Methylation of histone H3K23 blocks DNA damage in pericentric heterochromatin during meiosis
Figures
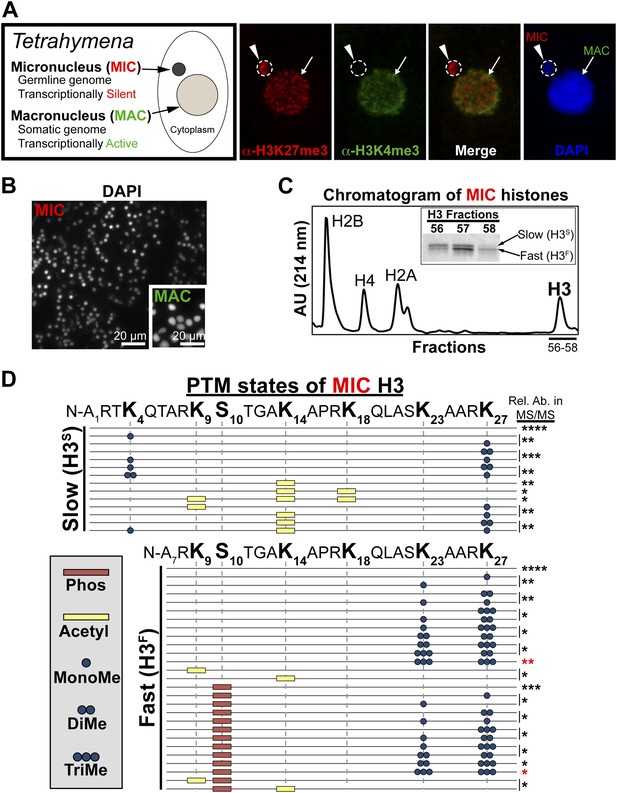
Purification of histone H3 from heterochromatic micronuclei and analysis of the associated PTM states.
(A) Co-immunofluorescence staining of Tetrahymena. (B) Following cell lysis, micronuclei were efficiently separated from macronuclei by differential centrifugation. Shown is a representative micronuclear fraction that was used in subsequent purification steps. (C) Chromatogram of micronuclear core histones resolved by RP-HPLC. Inset shows fractions containing two micronuclear histone H3 species, H3S (‘S’ for electrophoretically slow) and H3F (‘F’ for electrophoretically fast), which are stained with Coomassie following SDS-PAGE. (D) Summary of modified forms of micronuclear histone H3 N-terminal peptides (residues 1–50 for H3S and 7–50 for H3F) from vegetatively growing Tetrahymena as detected by HILIC-MS/MS. Sequences of H3S and H3F are shown up to K27 since no modifications were detected on residues 28–50. Each row in the table represents a single, discrete, and uniquely modified H3 N-terminal peptide, and each dotted horizontal line beneath a bolded residue represents a site where the specified modification was identified. The relative abundance of each peptide, or multiple isobaric peptides identified within the same spectrum (grouped by brackets on the right), is highlighted by the number of asterisks. For example, peptides with four asterisks are more abundant than peptides with one asterisk. Forms of the H3S(1–50) peptide with four, five, and six methyls were detected, but due to the isobaric character of each group of peptides and the low levels of these species it was not possible to determine the locations of the methyl modifications. See also Figure 1—figure supplement 1 for supporting antibody-based validation of the presence of H3K23 mono- and dimethylation in Tetrahymena, and Figure 1—source data 1 for supporting mass spectrometry data. Abbreviations: MonoMe, monomethylation; DiMe, dimethylation; TriMe, trimethylation.
-
Figure 1—source data 1
Shown are the tandem mass spectrometry data for the N-terminus of histone H3 in the unmodified, and dually modified H3K23me3/H3K27me3 states.
- https://doi.org/10.7554/eLife.02996.004

Presence of H3K23me1 and H3K23me2 in Tetrahymena.
(A) Western blot analysis of micronuclear and macronuclear acid-extracts and (B) indirect immunofluorescence staining using commercially available antibodies that recognize mono- or dimethylated H3K23. Black arrowheads (in part A) point to H3F.
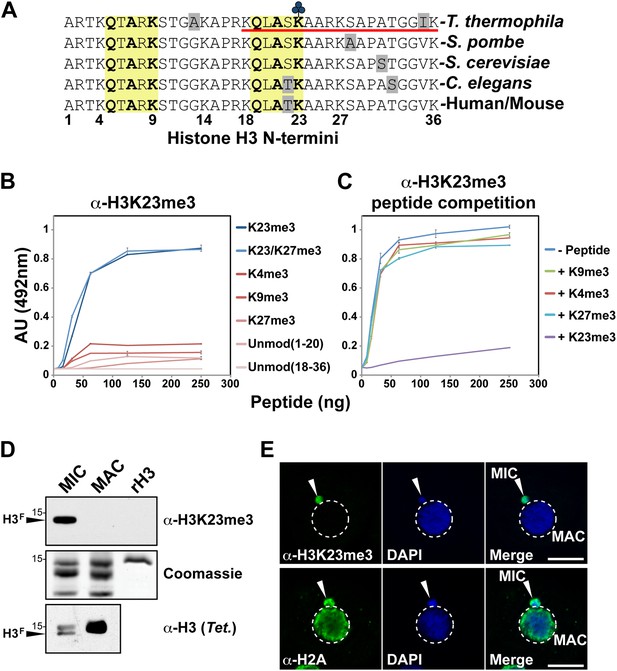
Development of an H3K23me3-specific antibody.
(A) Alignment of the first 36 residues of histone H3 from the indicated organisms. An H3 peptide spanning amino acids 18–36 (red underline) and trimethylated on K23 (blue clover) were synthesized and used as the immunogen for antibody generation in rabbits. Yellow boxes highlight the conserved QXAXK motif associated with H3K9 and H3K23. (B) ELISA analysis of α-H3K23me3 binding to the indicated peptides. See also Figure 2—figure supplement 1. (C) ELISA analysis of α-H3K23me3 binding to H3K23me3(18–36) peptide in the presence of excess competing peptides. (B–C) Data points represent mean ± SD of triplicate samples from a representative experiment. (D) Western blot analysis of α-H3K23me3 specificity using micronuclear and macronuclear extracts, and recombinant human H3 (rH3). (E) Indirect immunofluorescence staining of vegetatively growing Tetrahymena using either α-H3K23me3 or α-H2A (control). White arrowheads point to micronuclei. Scale bar, 10 μm.
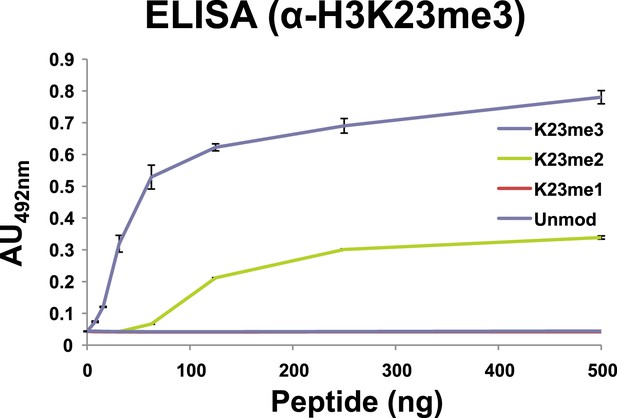
Affinity of α-H3K23me3 for distinct methylation states of H3K23.
ELISA analysis of α-H3K23me3 binding to H3unmod, H3K23me1, H3K23me2, and H3K23me3 peptides. Note the preference of α-H3K23me3 for the trimethylated epitope. Data points represent mean ± SD of triplicate samples from a single, representative experiment.
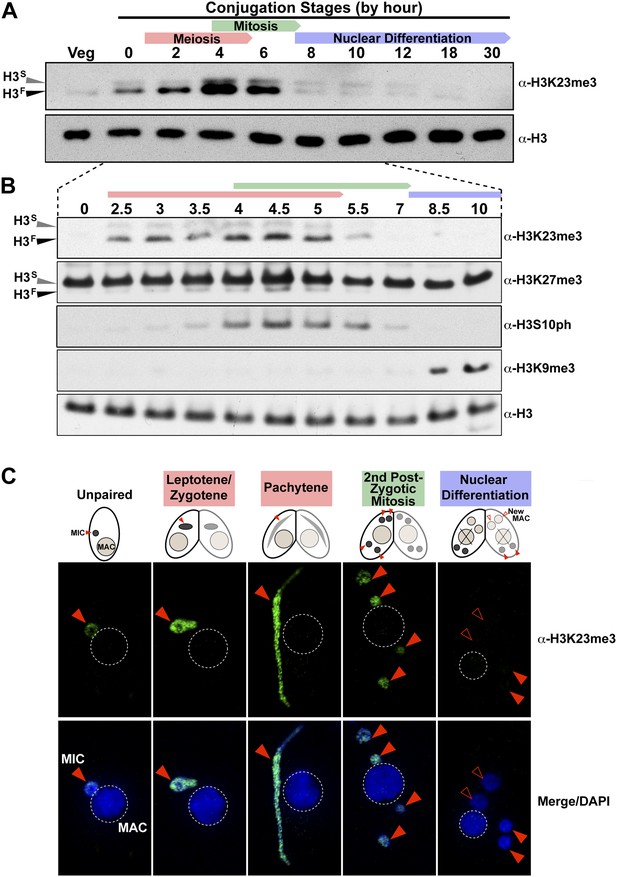
H3K23me3 levels are dramatically increased during early meiosis.
(A and B) Whole-cell-extracts of wild type cells were collected at the indicated time points during conjugation, resolved on SDS-PAGE gels, and analyzed by Western blotting. Color-coded arrows highlight the period of the specified developmental stage. See also Figure 3—figure supplement 1A. (C) Indirect immunofluorescence analysis of conjugating Tetrahymena using α-H3K23me3. Red arrowheads point to micronuclei. During nuclear differentiation, the parental macronucleus (labeled with ‘X’ in the cartoon) degrades while two post-zygotic nuclei begin to differentiate to new macronuclei (open red arrowheads) and the other two post-zygotic nuclei remain as micronuclei. See also Figure 3—figure supplement 1B.
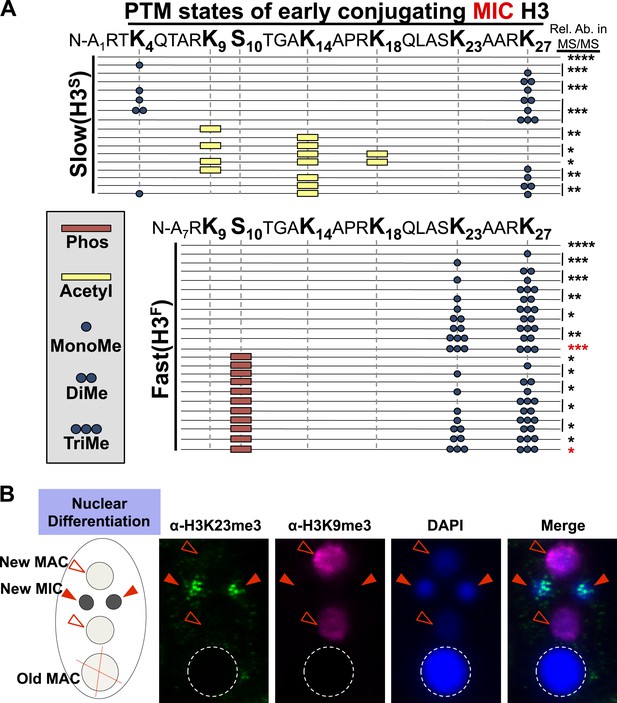
Combinatorial modification states of micronuclear H3 from conjugating Tetrahymena.
(A) Summary of modified forms of histone H3 N-terminal peptides, as detected by HILIC-MS/MS, from Tetrahymena micronuclei of hour 5 conjugating cells. As in main Figure 1D, each row represents a uniquely modified H3 N-terminal peptide species (residues 1–50 for H3S and 7–50 for H3F), and each dotted horizontal line beneath a bolded residue represents a site where the specified modification was identified. No modifications were detected on residues 28–50 on H3S and H3F. The relative abundance of each peptide, or multiple isobaric peptides identified within the same spectrum (grouped by brackets on the right), is highlighted by the number of asterisks. For example, peptides with four asterisks are more abundant than peptides with one asterisk. Red asterisks highlight the dually modified H3K23me3/H3K27me3 state enriched during early conjugation. Forms of the H3S(1–50) peptide with four, five, and six methyls were detected, but due to the isobaric character of each group of peptides and the low levels of these species it was not possible to determine the locations of the methyl modifications. Also, an H3F(7–50) peptide with one acetylation and one phosphorylation modification was detected by accurate mass, but no MS/MS spectra were acquired and therefore location of these modifications could not be determined. Abbreviations: MonoMe, monomethylation; DiMe, dimethylation; TriMe, trimethylation; Acetyl, acetylation; Phos, phosphorylation. (B) Co-immunofluorescence staining of conjugating Tetrahymena (at nuclear differentiation stage) using α-H3K23me3 and α-H3K9me3 antibodies. Red arrowheads point to micronuclei and open red arrowheads point to new developing macronuclei. The parental macronucleus (Old MAC) is marked by an ‘X’ in the cartoon and by dotted circles in the immunofluorescence images.
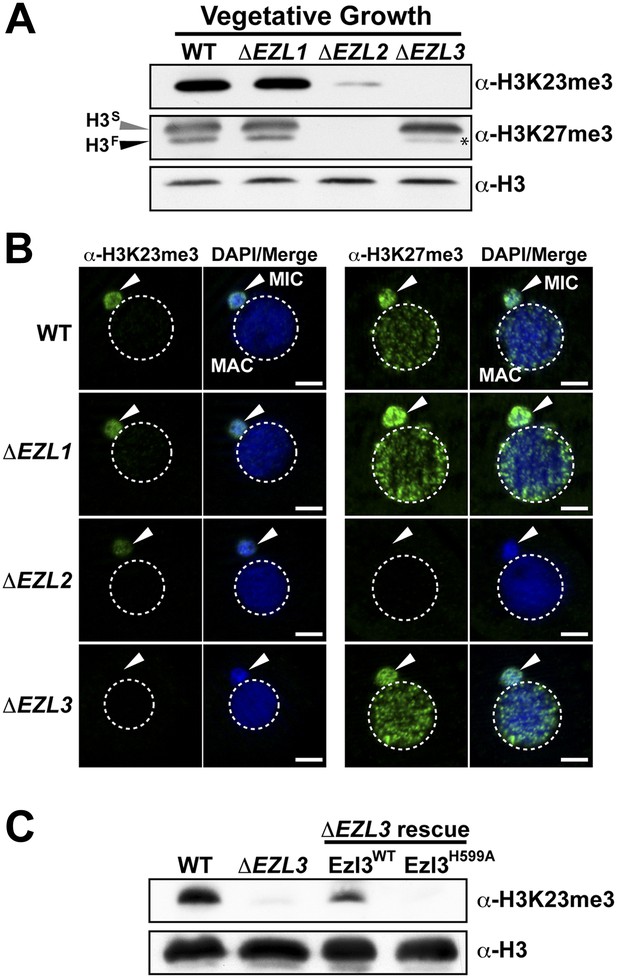
An Enhancer of zeste homolog is required for H3K23 methylation in Tetrahymena.
(A) Western blot analysis of whole-cell-extracts and (B) indirect immunofluorescence staining of wild type (WT), ΔEZL1, ΔEZL2, and ΔEZL3 cells grown under vegetative conditions. The asterisk highlights the reduction of H3FK27me3 in ΔEZL3 cells. White arrowheads point to micronuclei. Scale bar, 5 μm. (C) Ezl3p expression was reconstituted in ΔEZL3 cells. As a control, a catalytically inactive mutant generated by a His (599) to Ala mutation was included. See also Figure 4—figure supplement 1.
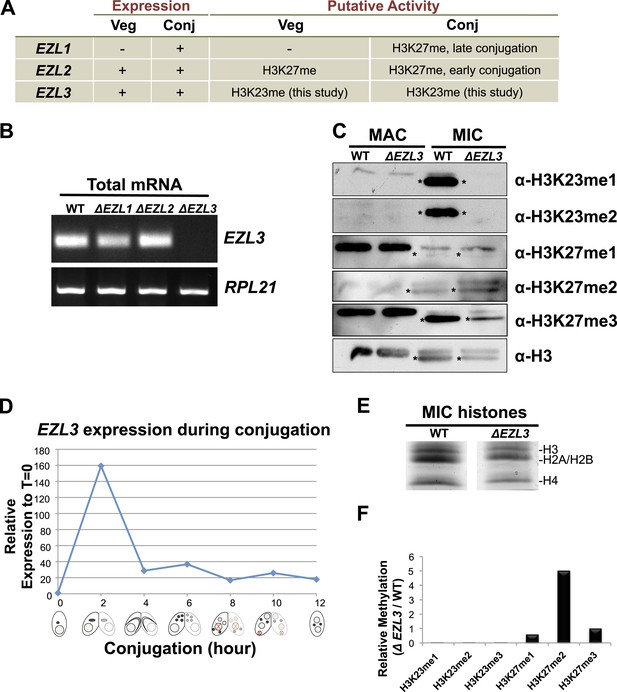
Characterization of EZL3 in vegetatively growing and conjugating Tetrahymena.
(A) Expression and putative enzyme activity of Ezl (Enhancer of zeste-like) proteins during vegetative growth and conjugation. (B) Total mRNA from the indicated cells (growing vegetatively) was purified and analyzed for the presence of EZL3 mRNA. RPL21 is a constitutively transcribed ribosomal gene used as a positive control. (C) Western blot analysis of micronuclear and macronuclear acid-extracts from vegetatively growing wild type and ΔEZL3 cells. Asterisks point to the location of H3F. (D) EZL3 expression during conjugation as determined by qPCR. Normalization was carried out by comparison to RPL21 expression, which is constitutive throughout conjugation. EZL3 transcript levels were plotted relative to their levels at time 0. This trend is similar to the one reported in the Tetrahymena gene expression database (http://tfgd.ihb.ac.cn/; gene ID# TTHERM_00499660). (E) MIC histones from wild type or ΔEZL3 cells were resolved by SDS-PAGE and visualized by Coomassie staining. These samples were used for quantitative mass spectrometry in Figure 4—figure supplement 1F. (F) Quantitative mass spectrometric analysis of histone lysine methylation levels identified a loss of H3K23 methylation and increase in H3K27me2 upon deletion of EZL3.
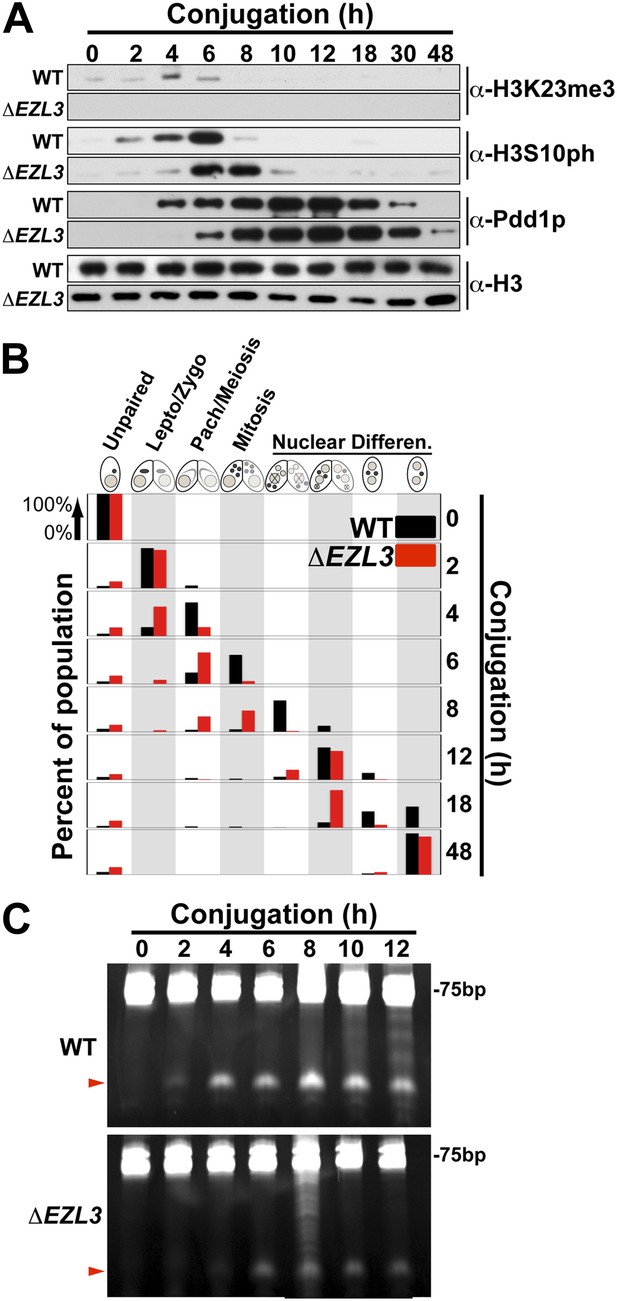
Meiotic progression is disrupted in Tetrahymena lacking Ezl3p and H3K23 methylation.
(A) Western blot analysis of whole-cell-extracts of wild type and ΔEZL3 cells progressing through conjugation. H3S10ph and Pdd1p were used as markers of early and late conjugation, respectively. (B) Conjugating wild type (black) and ΔEZL3 (red) cells was fixed at the indicated time points and analyzed by microscopy. Cytological hallmarks associated with each developmental stage (cartoons on top) were used to assess their progression through conjugation. This analysis revealed a developmental lag in early meiotic ΔEZL3 cells (4 hr). (C) Total RNA, extracted from wild type and ΔEZL3 cells during conjugation, was resolved on denaturing acrylamide gels and stained with ethidium bromide. Red arrowheads point to the location of piRNA-like scanRNA. See also Figure 5—figure supplement 1 for additional ΔEZL3 phenotype information.
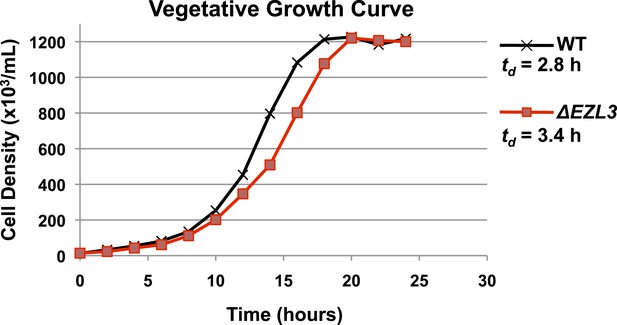
Vegetatively growing ΔEZL3 cells have a slow growth phenotype.
Growth curve of wild type and ΔEZL3 cells grown under vegetative conditions in SPP media at 30°C. Doubling time (td) was calculated by measuring the growth rate of each strain at mid-log phase. Experiments were performed in triplicates, and similar doubling times were obtained in each case.
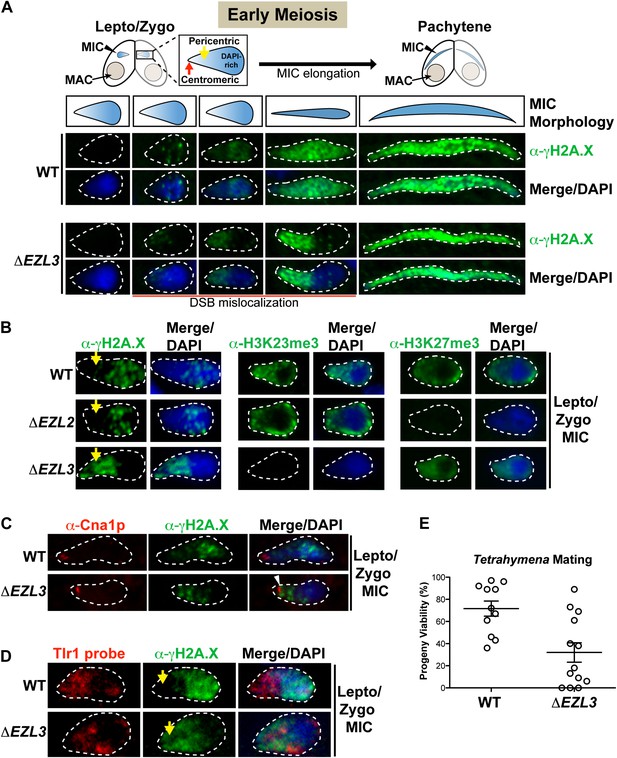
Loss of K3K23me3, but not H3K27me3, during meiosis leads to increased DSBs in pericentric heterochromatin.
(A) Cartoon depiction of the morphological changes to micronuclei during early meiotic prophase. At leptotene/zygotene, chromatin is spatially organized within the micronucleus with a DAPI-rich pole, a DAPI-poor pericentric region (yellow arrow), and a centromeric pole (red arrow). Below the illustrations are images of indirect immunofluorescence stains of leptotene/zygotene micronuclei using α-γH2A.X. Representative images are shown from at least three independent experiments. The micronuclei presented in this figure represent stages II–III/IV of meiotic prophase as reported previously (Sugai and Hiwatashi, 1974). (B) Indirect immunofluorescence staining of leptotene/zygotene micronuclei from wild type, ΔEZL2, and ΔEZL3 cells using α-γH2A.X, α-H3K23me3, and α-H3K27me3. Yellow arrows point to the pericentric region of meiotic micronuclei. (C) Images of leptotene/zygotene micronuclei from wild type and ΔEZL3 cells co-stained with α-Cna1p (centromeric H3) and α-γH2A.X. Note the white arrowhead at the tight, but non-overlapping junction between DSBs and the centromere in ΔEZL3 micronuclei. (D) ImmunoFISH staining using α-γH2A.X and a DNA probe against the pericentric marker Tlr1. See also Figure 6—figure supplement 1. (E) Percentage of genetic progeny resulting from mating of wild type lines vs isogenic ΔEZL3 lines. Shown in the graphs are mean ± SEM (p < 0.005 as calculated by a two-tailed t-test). See also Figure 6—source data 1 for the wild type and mutant crossing data, and Figure 6—figure supplement 2 for a model proposing how H3K23me3 may function during meiosis.
-
Figure 6—source data 1
Progeny viability is significantly reduced in ΔEZL3 cells.
Six independent ΔEZL3 mutant strains were generated in each of the B2086 and CU428 backgrounds (that are compatible for mating). Eight independent wild type lines were also generated in each of the B2086 and CU428 backgrounds. ΔEZL3 mutant lines were crossed to the other ΔEZL3 mutant lines of the opposite mating type. The proportions of viable progeny resulting from 11 wild type (B2086 × CU428) and 13 ΔEZL3 mating experiments are shown here and graphed in main Figure 6E.
- https://doi.org/10.7554/eLife.02996.015
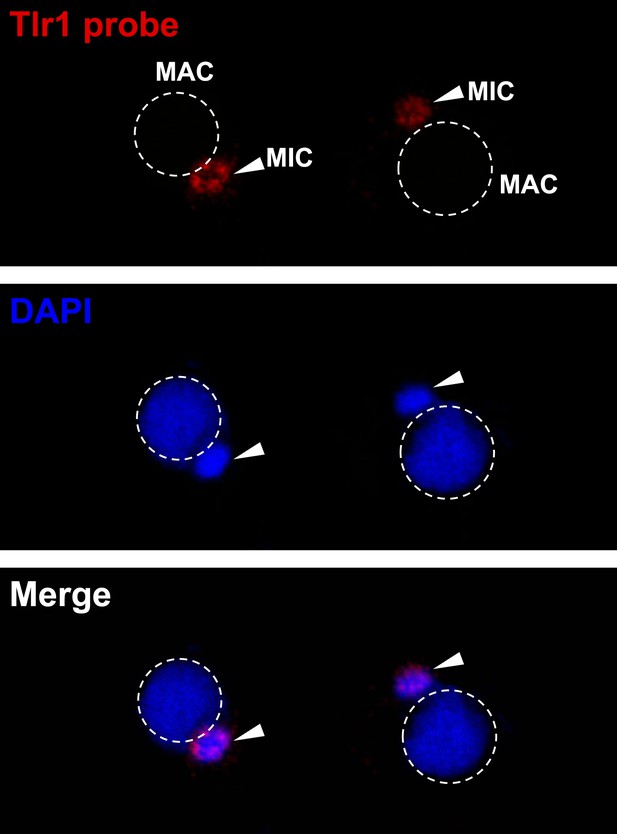
The Tlr1 probe is specific for germline chromatin.
Vegetative cells were starved overnight and subjected to FISH analysis using the Tlr1 probe.
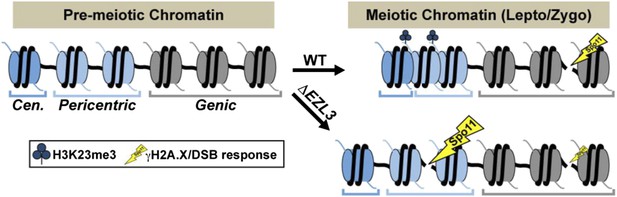
A model proposing that H3K23me3 mediates the formation of a higher-ordered heterochromatin structure to limit DSBs at pericentric sequences during early meiosis.
https://doi.org/10.7554/eLife.02996.017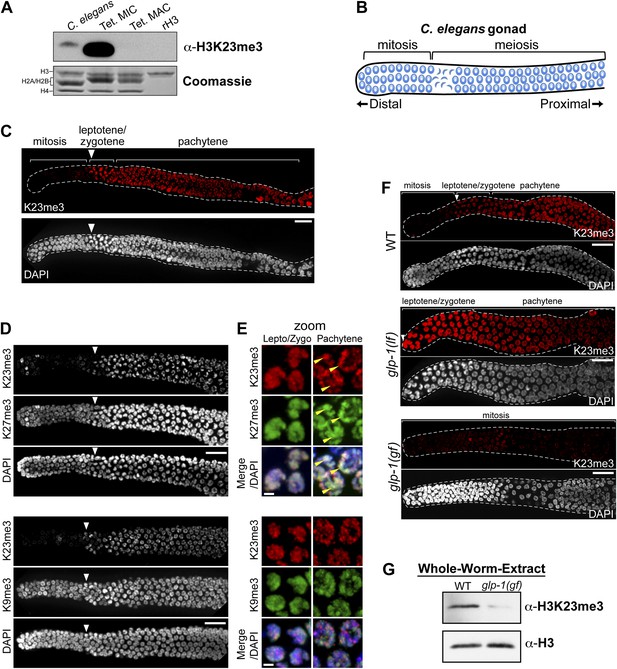
H3K23me3 is conserved in the C. elegans germline.
(A) Western blot analysis of nuclear acid-extracts using α-H3K23me3. C. elegans embryonic nuclei, Tetrahymena micro- or macronuclei, and recombinant human histone H3 (rH3) were resolved by SDS-PAGE and normalized by Coomassie staining. (B) Schematic of C. elegans adult hermaphrodite gonad. (C) Immunofluorescence staining of formaldehyde fixed C. elegans gonad stained with α-H3K23me3 and counterstained with DAPI. Scale bar, 10 μm (C, D, and F). White arrowheads point to the mitotic/meiotic transition zone (C, D, and F). (D) Co-immunofluorescence staining of the C. elegans gonad using α-H3K23me3 and either α-H3K9me3 or α-H3K27me3. (E) Zoomed images of germline nuclei co-stained with antibodies targeting the indicated H3 PTMs. Yellow arrowheads point to the location of the silenced X chromosomes. Scale bar, 2 μm. (F) H3K23me3 localization in loss-of-function and gain-of-function mutants (lf or gf, respectively) of the Notch-like receptor GLP-1. (G) Western blot analysis of whole-worm-extracts to evaluate total levels of H3K23me3 in glp-1(gf) worms with defective meiosis. See also Figure 7—figure supplement 1 for supporting information.
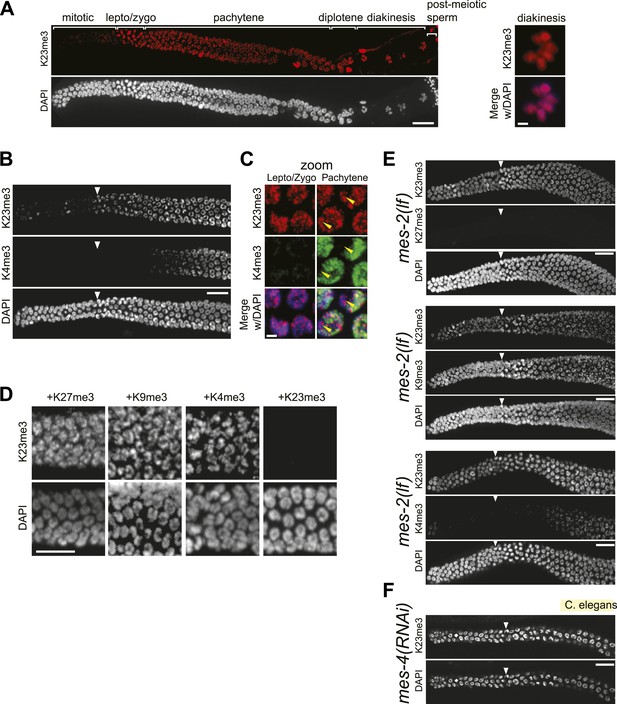
Conservation of H3K23me3 enrichment during meiosis in higher eukaryotes.
(A) An expanded view of the C. elegans gonad staining from main Figure 7C that includes germline nuclei undergoing diplotene and diakinesis (Scale bar, 10 μm). On the right, a zoomed image of a germline nucleus undergoing diakinesis (Scale bar, 3 μm). (B) Co-immunofluorescence staining using α-H3K23me3 and α-H3K4me3 antibodies in the C. elegans gonad. Scale bar, 10 μm. (C) Zoomed images (Scale bar, 2 μm) of early meiotic germline nuclei co-stained with α-H3K23me3 and α-H3K4me3. Yellow arrowheads point to the location of the silenced X chromosomes. White arrowheads indicate the region where meiotic (leptotene/zygotene) nuclei are detected (B, E, and F). (D) Worm germline immunostained with α-H3K23me3 antibodies preabsorbed with the indicated peptides (on top). Scale bar, 10 μm. (E–F) Immunofluorescence staining of H3K23me3 levels in the germline of C. elegans lacking MES-2 (E), the only worm homolog of Enhancer of zeste, and MES-4 (F). Scale bar, 10 μm.





