size of D. discoideum ErkB reproducibly increased in intensity over time in both strains, but more intensely in WT cells than NF1 mutants, perhaps reflecting starvation-induced development. A band of the approximate expected size of ErkA varied in its pattern of intensity in different experiments, but showed no tendency to be more intense in mutants. A single representative experiment is shown.
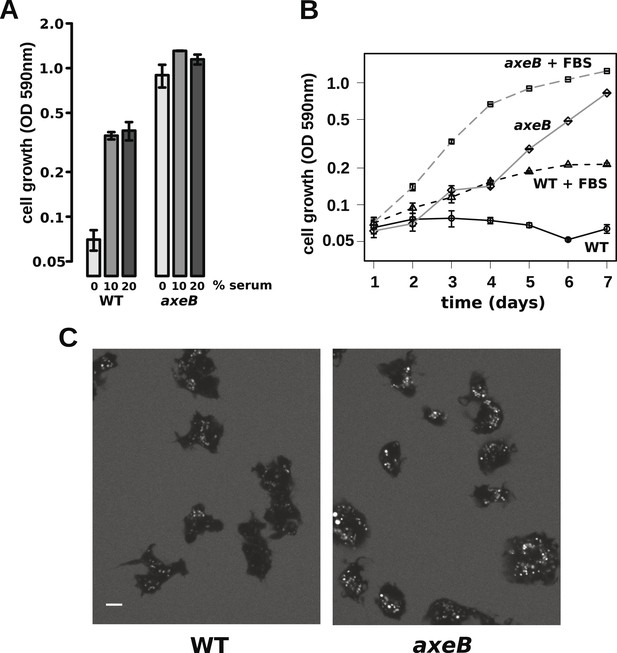
Wildtype amoebae can grow axenically in medium supplemented with bovine serum.
(A) Wildtype (DdB) and NF1 mutant (HM1591) cells were incubated in HL5 medium supplemented with vitamins and microelements without further additions or with 10% or 20% foetal bovine serum (FBS and filter-sterilised HL5 mixed in 1:9 or 1:4 ratios) in 24-well tissue culture dishes at a starting density of 5 × 104 cells per well. After 7 days growth was measured using the crystal violet assay. FBS stimulated growth of both wildtype and NF1 mutant cells, with mutants having a growth advantage in all axenic conditions. (B) Time courses of growth in the presence and absence of 10% FBS in the same conditions as above except that the HL5 medium was dissolved in 10% FBS or in water, then filter-sterilised. Data are means plus and minus standard errors of three (A) or four (B) independent experiments. (C) Wildtype amoebae retain their normal vegetative morphology after growth in serum-supplemented HL5 medium and NF1 mutants are still distinguished by a more flattened appearance. Cells were grown in HL5 plus 10% FBS for 4 days before being washed and placed into Loflo plus 10% FBS in presence of TRITC-dextran. After 30 min, the cells were imaged by confocal microscopy. Scale = 5 μm. See also Figure 6—figure supplements 1, 2.
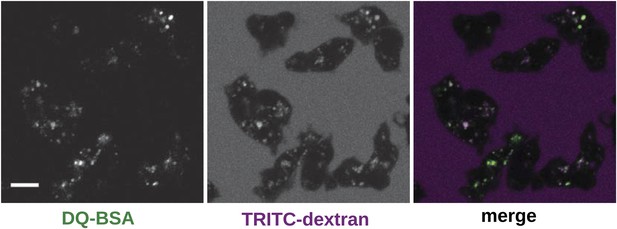
Wildtype cells degrade extracellular protein effectively after growth in rich axenic media.
DdB cells were grown in HL5 medium plus 10% foetal bovine serum for 3 days before being washed, resuspended in Loflo medium plus 50 µg/ml DQ Green BSA and 2 mg/ml TRITC dextran. Images were taken after 20 min. Scale = 5 μm.
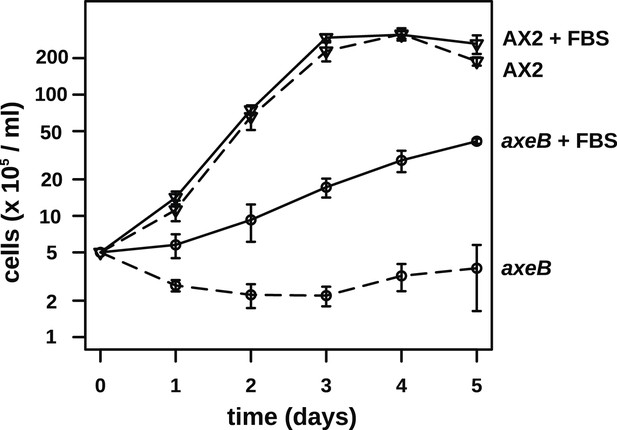
NF1 mutants are able to grow in suspension in rich axenic media.
The established axenic mutant strain Ax2 was selected for high rates of growth in HL5 medium in shaken suspension; deleting axeB in the wildtype background only partially recapitulated this pheontype, the cells only being able to grow well in HL5 when attached to a substratum, indicating that at least one other mutation is required to account for the full axenic phenotype. However the single mutant is able to grow in shaken suspension when HL5 is supplemented with 10% foetal bovie serum. Cells were inoculated at a starting density of 5 × 105 cells per ml in 50 ml of medium in 250 ml Erlenmeyer flasks at 22°C and counted at the stated intervals using a haemocytometer. The means and standard errors are given for three independent experiments.
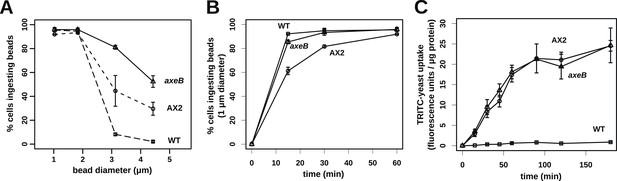
NF1 mutants can phagocytose larger particles than wildtypes.
(A) Axenic mutants ingest small bacterium-sized beads at a similar rate as wildtypes, but wildtype cells are dramatically less efficient at ingesting beads greater than 2 μm in diameter. Cells were harvested from bacterial growth plates, washed, then shaken with fluorescent microspheres of the indicated diameter, then after 1 hr scored for the presence of internalised beads. (B) The Ax2 mutant accumulated small 1.0 μm beads more slowly than the wildtype DdB or the axeB deletion mutant. (C) Axenic mutants can ingest fluorescently labelled budding yeast cells much more easily than wildtype cells. All data are mean ± standard error for three independent experiments. See also Figure 7—figure supplements 1–3.
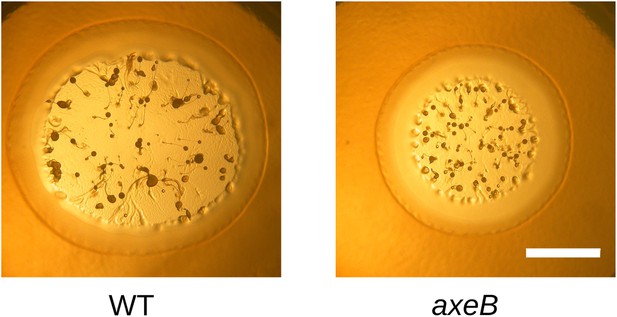
NF1 mutants grow and develop when grown on bacterial lawns.
DdB (WT) and HM1591 (axeB null) spores were plated clonally on SM agar plates in association with Klebsiella pneumoniae. After 5 days, plaques of amoebae growing outwards on the bacterial lawn were photographed; aggregates and fruiting bodies are visible where the bacteria have been cleared causing the amoebae to enter their asexual developmental cycle (scale = 5 mm). Fruiting bodies in the mutant tend to be smaller than wildtype, but otherwise do not show obvious defects.
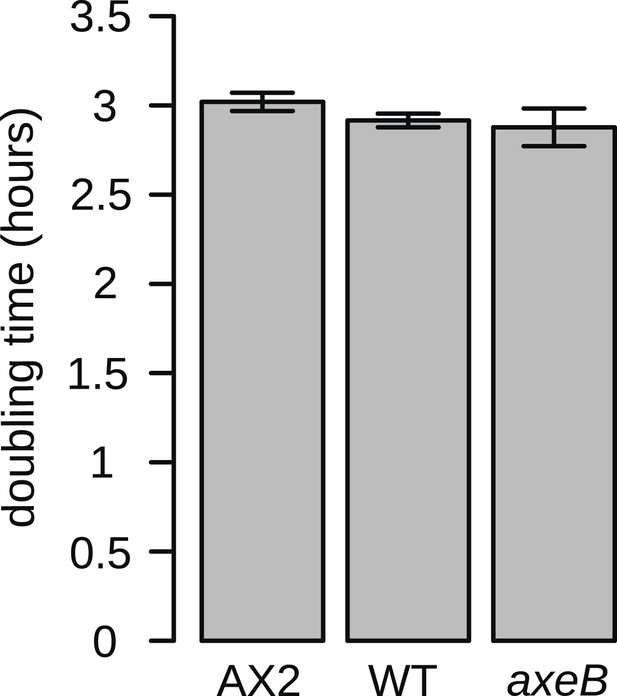
NF1 mutants grow normally when shaken in suspensions of dead bacteria.
To quantify cell growth without complicating factors such as cell motility and susceptibility to harmful bacterial metabolites, cells were grown on heat-killed Escherichia coli strain B/r in shaking suspension. The established axenic strain Ax2 grows consistently more slowly than DdB and HM1591, with a doubling time on average approximately 5% greater. Data are means plus and minus standard error for three independent experiments.
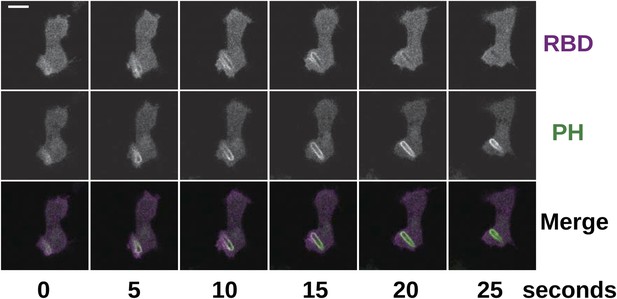
Phagocytosis is accompanied by Ras and PI3K activity in the same way as in macropinocytosis.
NF1 mutants (HM1591) were transformed with an expression construct containing both mCherry-Raf1-RBD and PH(CRAC)-GFP and imaged in the presence of Klebsiella cells; the initial engulfment occurred out of the plane of acquisition, but Ras and PI3K activity remained visible as the nascent phagosome moved into view. Scale = 5 μm.
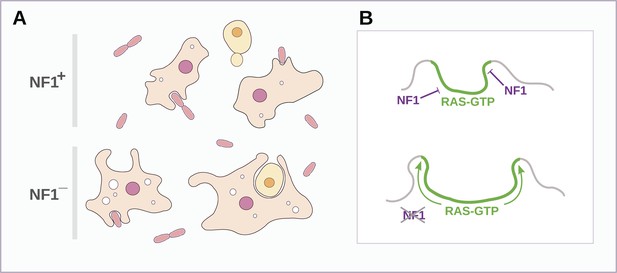
Schematic model of NF1 function in Dictyostelium.
(A) While wildtype NF1+ amoebae ingest bacteria most readily, NF1− cells are also able to ingest larger particles such as yeast cells, and accumulate more fluid in macropinosomes. (B) The large concave membrane ruffles formed during phagocytosis and macropinocytosis both are marked by intense Ras signalling (green); NF1 localises dynamically to these regions, stimulating the GTPase activity of Ras proteins there, inactivating them and thereby limiting the expansion and spread of the ‘activated’ membrane domain.
Tables
Mutations in the axeB gene in Dictyostelium discoideum axenic mutants
| Strain | Mutation | Effect on Dd NF1 protein | Position in human NF1 protein |
|---|---|---|---|
| Ax2 | c.-1954_6926delinsCM000150.2:1390060_1390808 | Deletion to amino acid 2309 | to 2358 |
| AX4 | c.-1954_6926delinsCM000150.2:1390060_1390808 | Deletion to amino acid 2309 | to 2358 |
| HM557 | c.226_230del | Deletion, frameshift | 66 |
| HM587 | c.1015A > T | Nonsense | 315 |
| HM591 | c.3033_3040del | Deletion, frameshift | ∼1060 (in insertion relative to Human) |
| NP73 | c.3508del | Deletion, frameshift | 1228 |
| HM589 | c.4113G > T | K > N | 1423 |
| HM590 | c.4227_4459del | Deletion, frameshift | 1461–1533 |
| HM558 | c.6393_6413inv | DPVVSAIL > EELQKPND | 2182–2189 |
| HM586 | c.6833_7077del | Deletion, frameshift | 2325–2481 |
| HM559 | c.7137_7143del | Deletion, frameshift | 2525–2529 |
-
Strains are described fully in Table 2. Description of changes to the coding sequence of the axeB gene follow the recommendations of the Human Genome Variation Society (den Dunnen and Antonarakis, 2000); the effect on the protein sequence is indicated, using the IUPAC one-letter code for amino-acid substitutions. All changes except one are predicted to inactivate the protein either through the introduction of premature stop codons or the substitution of a conserved residue known to be important for function in the human version of the protein. Approximate corresponding locations in the amino-acid sequence of the human orthologue are also indicated.
Strains used in this study
| Strain | Parent | Genotype | Reference |
|---|---|---|---|
| Ax2 | DdB | axeA2 axeB2 axeC2 | (Watts and Ashworth, 1970) |
| AX4 | DdB | axeA1 axeB1 axeC1 | (Knecht et al., 1986) |
| DdB | NC4 | Wildtype | (Bloomfield et al., 2008, as ‘DdB(Wel)’) |
| NP73 | V12 | axeB3 | (Williams, 1976) |
| HM557 | DdB | axeB(GB1) | This study |
| HM558 | DdB | axeB(GB2) | This study |
| HM559 | DdB | axeB(GB3) | This study |
| HM586 | DdB | axeB(GB4) | This study |
| HM587 | DdB | axeB(GB5) | This study |
| HM589 | DdB | axeB(GB6) | This study |
| HM590 | DdB | axeB(GB7) | This study |
| HM591 | DdB | axeB(GB8) | This study |
| HM1591 | DdB | axeB(GB9) neoR | This study |
| HM1709 | DdB | nfaA(GB1) hygR | This study |
| HM1710 | HM1591 | nfaA(GB1) axeB(GB9) neoR hygR | This study |
-
The generally accepted genotype of Ax2 and AX4 is given, although the true number of mutations contributing substantially to their fast axenic growth phenotype remains unknown. AX4 derives from another axenic mutant, AX3 (or A3), which was isolated from wildtype cells independently from Ax2 (Loomis, 1971). Extant AX3 and AX4 strains share a large inverted duplication on chromosome 2 (Eichinger et al., 2005) that is not present in Ax2. However, the mutation in axeB in Ax2 and AX4 is identical, suggesting that the extant lines of these strains, along with AX3, had a common ancestor that was axenic. It might not be possible to determine the reason for this discrepancy with the literature; one possibility is that very early in these strains' contemporaneous history one line was contaminated with the other and the slower-growing of the two then lost. In formally numbering alleles we have retrospectively assigned allele number ‘3’ to the axeB mutation in the historic strain NP73, but follow recent recommendations (http://dictybase.org/Dicty_Info/nomenclature_guidelines.html) for new strains, and use the same number for gene disruptions using the same knockout construct.
Additional files
-
Source code 1
MatLab script for quantification of active regions of the cell perimeter.
- https://doi.org/10.7554/eLife.04940.032





