Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins
Figures
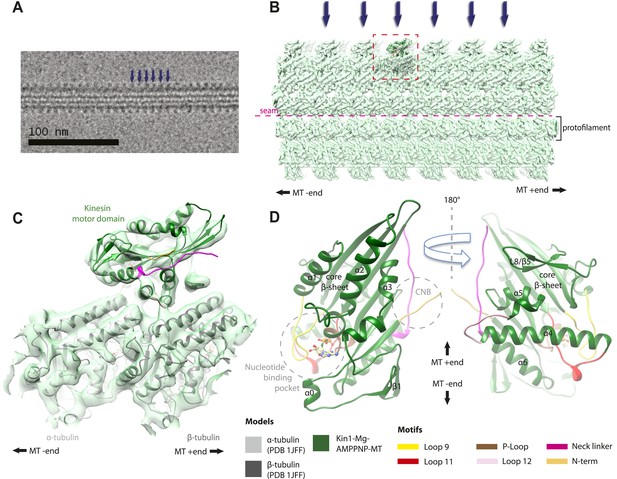
Overview of MT-bound kinesin motor domain cryo-EM reconstructions.
(A) Example cryo-EM image of kinesin-decorated MT (Kin1-Mg-AMPPNP); blue arrows indicate individual Kin1 motor domains. (B) Example of cryo-EM reconstruction of 13 protofilament, kinesin-decorated MT (Kin1-Mg-AMPPNP); blue arrows indicate individual Kin1 motor domains, and the dotted red box shows an asymmetric unit. A single protofilament is indicated along with the position of the lattice seam. (C) Example of an individual asymmetric unit (Kin1-Mg-AMPPNP), contoured to show secondary structural elements. (D) Two views, related by 180°, of an exemplar pseudo-atomic model (Kin1-Mg-AMPPNP) calculated using our cryo-EM reconstruction. The major mechanochemical elements discussed in the text are colour-coded as indicated in the key.
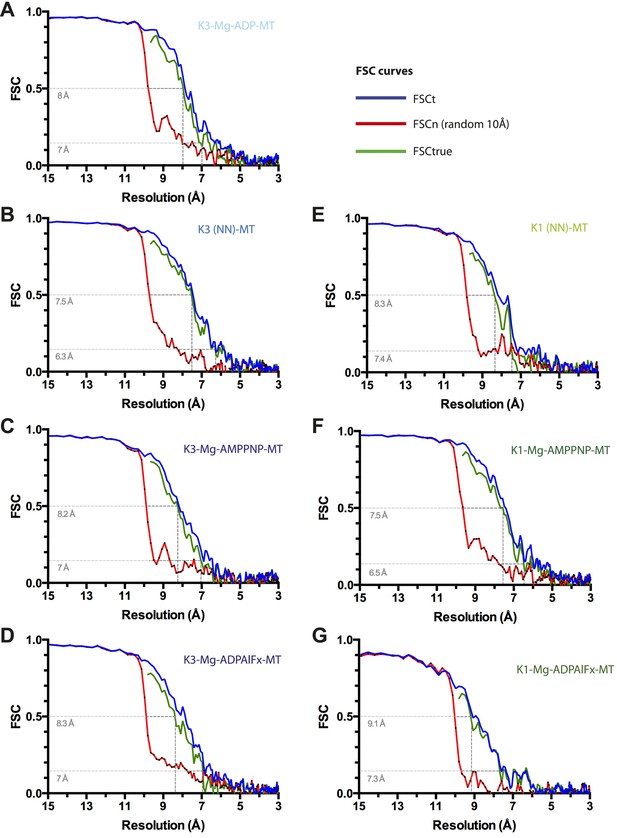
Resolution estimation for cryo-EM reconstructions.
For each reconstruction, three Fourier Shell Correlation (FSC) curves are plotted: standard FSCt (blue) between two half data sets, FSCn (noise substitution cutoff 10 Å, red) and FSCtrue (green, see Chen et al., 2013). (A) Kin3-Mg-ADP-MT, (B) Kin3-NN-MT, (C) Kin3-Mg-AMPPNP-MT, (D) Kin3-Mg-ADPAlFx-MT, (E) Kin1-NN-MT, (F) Kin1-Mg-AMPPNP-MT, (G) Kin1-Mg-ADPAlFx-MT. Dotted lines indicate estimated resolution by FSCtrue at 0.143 (considered appropriate for FSCtrue) and 0.5 criteria. The overall good agreement between FSCt and FSCtrue curves demonstrates that minimal over-fitting occurred during refinement of the cryo-EM data.
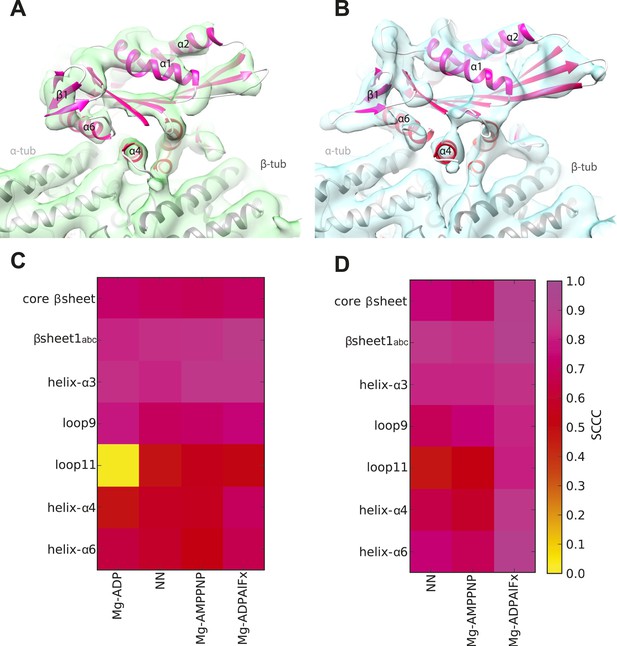
Local assessment of fit quality of the pseudo-atomic models within the cryo-EM density.
Following flexible fitting of each kinesin motor domain, the local fit quality of specific elements was calculated. (A and B) NN cryo-EM density for (A) Kin3 and (B) Kin1 is shown with their respective docked pseudo-atomic model colour-coded according to segment based cross correlation coefficient (SCCC, see colour key; Pandurangan et al., 2014). (C and D) Heat map showing the quality of the local fit for specific elements of the motor domain in different nucleotide states for (C) Kin3 and (D) Kin1. The colour (see key) denotes the SCCC score as calculated with TEMPy (Farabella et al., 2014). This analysis shows the quality of the fits and provides confidence in our interpretation of conformational changes in these regions. In particular, it shows that loop9 and loop11 have similar (good) quality of fit compared to the α-helices, apart from loop11 in the Kin3-Mg-ADP reconstruction, for which cryo-EM density was not seen.
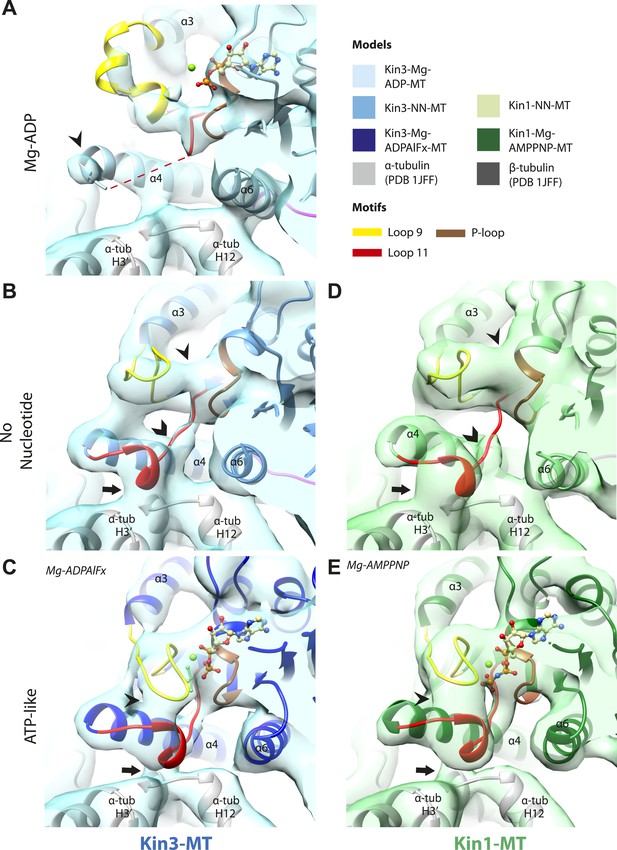
Conserved conformations at the nucleotide-binding pocket in Kin3s and Kin1s.
(A–C) The nucleotide-binding pocket of MT-bound Kin3 reconstructions (shown as blue transparent density) in (A) Mg-ADP, model shown in light blue; the arrowhead indicates residual flexibility in the helix-α4 N-terminus and the region of loop11 for which density is missing is depicted by a dotted red line; (B) no nucleotide (NN), model shown in mid-blue; density connects the C-terminal helical turn of loop11 with the MT (arrow), density corresponding to the rest of loop11 is seen (chevron) and density now connects the extended loop 9 and the P-loop (arrowhead); (C) Mg-ADPAlFx, model shown in dark blue; the C-terminal helical turn of loop11 has moved away from the MT (arrow) and strong density is seen connecting it, helix-α4 and loop9 around the bound nucleotide. (D–E) The nucleotide-binding pocket of MT-bound Kin1 reconstructions (shown as green transparent density) in (D) no nucleotide, model shown in light green; density connects the C-terminal helical turn of loop11 with the MT (arrow), density corresponding to the majority of loop11 is seen (chevron) and density now connects the extended loop 9 and the P-loop (arrowhead); (E) Mg-AMPPNP, model shown in dark green; the C-terminal helical turn of loop11 has moved away from the MT (arrow) and strong density is seen connecting it, helix-α4 and loop9 around the bound nucleotide. In all reconstructions, density for the motor domain was contoured to an equivalent volume.
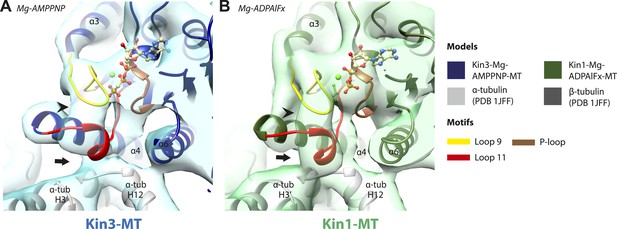
Conserved conformations at the nucleotide-binding pocket in Kin3 and Kin1 alternative ATP-like states.
(A) The nucleotide-binding pocket of the MT bound Kin3-Mg-AMPPNP (blue transparent density and navy blue model). (B) The nucleotide-binding pocket of the MT bound Kin1-Mg-ADPAlFx reconstruction (green transparent density and olive green model). The major features are shared by all the ATP-like reconstructions: in Kin3-Mg-AMPPNP the C-terminal helical turn of loop11 has moved away from the MT (arrow) and strong density (arrowhead) is seen connecting it, helix-α4 and loop9 around the bound nucleotide. The Kin1-Mg-ADPAlFx reconstruction is lower resolution (FSCtrue, 0.143 = 7.7), which may explain why residual density connects the C-terminal helical turn of loop11 with the MT (arrow); however strong density is seen connecting it, helix-α4 and loop9 around the bound nucleotide. In all reconstructions, density for the motor domain was contoured to an equivalent volume.
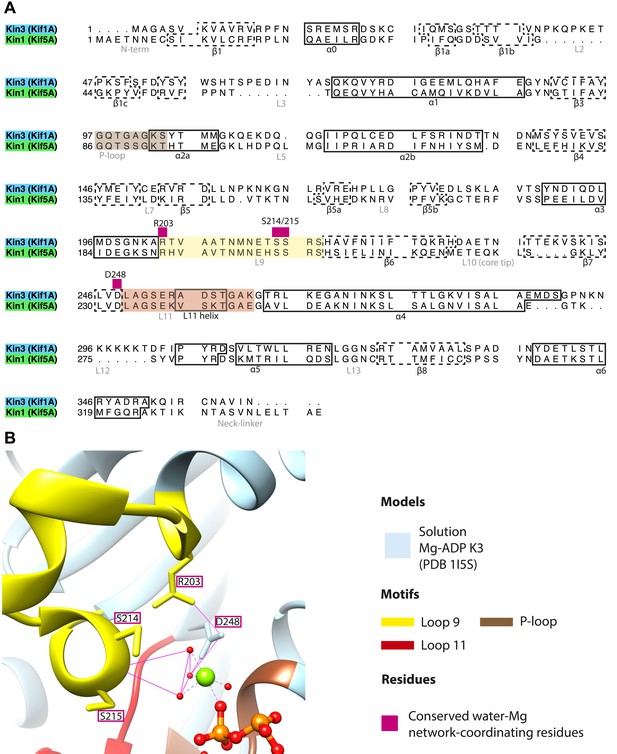
Coordination of Mg-ADP cluster by loop9 and loop11.
(A) Sequence alignment of Kin3 and Kin1 highlighting conserved Mg-water ‘cap’ coordinating residues (magenta squares above residue letters) in loop9 (yellow shading) and near loop11 (red shading). (B) The crystal structure of Kin3-Mg-ADP (Kif1A; PDB 1I5S; Kikkawa et al., 2001) showing the side chains of the residues (Kin3: Arg203, Ser214, Ser215, Asp248) indicated in panel A. Putative hydrogen bonds (displayed with FindHBond Chimera plugin) between these residues and the Mg-water cap are shown as solid magenta lines. Water molecules and Mg are shown as red and green spheres respectively. We propose that MT-triggered displacement of loop9 leads to destabilization of the Mg-water cap and consequent Mg-ADP release from the nucleotide pocket.
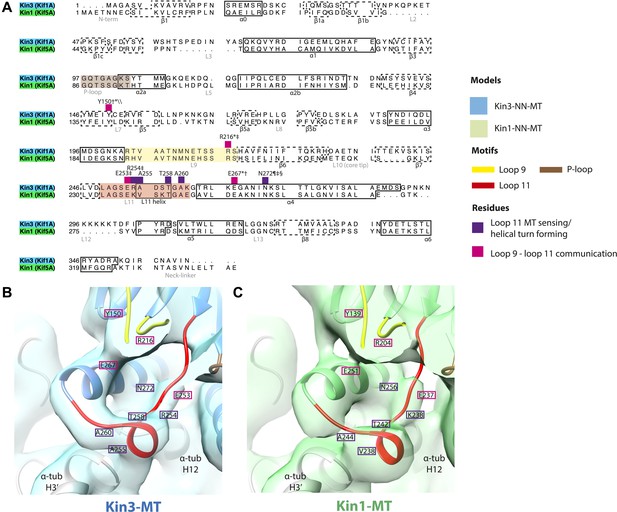
Conserved residues involved in MT-mediated stimulation of Mg-ADP release.
(A) Sequence alignment of Kin3 and Kin1 highlighting residues likely to be important in MT-mediated stimulation of Mg-ADP release. Residues involved in MT sensing and stabilization of loop11 are indicated by purple squares above residue letters (Kin3 residue number), whereas those involved in communication between loop11 (at the MT) and loop9 (water-Mg-ADP coordination) are indicated by magenta squares. Loop9 is indicated by yellow shading, loop11 by red shading, and the P-loop by brown shading. (B and C) Location of these residues in the NN-MT-bound models of (B) Kin3 (mid blue) within the equivalent reconstruction (blue transparent density) and (C) Kin1 (light green) within the equivalent reconstruction (green transparent density), contoured at equivalent volumes. We propose that MT binding reduces the conformational freedom of loop11, stabilizing a helical turn that involves Kin3 Ala255 (Kin1 Val238) and Ala260 (Kin1 Ala244), and Kin3 Thr258 (Kin1 Thr242) above α-tubulin's H3ʹ. Kin3 helix-α4 Asn272 (Kin1 Asn256) sits at the interface of α-tubulin and loop11, likely interacting with both (Gigant et al., 2013) and presumably stabilizing loop11. Kin3 loop11 Arg254 (Kin1 Lys238) may help stabilize loop11 through its interaction with the acidic tip of α-tubulin's H12 (Gigant et al., 2013). Communication between loop11 and loop9 likely occurs via a salt bridge between Kin3 loop Glu253 (Kin1 Glu237) and loop9 Arg216 (Kin1 Arg204) as reported in hydrolysis-competent conformation ATP-like crystal structures (Chang et al., 2013; Gigant et al., 2013; Parke et al., 2010). Kin3 helix-α4 Glu267 (Kin1 Glu251) also interacts with loop9 Arg216 (Kin1 Arg204), an interaction that also involves loop7 Tyr150 (Kin1 Tyr139; Liu et al., 2012a). Evidence for these residues involvement in MT-mediated Mg-ADP release is provided by structural and biochemical studies and disease-causing patient mutations (*Nitta et al., 2008; †Woehlke et al., 1997; ‡Yun et al., 2001; §Ebbing et al., 2008; ¶Song and Endow, 1998; \\Liu et al., 2012a).
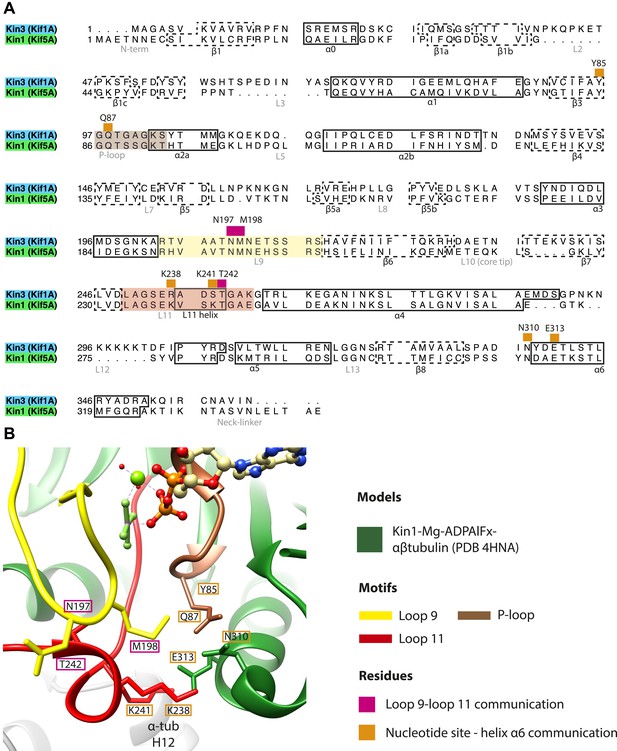
Structural routes of communication between the nucleotide-binding pocket and helix-α6 for mechanochemical coupling.
(A) Sequence alignment of Kin3 and Kin1 highlighting residues involved in communication from the nucleotide-binding pocket to helix-α6. Residues involved in loop9-loop11 communication are indicated by magenta squares above residue letters and loop11-helix-α6 communication by orange squares above residue letters. Residue numbers for Kin1 (Kif5A) are indicated. Loop9 is indicated by yellow shading, loop11 by red shading, and the P-loop by brown shading. (B) The crystal structure of tubulin dimer-bound Kin1-Mg-ADPAlFx (Kif5B; PDB 4HNA) focusing on the residues indicated in panel A. Residue numbers for Kif5A are indicated. The close association of loop9 and loop11 in ATP-like crystal structures (Chang et al., 2013; Gigant et al., 2013; Parke et al., 2010) involves backbone hydrogen bonds between loop9 Asn197 and loop11 Thr242, and also involves Met198. Residues in loop11 (Lys241, Lys238 in Kin1, Arg264 in Kin3) interact with the base of helix-α6 (Asn310, Glu313 in Kin1, Asn337, Glu340 in Kin3). P-loop residues in Kin1 (Tyr85, Gln87; Kin3 Tyr96, Gln98) also interact with helix-α6. We propose that these interactions will form in the transition from NN to Mg-ATP bound (Figure 2) and will contribute to mechanical transmission (Figure 3).
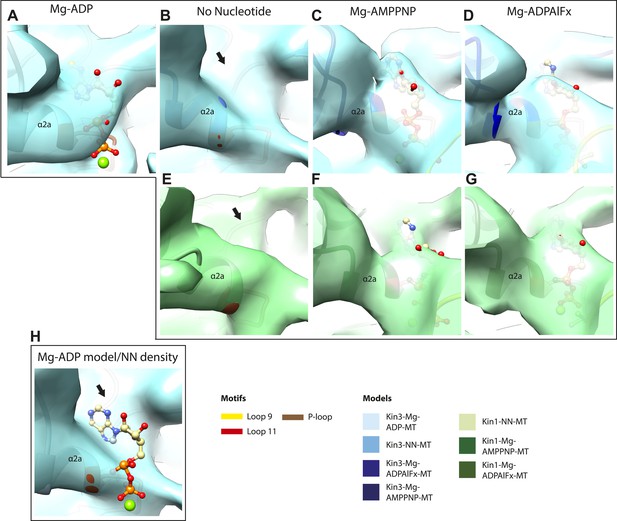
Occupancy of the nucleotide pocket.
Similar views of the nucleotide-binding pocket aligned on the P-loop are shown for each reconstruction, with the corresponding model fitted into density; (A) Kin3-Mg-ADP, (B) Kin3-NN, (C) Kin3-Mg-AMPPNP, (D) Kin3-Mg-ADPAlFx, (E) Kin1-NN, (F) Kin1-Mg-AMPPNP, (G) Kin1-Mg-ADPAlFx. The presence or absence of density in the nucleotide-binding pocket is consistent with the sample preparation used for each reconstruction and supports their interpretation. (H) The Kin3-Mg-ADP model is shown in the Kin3-NN reconstruction, clearly demonstrating the lack of density in the nucleotide-pocket to accommodate Mg-ADP (arrow) and supporting our assignment of this structure as nucleotide-free. The opacity of all reconstructions in this figure has been increased in order to more clearly illustrate the boundary of the EM density compared to the docked model. The contouring is the same as in all other figures.
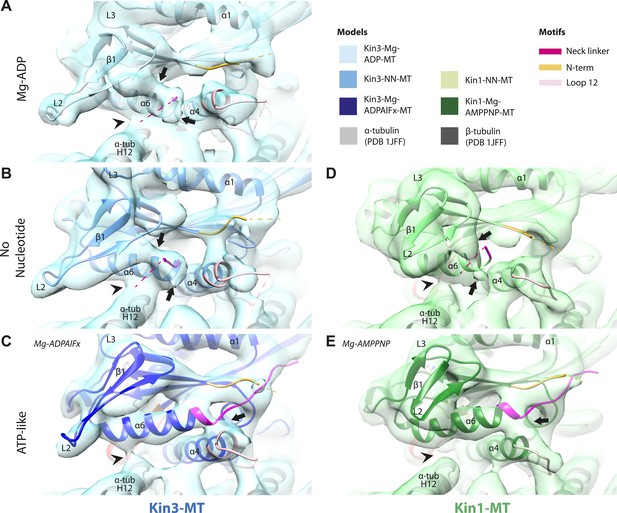
Conserved conformational changes of helix-α6 alter MT connectivity and allow neck linker docking on Mg-ATP binding.
(A–C) View of helix-α6 and the neck linker (in fuchsia) of MT-bound Kin3 reconstructions (shown as blue transparent density) in (A) Mg-ADP, model shown in light blue, (B) no nucleotide (NN), model shown in mid-blue, (C) Mg-ADPAlFx, model shown in dark blue; (D–E) View of helix-α6 and the neck linker (in fuchsia) of MT-bound Kin1 reconstructions (shown as green transparent density) in (D) no nucleotide, model shown in light green, (E) Mg-AMPPNP, model shown in dark green. In Mg-ADP (Kin3) and NN states (both motors), helix-α6 contacts the surface of α-tubulin (arrowhead) and its orientation with respect to helix-α4 ensures that the neck linker cannot dock. Regions of density at the C-terminal end of helix-α6 likely representing conformers of the N-terminal portion of the neck linker are observed (arrows), although the majority is not visible, presumably due to flexibility. In both motors, peeling of the motor domain β-sheet core away from helix-α4 upon Mg-ATP binding allows rotation and extension of helix-α6, drawing it away from the MT surface (arrowhead), and allowing it to occupy the space between helix-α4 and the β-sheet core. The neck linker docks towards the MT plus end (arrow) and forms the CNB with the N-terminus (in orange). In all reconstructions, density for the motor domain was contoured to an equivalent volume.
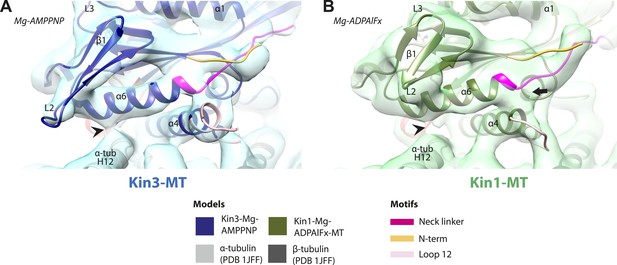
Conserved conformation of helix-α6 allows neck linker docking on Mg-ATP binding in Kin3 and Kin1 alternative ATP-like states.
(A) View of helix-α6 and the neck linker (in fuchsia) of MT bound Kin3-Mg-AMPPNP (blue transparent density and navy blue model). (B) View of helix-α6 and the neck linker (in fuchsia) of MT bound Kin1-Mg-ADPAlFx reconstruction (green transparent density and olive green model). The major features are shared by all the ATP-like reconstructions: in both motors, peeling of the motor domain β-sheet core on Mg-ATP binding allows rotation and extension of helix-α6, drawing it away from the MT surface (arrowhead). The neck linker docks towards the MT plus end (arrow) and forms the CNB with the N-terminus (in orange). In all reconstructions, density for the motor domain was contoured to an equivalent volume.
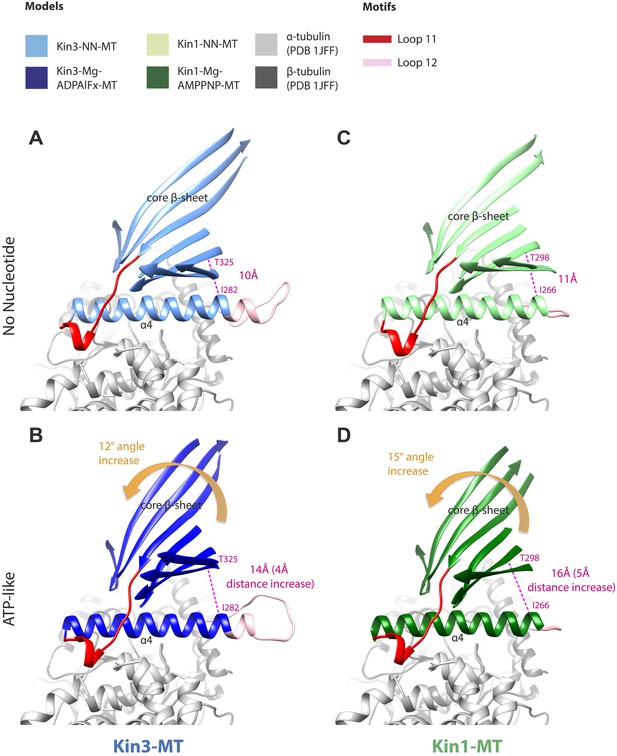
Tilting of the core β-sheet on Mg-ATP binding in Kin1 and Kin3 causes peeling of the β-sheet from the C-terminus of helix-α4 to allow movement and extension of helix-α6 and neck linker docking.
In each panel, a stripped-down depiction of each pseudo-atomic model is presented showing helix-α4, adjacent loops (shown for orientation) and the core β-sheet, viewed from the MT minus end. (A) MT bound Kin3-NN; (B) MT bound Kin3-ATP-like; (C) MT bound Kin3-NN; (D) Kin-ATP-like. In each case, the distance between the backbone Cα of conserved residues at the helix-α4 C-terminus and the immediately overlying β-sheet region were measured in Chimera (indicated in pink). The tilt of each β-sheet upon ATP-analogue binding was calculated by measuring the change in angle between helix-α4 and the β-sheet using the Axes/Planes/Centroids tool in Chimera.
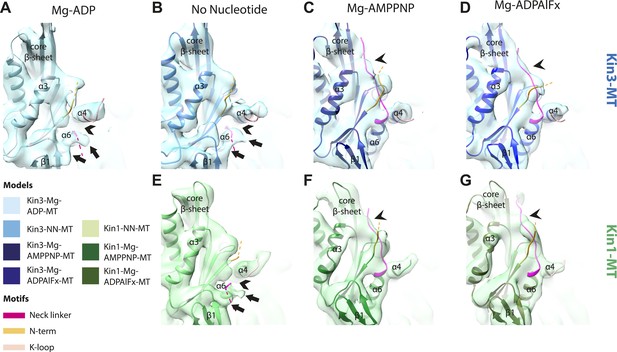
Conserved conformational changes of helix-α6 relative to helix-α4 control neck-linker docking along the core β-sheet when Mg-ATP binds.
(A–D) View towards the MT with the plus end towards the top of MT-bound Kin3 reconstructions (shown as blue transparent density) in (A) Mg-ADP, model shown in light blue, (B) no nucleotide (NN), model shown in the mid-blue, (C) Mg-AMPPNP, model shown in navy blue, and (D) Mg-ADPAlFx, model shown in dark blue; (E–G) Same view of MT-bound Kin1 reconstructions (shown is green transparent density in E) no nucleotide (NN), model shown in light green, (F) Mg-AMPPNP, model shown in dark green, (G) Mg-ADPAlFx, model shown in olive green. In Mg-ADP/NN states of Kin3 (A and B) and the NN state of Kin1 (E) helix-α6 terminates before helix-α4 leaving a gap (chevrons). Additional regions of density (arrows) at the helix-α6 C-terminus likely represent conformers of the initial portion of the neck linker (fuchsia), most of which is invisible and presumably flexible. However, in AMPPNP/ADPAlFx states of both Kin3 (C and D) and Kin1 (F and G), tilting of the motor domain allows helix-α6 to extend, closing the gap between helix-α4 and allowing neck linker docking, for which extra density is seen alongside the core β-sheet (arrowheads). Neck linker docking allows CNB formation with the N-terminus (orange). In all reconstructions, density for the motor domain was contoured to an equivalent volume.
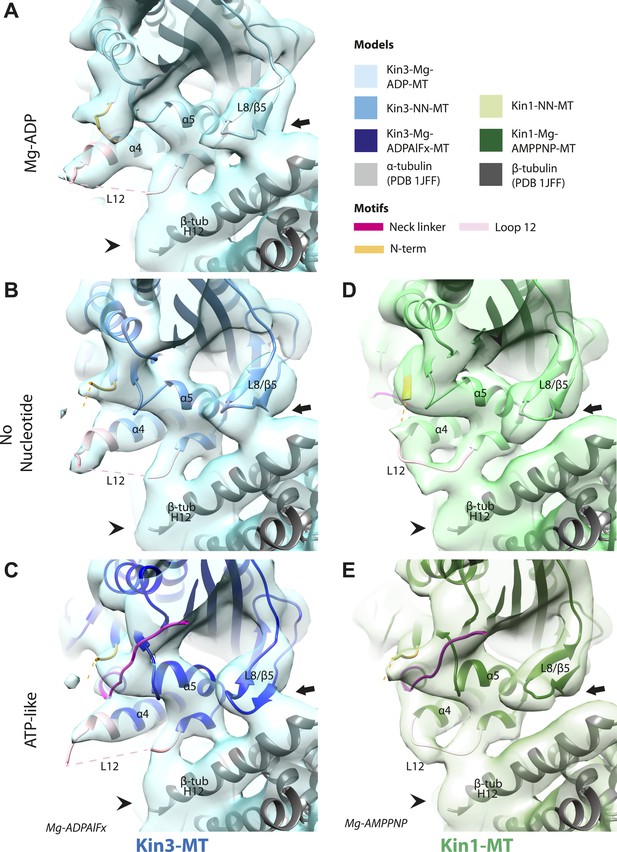
Nucleotide-independent interactions between the kinesin motor domain and the MT surface.
(A–C) View from the MT plus end of the motor domain-MT interface in MT-bound Kin3 reconstructions (shown as blue transparent density) in (A) Mg-ADP, model shown in light blue, (B) no nucleotide (NN), model shown in mid-blue, (C) Mg-ADPAlFx, model shown in dark blue, in which the CNB is formed between the neck linker (fuchsia) and N-terminus (orange). The N-terminus of loop12 (light pink) extends helix-α4 by a turn but the central, lysine-rich portion of this loop is not visible (dotted pink line), nor is the β-tubulin CTT (arrowhead) with which it is known to interact. Loop8/strand-β5 forms a clear connection to the MT surface (arrow). (D–E) The same view of the motor domain-MT interface in MT-bound Kin1 reconstructions (shown as green transparent density) in (D) no nucleotide, model shown in light green, (E) Mg-AMPPNP, model shown in dark green, in which the CNB is formed between the neck linker (fuchsia) and N-terminus (orange). The shorter Kin1 loop12 is clearly visualised and contacts the MT surface while loop8/strand-β5 are not connected by density to the MT surface (arrow). In all reconstructions, density for the motor domain was contoured to an equivalent volume.
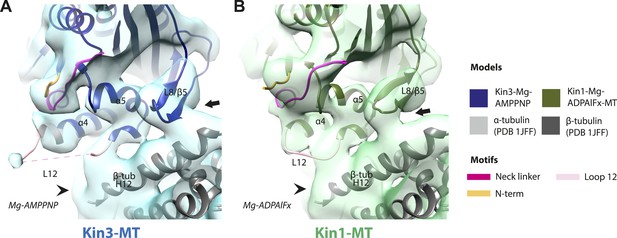
Conserved conformations at the kinesin motor domain and the MT surface in Kin3 and Kin1 alternative ATP-like states.
(A) View from the MT plus end of the motor domain-MT interface in the MT bound Kin3-Mg-AMPPNP (blue transparent density and navy blue model). (B) View from the MT plus end of the motor domain-MT interface in the MT bound Kin1-Mg-ADPAlFx reconstruction (green transparent density and olive green model). The major features are shared by all the ATP-like reconstructions: the CNB is formed between the neck linker (fuchsia) and N-terminus (orange). The N-terminus of loop12 (light pink) extends helix-α4 by a turn but the central, lysine-rich portion of this loop is not visible (dotted pink line), nor is the β-tubulin CTT (arrowhead) with which it is known to interact. Loop8/strand-β5 forms a clear connection to the MT surface (arrow). The Kin1-Mg-ADPAlFx reconstruction is lower resolution (FSCtrue, 0.143 = 7.7), which may explain why residual density connects Loop8/strand-β5 and the MT surface, which is not the case in the Kin1-Mg-AMPPNP reconstruction (Figure 4E). In all reconstructions, density for the motor domain was contoured to an equivalent volume.
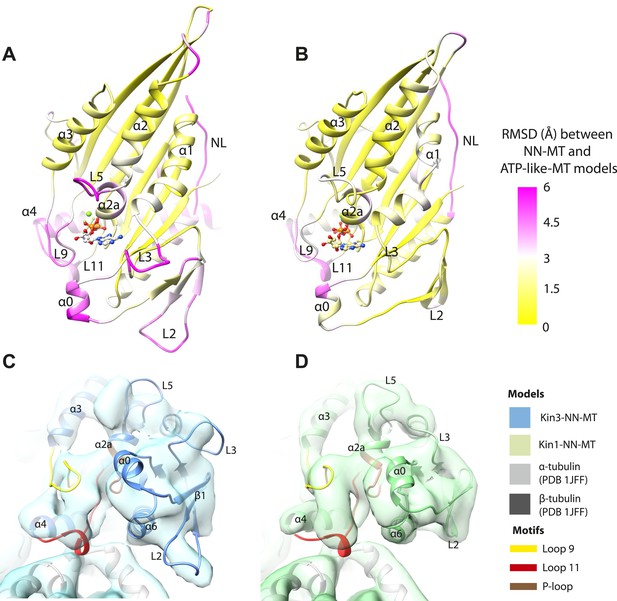
Transmission of force generation across the motor domain on Mg-ATP binding.
(A and B) Conformational changes relative to superposition of the core β-sheet of Kin3 (A) and Kin1 (B) showing the RMSDs due to Mg-ATP binding coloured from yellow (no change) to pink (large change), depicted on the Mg-ATP-like structures. Note, because the core β-sheet moves relative to helix-α4, which is held at the MT interface, alignment of the β-sheet artificially shows large displacements of helix-α4 and other nucleotide-invariant MT contacts at the back of this view. (C and D) Comparison of the nucleotide-binding site before and after Mg-ATP binding in Kin3 (C) and Kin1 (D). In each case, the NN model is depicted within the Mg-ATP cryo-EM density and shows that the regions of the largest RMSDs (pink in panels A and B) correspond to regions of the models that clearly do not fit in the density, that is, that undergo conformational changes when Mg-ATP binds.
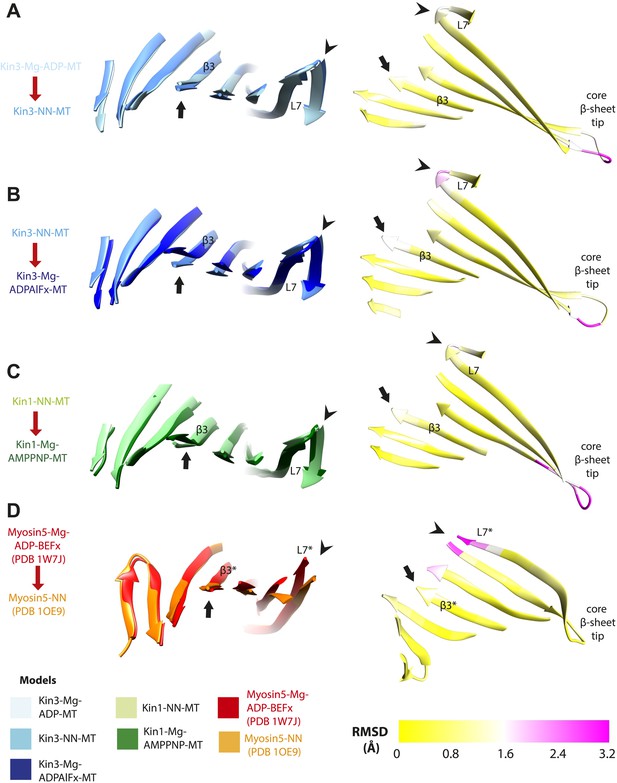
Limited β-sheet flexure during kinesin ATPase cycle compared to myosin5.
Superposition of the core β-sheets of motor domains in different nucleotide states reveals subtle differences at their edges, indicating β-sheet flexure at each transition. On the left of each panel, the core β-sheets of (A) Kin3-Mg-ADP-MT and Kin3-NN-MT, (B) Kin3-NN-MT and Kin3-Mg-ADPAlFx-MT, (C) Kin1-NN-MT and Kin1-Mg-AMPPNP-MT models are shown superimposed, viewed from the MT minus end. (D) For comparison Myosin5-NN (PDB 1OE9) and Myosin5 Mg-ADP-BeFx ATP-like (PDB 1W7J) crystal structures are shown superimposed, where β-sheet flexure has been shown to occur (Coureux et al., 2003; Reubold et al., 2003). Arrowheads indicate the tip of loop7 and arrows indicate strand-β3 (which connects to the P-loop), or the structurally equivalent region in the Myosin motor domain (indicated with *). On the right of each panel, the corresponding RMSDs of each overlay are shown, displayed using a scale from 0 (yellow) to pink (3.2 Å). The motor domain MT minus end is to the left and plus end, that contains the flexible loop10, to the right. (A) Kin3 Mg-ADP release: maximum loop7 RMSD ∼1.6 Å; (B) Kin3 Mg-ATP binding: loop7, RMSD ∼2.5 Å, strand-β3: RMSD ∼1.7 Å; (C) Kin1 Mg-ATP binding: loop7, RMSD ∼1.8 Å, strand-β3: RMSD ∼1.2 Å; (D) Myosin5 Mg-ADP release: loop7* maximum RMSD ∼3.3 Å (Coureux et al., 2003; Reubold et al., 2003).
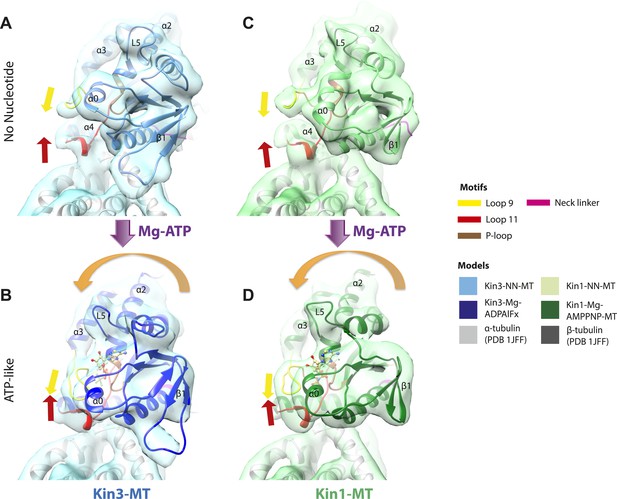
Pincer-like closure of loop9 and loop11 contributes to motor domain tilt when ATP binds.
(A) MT binding and Mg-ADP release in the Kin3-NN-MT, viewed from the MT minus end, induce an ordered loop9 and loop11 conformation; (B) ATP-binding induces loop9 and loop11 to move together contributing to motor domain tilting towards the bound nucleotide, thereby enabling neck linker docking. (C and D) The same conformational changes are seen in Kin1. Red and yellow arrows represent the ‘pincer’-like movement of loop9 and loop11 towards each other that produces the new density connection between them. Tilting of the motor domains relative to helix-α4 is indicated with orange-curved arrows.
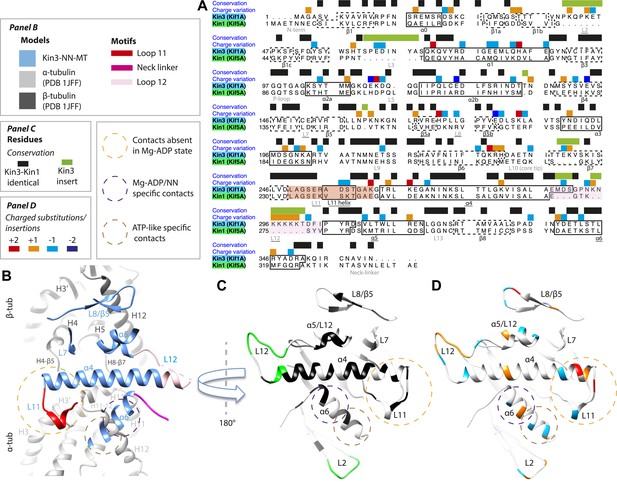
Comparison of Kin3 and Kin1.
(A) Sequence alignment of Kin3 (Kif1A) and Kin1 (Kif5A) motor domains showing secondary structural elements within the domains, annotated according to sequence and charge conservation. Elements depicted in other panels are underlined. (B) Longitudinal slice through the Kin3-NN model viewed from the front showing the MT contact elements and the underlying structural regions in αβ-tubulin. (C) MT binding surface of Kin3-NN model viewed from the MT surface (180° rotated compared to B) annotated by sequence identity (black) between Kin3 and Kin1 and sequence insertions (green). Structural elements in the MT are removed in this view to most clearly show elements in the motor domain. (D) MT binding surface of Kin3-NN model showing the differences in charge (blue: Kin3 more acidic than Kin1; red: Kin3 more basic than Kin1); same view as in C.
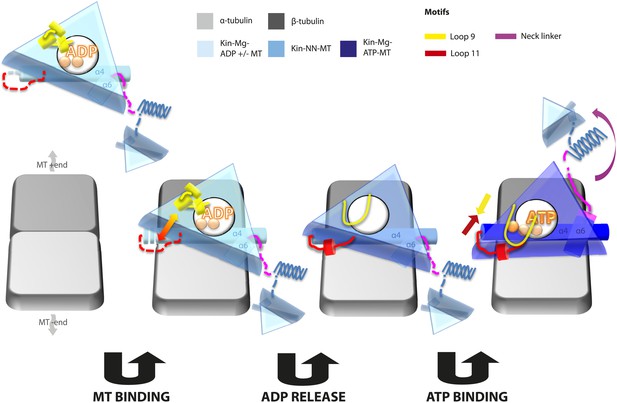
Model of conserved MT-bound kinesin mechanochemistry.
Loop11/N-terminus of helix-α4 is flexible in ADP-bound kinesin in solution, the neck linker is also flexible while loop9 chelates ADP. MT binding is sensed by loop11/helix-α4 N-terminus, biasing them towards more ordered conformations. We propose that this favours crosstalk between loop11 and loop9, stimulating ADP release. In the NN conformation, both loop11 and loop9 are well ordered and primed to favour ATP binding, while helix-α6—which is required for mechanical amplification–is closely associated with the MT on the other side of the motor domain. ATP binding draws loop11 and loop9 closer together; causing (1) tilting of most of the motor domain not contacting the MT towards the nucleotide-binding site, (2) rotation, translation, and extension of helix-α6 which we propose contributes to force generation, and (3) allows neck linker docking and biases movement of the 2nd head towards the MT plus end.
Tables
Data set size and estimated reconstruction resolutions
| Kinesin and nucleotide state | Number of AU | FSCt 0.5 (0.143) | FSCtrue 0.5 (0.143) | Rmeasure 0.5 (0.143) | EMDB accession number |
|---|---|---|---|---|---|
| Kin3-Mg-ADP | 181,311 | 7.9 (6.3) | 8 (7) | 8.1 (7.5) | EMD-2768 |
| Kin3-NN | 187,538 | 7.4 (6.3) | 7.5 (6.3) | 7.8 (6.9) | EMD-2765 |
| Kin3-Mg-AMPPNP | 97,877 | 8.1 (6.9) | 8.2 (7.0) | 8 (7.3) | EMD-2766 |
| Kin3-Mg-ADPAlFx | 156,845 | 7.9 (6.8) | 8.3 (7.0) | 8 (7.3) | EMD-2767 |
| Kin1-NN | 168,974 | 8.2 (7.2) | 8.3 (7.4) | 8.3 (7.3) | EMD-2769 |
| Kin1-Mg-AMPPNP | 186,329 | 7.3 (6.0) | 7.5 (6.5) | 7.7 (6.9) | EMD-2770 |
| Kin1-Mg-ADPAlFx | 65,572 | 9 (7.3) | 9.1 (7.7) | 9.1 (8.1) | EMD-2771 |
-
For each reconstruction, the motor domain and nucleotide state, number of asymmetric units (AU) in the final reconstruction, the resolutions at a cut-off of 0.5 and 0.143 estimated by standard FSC (FSCt) and that corrected with the HRnoise substitution test (FSCtrue) (Chen et al., 2013) and by Rmeasure (Sousa and Grigorieff, 2007) and the EMDB accession number are given.
Calculation of pseudo-atomic models
| Kinesin and nucleotide state | Models used | CCC initial model | CCC final model | PDB code |
|---|---|---|---|---|
| Kin3-Mg-ADP | 1VFZ (Nitta et al., 2004) | 0.66 | 0.68 | 4uxs |
| 1I5S (Kikkawa et al., 2001 | ||||
| 4AQW (Goulet et al., 2012) | ||||
| Kin3-NN | 1VFZ/1I5S/4HNA (Gigant et al., 2013)/4AQW | 0.63 | 0.68 | 4uxo |
| Kin3-Mg-AMPPNP | 1VFV (Nitta et al., 2004) | 0.72 | 0.75 | 4uxp |
| 4HNA | ||||
| Kin3-Mg-ADPAlFx | 1VFV/4HNA | 0.74 | 0.75 | 4uxr |
| Kin1-NN | 1BG2 (Kull et al., 1996)/4HNA/4AQW | 0.71 | 0.73 | 4uxt |
| Kin1-Mg-AMPPNP | 4HNA | 0.73 | 0.76 | 4uxy |
| Kin1-Mg-ADPAlFx | 4HNA | 0.69 | 0.72 | 4uy0 |
-
A set of starting models were used for each nucleotide state of each motor. Flexible fitting and further refinement were performed using Flex-EM and Modeller (see ‘Materials and methods’). Global CCCs of models with their respective reconstructions were calculated using the Fit In Map tool in Chimera. PDB accession codes for the final models are also shown.
Steady-state MT-activated ATPase parameters of our Kin3 and Kin1 motor domain constructs
| Kin3 (Kif1A) | Kin1 (Kif5A) | |
|---|---|---|
| kcat (s−1) | 43.4 ± 1.0 | 34.2 ± 5.7 |
| K0.5ATP (μM) | 30 ± 10 | 25 ± 5 |
| K0.5 MT (nM) | 53.7 ± 5.7 | 12,745 ± 4041 |
Final protein concentrations used for cryo-EM sample preparation
| Kinesin and nucleotide state | [MT] (μM) | [Motor domain] (μM) |
|---|---|---|
| Kin3 MgADP | 5 | 10 |
| Kin3 NN | 5 | 5 |
| Kin3 Mg-AMPPNP | 5 | 5 |
| Kin3 Mg-ADP.AlFx | 5 | 5 |
| Kin1 NN | 5 | 100 |
| Kin1 Mg-AMPPNP | 5 | 50 |
| Kin1 Mg-ADP.AlFx | 5 | 50 |
-
Kin1 samples required higher concentrations than Kin3 to achieve good MT occupancy.





