A conserved MCM single-stranded DNA binding element is essential for replication initiation
Figures
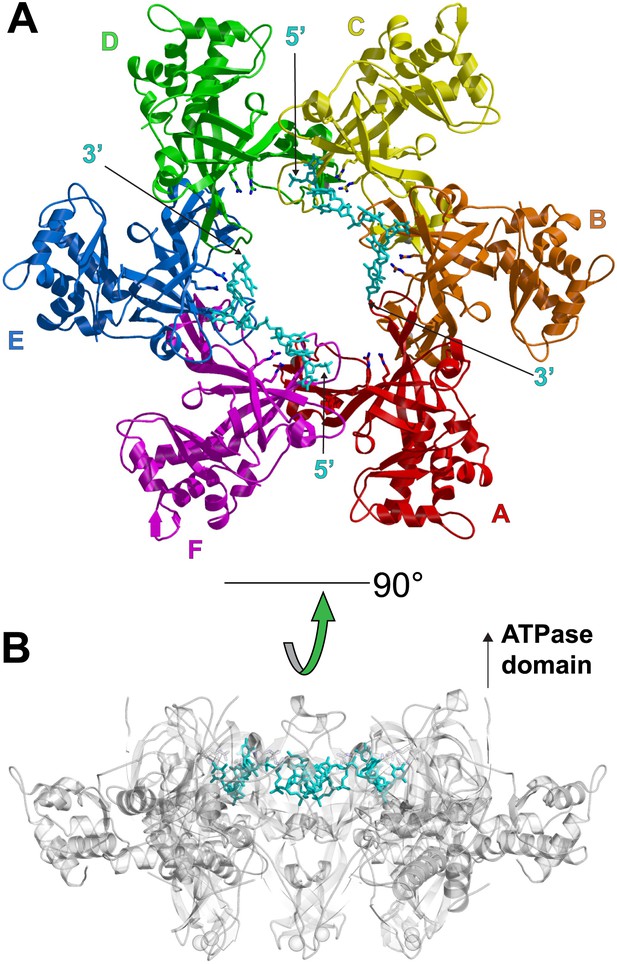
One crystallographically unique hexamer viewed parallel (A) and perpendicular (B) to the channel.
The ssDNA is colored cyan. (A) Each subunit is uniquely colored and labeled. The side-chains of the two MSSB arginine residues that bind ssDNA are represented in stick. The Zn-binding domains project into the page. The ATPase domains, not present in the crystal structure, would project out of the page. (B) The protein is represented in transparent grey to highlight that the ssDNA runs perpendicular to the channel. The Zn-binding domains are at the bottom, and the ATPase domains would be located at the top.
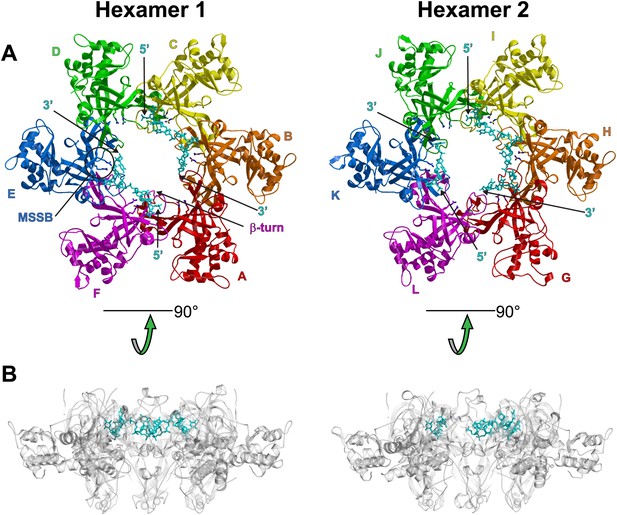
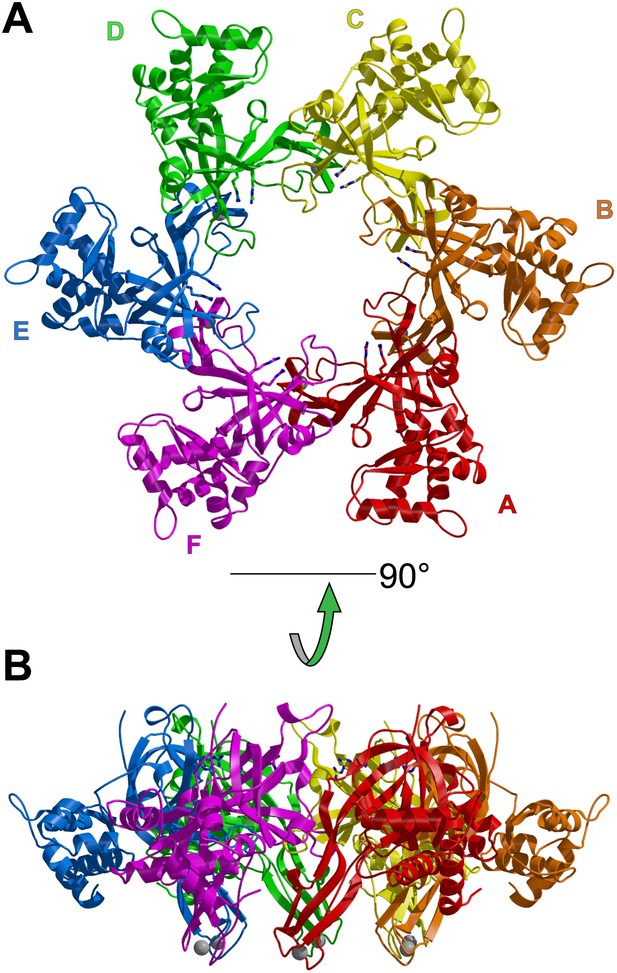
Views of the two hexamers of the crystallographic asymmetric unit parallel (A) and perpendicular (B) to the channel.
The ssDNA is colored cyan. (A) Each subunit is uniquely colored and labeled. For hexamer 1, an example MSSB and β-turn are labeled. The Zn-binding domains are projected into the page. The ATPase domains (not present in the crystal structure) would project out of the page. The 5′ and 3′ ends of the ssDNA are marked. (B) The protein is represented in transparent grey to highlight that the ssDNA runs perpendicular to the channel. The Zn-binding domains are at the bottom, and the ATPase domains (not present) would be at the top.
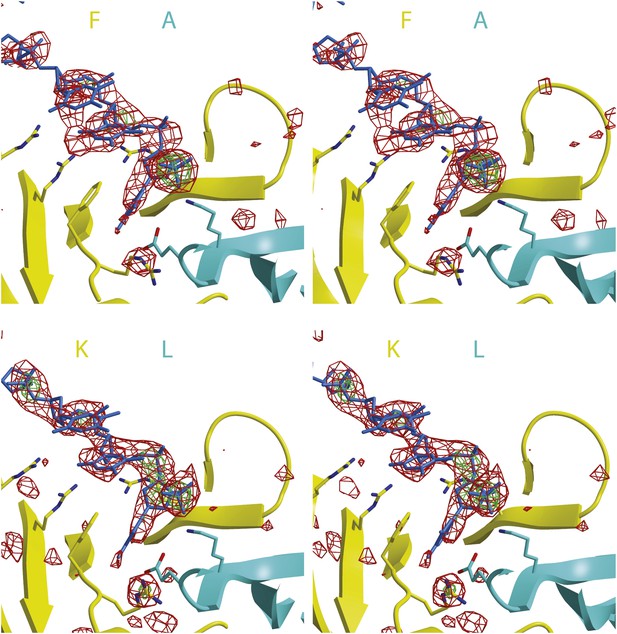
Stereoimages of one ssDNA binding PfMCMN subunit interface of each hexamer with Fo-Fc electron density calculated prior to including any DNA in the model.
The final model is displayed with the 2 subunits colored and labeled in yellow and cyan and the DNA colored blue. The Fo-Fc electron density is contoured at 3-sigma (red) and 5-sigma (green). The DNA backbone is visible at 3-sigma, and the phosphates are visible at 5-sigma.
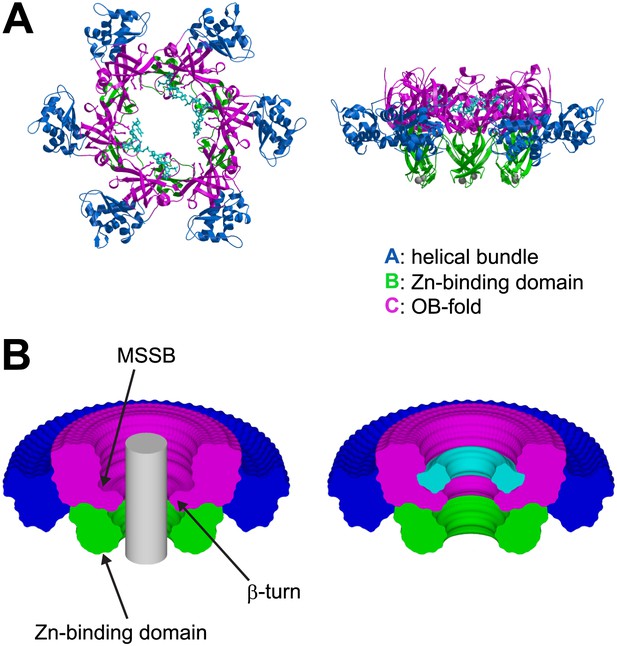
The ssDNA binds to the OB-fold subdomain.
(A) The individual subdomains are color-coded with the helical bundle in blue, the Zn-binding subdomain in green, the OB-fold subdomain in magenta, and the ssDNA in cyan. (B) Cylindrical merge showing how closely MCMN approaches the channel center at each position along the channel axis, and that the greatest available volume in the MCMN channel is at the OB-fold above the β-turn. The hexamer was rotated 360° about the channel axis in 5° increments. All of the models were superimposed, and the Cα positions of each subdomain were used to generate surfaces with MSMS (Sanner et al., 1996). The surfaces were uniquely colored as in (A), rendered simultaneously with Raster3D (Merritt and Bacon, 1997), and clipped with a vertical plane through the center to show the extent of projection into the channel for each part of the hexamer. A grey cylinder (unclipped) with 20 Å diameter was placed in the center to indicate the volume for a hypothetical B-form DNA. A similar 360° cylindrical merge was constructed for one of the contiguous ssDNA molecules, and a surface was constructed over all ssDNA atoms. The ssDNA surface was clipped with a vertical plane through the center, and is represented in cyan (right).
Crystal structure of PfMCMN in the absence of DNA viewed parallel (A) and perpendicular (B) to the channel.
Each subunit is uniquely colored and labeled. (A) The side-chains of the two arginine residues of the MSSB are represented in stick, and the Zn-binding domains are projected into the page. The ATPase domains, not present in the crystal structure, would project out of the page. (B) The Zn-binding domains are at the bottom, and the ATPase domains would be located at the top.
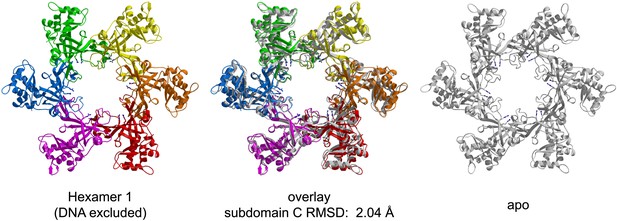
Comparison of the crystal structures of PfMCMN bound to ssDNA (left, in color) and in the absence of DNA (right, transparent grey).
The MSSB arginines are shown in stick representation. The two hexamers are superimposed based upon least-squares alignment of the six C-subdomains (middle). The oval shape of the ssDNA-bound ring is apparent at the red (chain A) and green (chain D) subunits, which are further from the channel center than in the DNA-free structure.
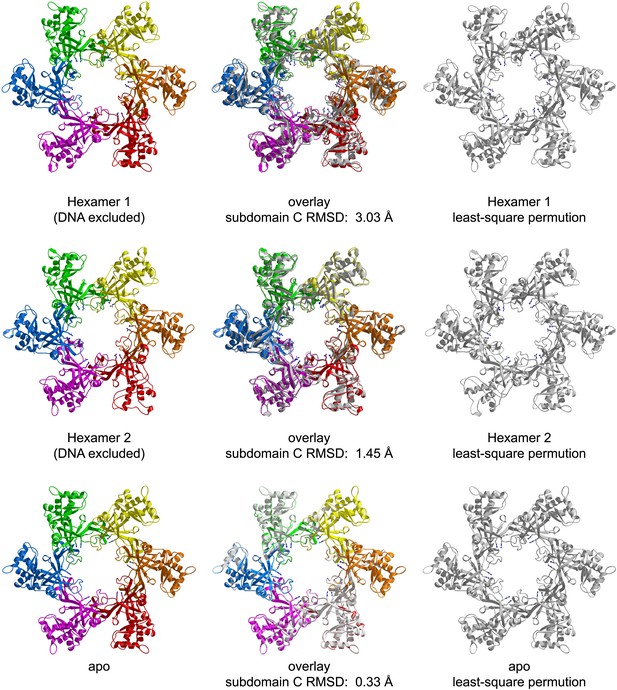
RMSD from sixfold symmetry for each crystallographic hexamer.
For each hexamer, the least-squares superposition of all six subunits upon the permuted configuration (chains ABCDEF superimposed upon BCDEFA) was calculated based upon the C-subdomains.
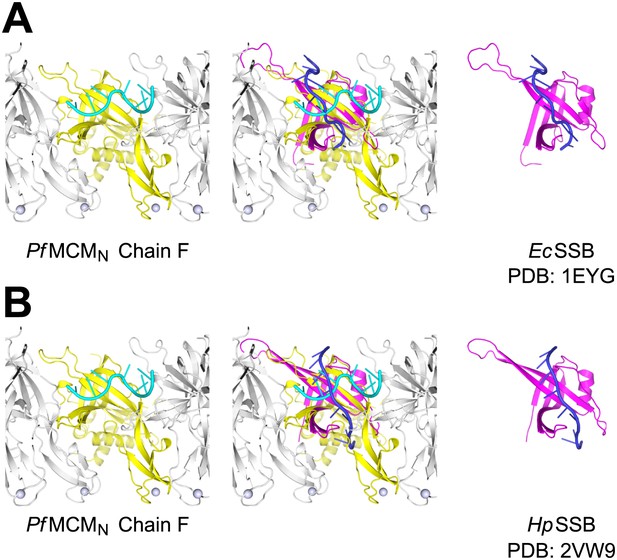
Comparison of ssDNA binding by the PfMCMN OB-fold subdomain C and by a prototypical OB-fold protein, SSB.
Left panels show one monomer of PfMCMN (chain F) colored yellow, and the other subunits of the hexamer colored grey. The ssDNA bound by PfMCMN is in cyan. (A) One monomer of E. coli SSB (Raghunathan et al., 2000) is shown in magenta and its associated ssDNA in blue in the right panel. An overlay with ssDNA bound PfMCMN is shown in the middle. (B) Comparison of PfMCMN:ssDNA with one monomer of H. pylori SSB (Chan et al., 2009) in magenta and its associated ssDNA in blue. Note the ∼90° change in direction of ssDNA for the PfMCM compared to the SSB structures.
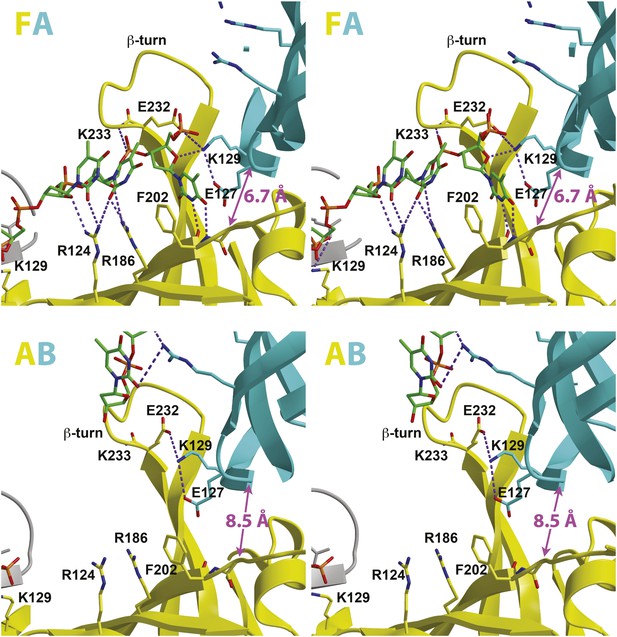
Stereoviews of the protein-DNA interaction details for two subunit interfaces.
The binding predominantly involves residues on the face of the OB-fold of one subunit, yellow, including an interaction between a thymidine base and main-chain atoms of the β-strand. This thymidine is sandwiched between F202 of one subunit and E127 of the adjacent subunit in cyan. Lysine 129 of the neighboring subunit (cyan) interacts with both the DNA and the yellow subunit. The specific interfaces depicted are (top) between chains F (yellow) and A (cyan) and (bottom) between chains A (yellow) and B (cyan). The structural details of DNA-binding appear highly similar at the other interfaces where DNA is observed (see Figure 2—figure supplement 1). The main interactions involve R124 and R186. The presence of ssDNA correlates with the proximity of the two subunits as defined by the distance between the R201 Cα and E127 Cα positions (magenta arrow).
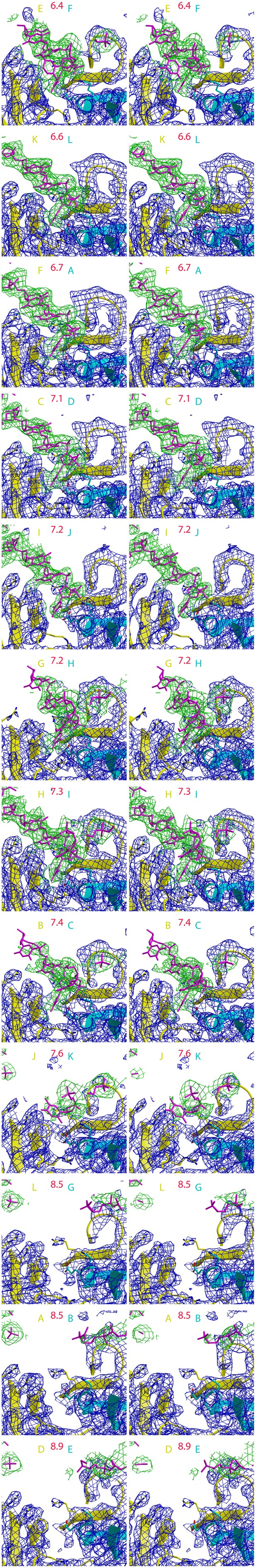
12 stereoimages of the PfMCM interfaces sorted by intersubunit distance to emphasize the correlation with DNA-binding.
Electron density following refmac refinement (refmac FWT map) is displayed around DNA in green, and around the protein in blue. The adjacent subunits are colored yellow and cyan, and the specific chains are noted with the same color scheme. The distance between the R201 Cα atom of the yellow subunit and the E127 Cα atom of the cyan subunit is displayed in red. Electron density for ssDNA is observed for each interface where the distance is less than 7.5 Å.
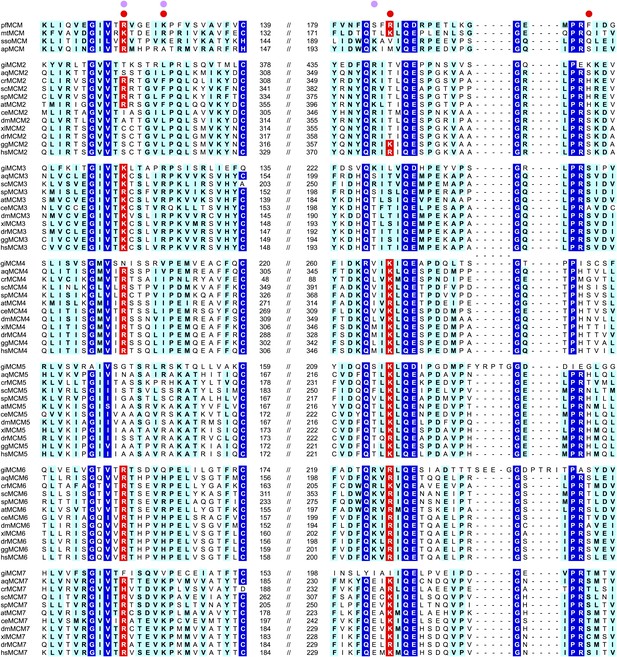
MCM family-specific sequence-alignment in the regions where the strongest interactions with ssDNA are observed.
Globally conserved residues are shaded dark blue, and family-specific conserved residues are shaded light blue. Residues identified to participate in DNA-binding from our structure (red dot) and prior work (Pucci et al., 2004) (lavendar dot) are noted above the sequences. Conserved residue positions for ssDNA binding are shaded red and correspond to R124 and R186 in PfMCM (Figure 2). pf = Pyrococcus furiosus; mt = Methanothermobacter thermautotrophicus; sso = Sulfolobus solfataricus; ap = Aeropyrum pernix; gi = Giardia lamblia; aq = Amphimedon queenslandica; cr = Chlamydomonas reinhardtii; sc = Saccharomyces cerevisiae; sp = Schizosaccharomyces pombe; at = Arabidopsis thaliana; ce = Caenorhabditis elegans; dm = Drosophila melanogaster; xl = Xenopus laevis; dr = Danio rerio; gg = Gallus gallus; hs = Homo sapiens.
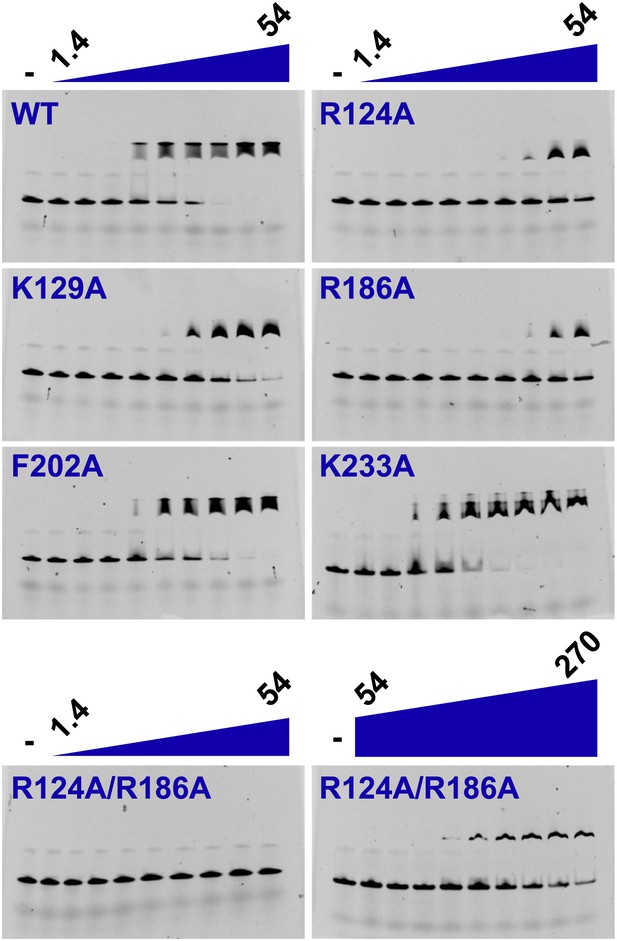
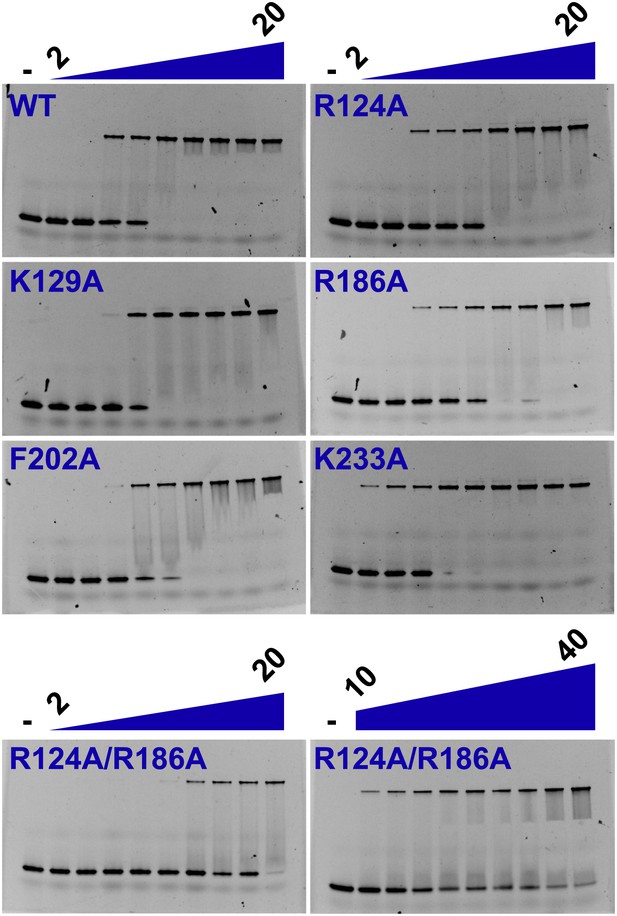
Electrophoretic mobility shift of 40-mer oligo-dT in the presence of PfMCMN.
The ssDNA, 160 nM with a 5′-fluorescein-label, was titrated with increasing concentrations (1.4, 2, 2.7, 6.8, 13.5, 20.3, 27, 40.5, 54 μM) of PfMCMN. The lane marked ‘−’ is loaded with control sample lacking protein. Mutation of residues R124 and R186 significantly impairs binding to ssDNA. The R124A/R186A double mutant was titrated with larger concentrations (54, 81, 108, 135, 162, 189, 216, 243, 270 μM) of PfMCMN in order to detect binding.
Electrophoretic mobility shift assay of a 26-mer dsDNA substrate in the presence of PfMCMN.
The dsDNA, 160 nM with a 5′-fluorescein-label, was titrated with increasing concentrations (2, 3, 4, 5, 7.5, 10, 12.5, 15, 20 μM) of PfMCMN. The lane marked ‘−’ is loaded with control sample lacking protein. The R124A/R186A double mutant was slightly impaired in binding dsDNA and was titrated with larger concentrations (10, 12.5, 15, 17.5, 20, 25, 30, 35, 40 μM) of PfMCMN.
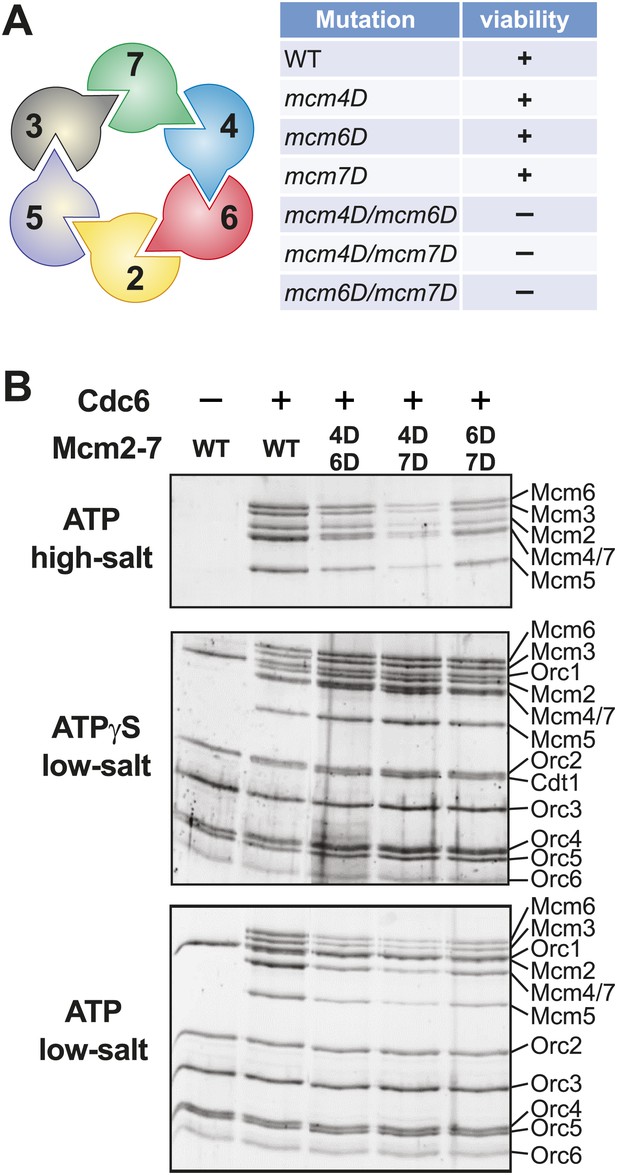
Mutation of two MSSB motifs is lethal and causes helicase loading defects. (A) Mutation of two Mcm4, 6, 7 MSSB motifs is lethal. Subunit arrangement in the Mcm2-7 ring viewed from the C-terminal side. The Mcm4, 6, and 7 subunits are adjacent to each other across from the Mcm2/5 gate. All pairwise combinations of the Mcm4, 6 and 7 MSSB mutants are lethal whereas the individual MSSB mutants are viable. (B) Helicase loading with the indicated MSSB double mutant Mcm2-7 complexes. Three forms of the assay are shown: following a high-salt wash to monitor completion of loading (top panel); in the presence of ATPγS instead of ATP to monitor the initial association of the helicase and all of the helicase loading proteins (ORC, Cdc6 and Cdt1, middle panel); and with ATP following a low salt-wash, allowing bound helicase loading proteins to be maintained (bottom panel). All loading was dependent on Cdc6 and proteins are detected after SDS-PAGE and fluorescent protein staining.
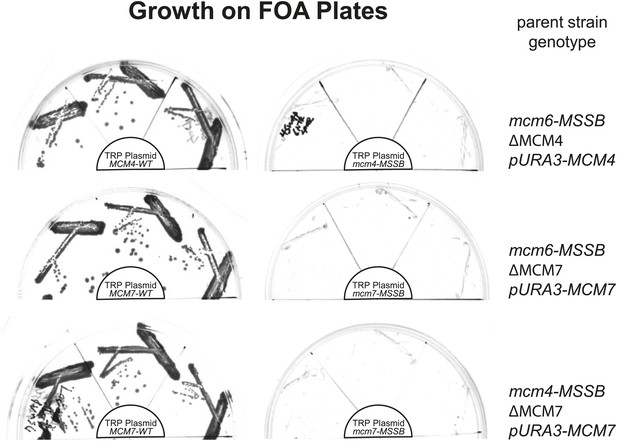
All pairwise combinations of mcm4D, mcm6D and mcm7D mutants were not viable.
The parent strains have an MSSB mutation in the indicated MCM gene. They are also deleted for the indicated MCM gene and depend on a URA+ plasmid expressing a wild-type copy of the same gene for viability. These strains were transformed with the indicated (in the center of each plate) TRP+ plasmid expressing wild-type MCM gene (left) or the MSSB mutant MCM gene (right). Complementation in the absence of the URA3/MCMX-WT plasmid was tested by growth on plates containing 5-FOA (which selects against cells containing the URA3 plasmid). Consistent with the single mcm4, mcm6 or mcm7-MSSB mutations being viable, we observe growth in the presence of the pTRP/MCMX-WT plasmid but no growth when the TRP plasmid contains an MSSB allele in the second MCM gene (creating MSSB mutations in two of the MCM4/6/7 genes).
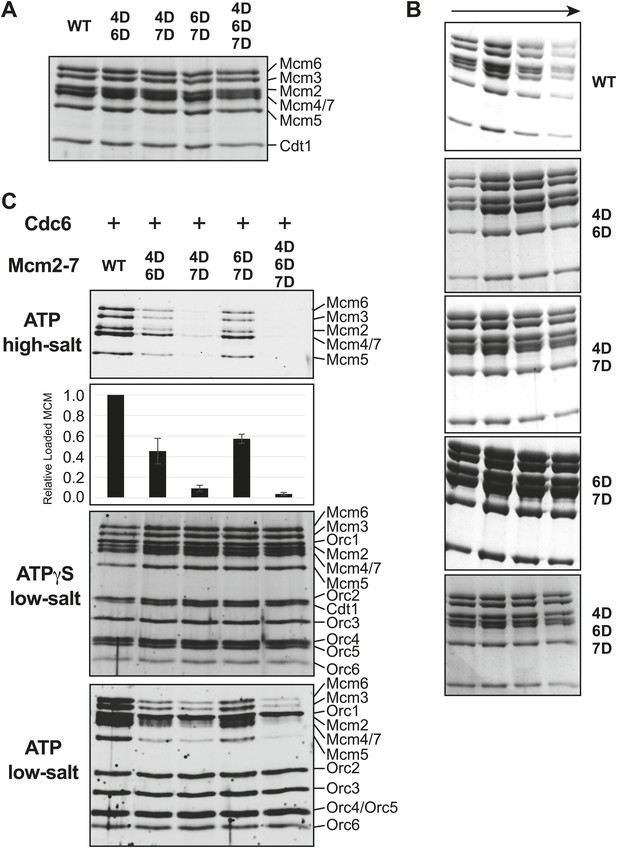
Comparison of wild-type and MSSB double- and triple-mutant Mcm2-7/Cdt1 complexes.
(A) Wild-type and MSSB mutant Mcm2-7/Cdt1 complexes have similar subunit composition. Purified Mcm2-7/Cdt1 complexes were separated by SDS-PAGE and stained with coomassie blue. Mcm2-7 and Cdt1 proteins are indicated. (B) Wild-type and MSSB mutant complexes have similar Stokes radii. Wild-type and mutant Mcm2-7/Cdt1 complexes were separated on a Superdex 200 gel filtration chromatography. Fractions 16–19 of each separation are shown after SDS-PAGE and coomassie blue staining. (C) Helicase loading with the indicated MSSB mutant Mcm2-7 complexes. Three forms of the assay are shown: following a high-salt wash to monitor completion of loading (top panel); in the presence of ATPγS instead of ATP to monitor the initial association of all of the helicase and all of the helicase loading proteins (ORC, Cdc6 and Cdt1, third panel); and with ATP following a low salt-wash, allowing bound helicase loading proteins to be maintained (fourth panel). The relative loading of the Mcm mutants compared to wild-type Mcm2-7 was measured based on three independent loading (high-salt wash) experiments (second panel). Error bars indicate the standard deviation.
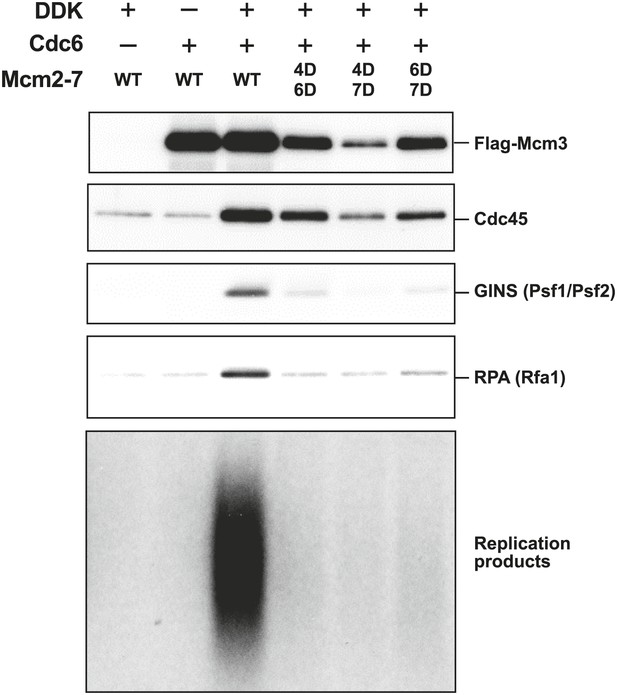
The Mcm2-7 MSSB double mutants are severely defective for in vitro DNA replication.
Proteins associated with the DNA template during DNA replication were analyzed by immunoblotting (top panels) and radiolabeled DNA replication products were analyzed by alkaline agarose electrophoresis (bottom panel). All of the mutants are strongly defective for DNA replication and GINS and RPA DNA template association relative to wild-type Mcm2-7. The levels of Cdc45 and Mcm2-7 (FLAG-Mcm3) association reflected the levels of helicase loading by the same MSSB double mutant Mcm2-7 complexes. Quantitation of these data is shown in Figure 6—figure supplement 1.
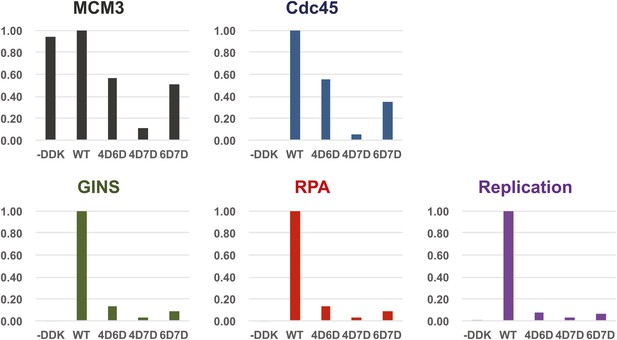
Quantitation of DNA template association of Mcm3, Cdc45, GINS and RPA and DNA replication products for the Mcm2-7 mutants relative to wild-type.
The level of Cdc45 DNA-association mirrored the level of Mcm3 DNA-association. All of the mutants were severely defective for GINS and RPA template association and in vitro replicaiton (bottom right). The levels of GINS and RPA template association (rather than Cdc45 association) correlate with the levels of replication observed.
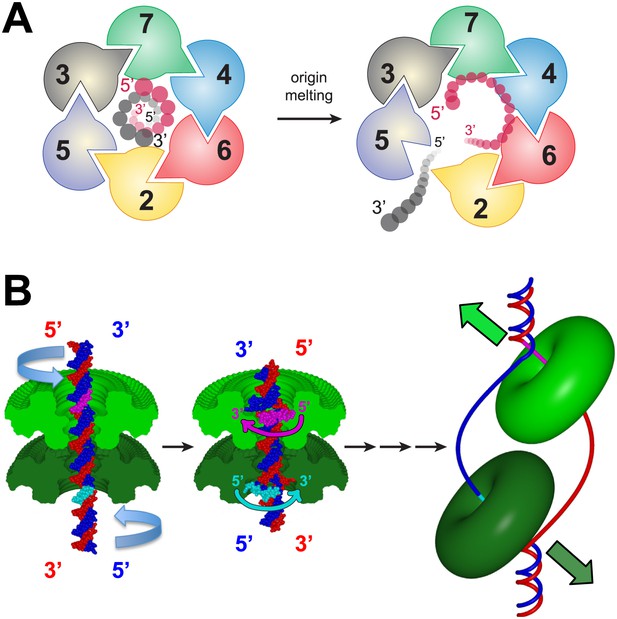
A model for MSSB-dependent selection of the translocating DNA strand during helicase activation.
(A) The defined polarity of ssDNA binding by the MCMN would preferentially bind the leading-strand DNA template. The Mcm2-7 complex N-terminus is shown from the C-terminal side of the complex. This is the side where DNA is expected to enter during translocation. Duplex DNA is first encircled by the ring (left). Only the red strand can readily attain the 5′→3′ clockwise polarity observed in the crystal structure. This strand passes through the ring 5′→3′ from the C- to the N-terminal side and thus is the correct polarity to serve as the translocating strand. We propose the grey, lagging strand DNA template will exit through the Mcm2/5 gate, possibly as a result of accumulation of ssDNA in the central channel (right). (B) A model for selecting the translocating strand during origin melting. Symmetric surfaces in different shades of green represent the two MCMN portions of a double hexamer. The dsDNA is first encircled by the MCM double hexamer (left panel). The dsDNA is driven toward the double hexamer interface by the dsDNA translocase activity of the AAA+ ATPase domains (not shown), which would be located above the light green surface and below the dark green surface. The dsDNA translocation creates strand separation where volume is available, enabling the MSSB to preferentially bind the strand with 5′→3′ clockwise polarity when viewed from the ATPase domain (middle panel). Importantly, the MSSB-bound strand corresponds to the strand upon which the MCM helicase will translocate during unwinding (right panel, magenta at top, cyan at bottom).
Videos
Crystal structure details for PfMCMN:dT30.
The video illustrates the asymmetric unit, which includes two MCM hexamers in a side-by-side orientation. Each subdomain is illustrated in Hexamer 1 to show that the ssDNA interacts with the OB-fold subdomain C. Finally, detailed views of the β-turn and the MCM Single-Stranded DNA binding motif (MSSB) are illustrated.
Animation of a model for MCM to select the translocating strand during origin melting.
Symmetric surfaces in different shades of green represent the two MCMN portions of a double hexamer. The dsDNA is first encircled by the MCM double hexamer. The dsDNA is driven toward the double hexamer interface by the dsDNA translocase activity of the AAA+ ATPase domains (not shown), which would be located above the light green surface and below the dark green surface. The dsDNA translocation creates strand separation where volume is available, enabling the MSSB to preferentially bind the strand with 5′→3′ clockwise polarity when viewed from the ATPase domain. Importantly, the MSSB-bound strand corresponds to the strand upon which the MCM helicase will translocate (magenta at top, cyan at bottom), as shown in Figure 7B, right panel.
Tables
Data collection and refinement statistics
| PfMCMN:dT30 | PfMCMN (no DNA) | |
|---|---|---|
| Data collection | ||
| Space group | P21 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 94.276, 113.397, 196.854 | 122.849, 103.064, 122.435 |
| α, β, γ (°) | 90, 101.354, 90 | 90, 119.85, 90 |
| Resolution (Å) | 50-3.20 (3.31–3.20) | 50-2.65 (2.74–2.65) |
| Rsym | 0.109 (0.786) | 0.100 (0.569) |
| I/σI | 13.4 (1.64) | 16.3 (2.26) |
| Completeness (%) | 100 (100) | 98.8 (98.2) |
| Redundancy | 4.1 (4.1) | 3.7 (3.7) |
| Refinement | ||
| Resolution (Å) | 50-3.20 (3.29–3.20) | 50-2.65 (2.72–2.65) |
| No. reflections | 63497/3376 (4453/218) | 72376/3839 (5183/285) |
| Rwork/Rfree | 0.257/0.294 (0.372/0.373) | 0.259/0.270 (0.484/0.502) |
| No. atoms | ||
| Protein | 24359 | 12258 |
| DNA | 584 | 0 |
| Zn2+ | 12 | 6 |
| Water | 0 | 0 |
| B-factors | ||
| Protein | 129 | 78 |
| DNA | 179 | N/A |
| Zn2+ | 204 | 145 |
| Water | N/A | N/A |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.008 | 0.011 |
| Bond angles (°) | 1.164 | 1.361 |
Yeast strains used in this study
| Strains | Genotype | Source |
|---|---|---|
| ySKM01 | ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 can1-100 bar1::HisG lys2::HisG pep4Δ::unmarked | This study |
| his3::pSKM004 (GAL1,10-MCM2, Flag-MCM3) leu::pSKM007 (GAL1,10-Cdt1-Strep, GAL4) | ||
| lys::pSKM002 (GAL1,10-MCM4, MCM5) | ||
| trp::pSKM003 (GAL1,10-MCM6, MCM7) | ||
| ySKM02 | ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 can1-100 bar1::HisG lys2::HisG pep4Δ::KanMX6 | This study |
| MCM4-V5 (NatMX4) MCM6-V5 (CaURA3MX4) MCM7-V5 (HphMX4) | ||
| his3::pSKM004 (GAL1,10-MCM2, Flag-MCM3) leu::pSKM007 (GAL1,10-Cdt1-Strep, GAL4) | ||
| lys::pSKM008 (GAL1,10-mcm4[R334A/K398A], MCM5) trp::pSKM009 (GAL1,10-mcm6[R296A/R360A], MCM7) | ||
| ySKM03 | ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 can1-100 bar1::HisG lys2::HisG pep4Δ::KanMX6 | This study |
| MCM4-V5 (NatMX4) MCM6-V5 (CaURA3MX4) MCM7-V5 (HphMX4) | ||
| his3::pSKM004 (GAL1,10-MCM2, Flag-MCM3) leu::pSKM007 (GAL1,10-Cdt1-Strep, GAL4) | ||
| lys::pSKM008 (GAL1,10-mcm4[R334A/K398A], MCM5) trp::pSKM010 (GAL1,10-MCM6, mcm7[R247A/K314A]) | ||
| ySKM04 | ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 can1-100 bar1::HisG lys2::HisG pep4Δ::KanMX6 | This study |
| MCM4-V5 (NatMX4) MCM6-V5 (CaURA3MX4) MCM7-V5 (HphMX4) | ||
| his3::pSKM004 (GAL1,10-MCM2, Flag-MCM3) leu::pSKM007 (GAL1,10-Cdt1-Strep, GAL4) | ||
| lys::pSKM002 (GAL1,10-MCM4, MCM5) | ||
| trp::pSKM011 (GAL1,10-mcm6[R296A/R360A], mcm7[R247A/K314A]) | ||
| ySKM05 | ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 can1-100 bar1::HisG lys2::HisG pep4Δ::KanMX6 | This study |
| MCM4-V5 (NatMX4) MCM6-V5 (CaURA3MX4) MCM7-V5 (HphMX4) | ||
| his3::pSKM004 (GAL1,10-MCM2, Flag-MCM3) leu::pSKM007 (GAL1,10-Cdt1-Strep, GAL4) | ||
| lys::pSKM008 (GAL1,10-mcm4[R334A/K398A], MCM5) | ||
| trp::pSKM011 (GAL1,10-mcm6[R296A/R360A], mcm7[R247A/K314A]) | ||
| ySKS10 | ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 can1-100 lys2::HisG pep4Δ::Hph cdc7-4 | This study |
| pol1-5xFlag (KanMX4) | ||
| leu::pSKS001 (GAL1,10-Cdc45-V5, Sld3-S) | ||
| lys::pSKS002 (GAL1,10-Dpb11-VSVG, Sld2-HSV) | ||
| ura::pSKS003 (Gal1,10-Cdc28, Clb5) | ||
| his::pSKS004 (Gal1,10-Sld7) | ||
| ySKS11 | ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 can1-100 lys2::HisG pep4Δ::Hph cdc7-4 | This study |
| pol2-5xFlag (KanMX4) | ||
| leu::pSKS001 (GAL1,10-Cdc45-V5, Sld3-S) | ||
| lys::pSKS002 (GAL1,10-Dpb11-VSVG, Sld2-HSV) | ||
| ura::pSKS003 (Gal1,10-Cdc28, Clb5) | ||
| his::pSKS004 (Gal1,10-Sld7) | ||
| ASY1059.1 | MatA, ade2-1, ura3-11, his3-11,15, leu2-3,12, can-100, trp1-1 | (Schwacha and Bell, 2001) |
| mcm4 Δ::hisG/pAS412 (ARS/CEN URA+ PMCM5-MCM4-HA/HIS) | ||
| ASY2157 | MatA, ade2-1, ura3-11, his3-11,15, leu2-3,12, can-100, trp1-1, lys2::hisG, bar1::hisG, PEP4 Δ::KANMX4, | (Schwacha and Bell, 2001) |
| MCM6 Δ::HISMX6/pAS452 (ARS/CEN URA+ PMCM5-MCM6-HA/HIS) | ||
| ASY1050.1 | MatA, ade2-1, ura3-11, his3-11,15, leu2-3,12, can-100, trp1-1 | (Schwacha and Bell, 2001) |
| mcm7Δ::hisG/pGEMCDC47 (ARS/CEN URA+ MCM7) | ||
| ySKC01 | MatA, ade2-1, ura3-11, his3-11,15, leu2-3,12, can-100, trp1-1 | This study |
| mcm4 Δ::hisG/pAS412 (ARS/CEN URA+ PMCM5-MCM4-HA/HIS) mcm6::LEU2-PMCM5-mcm6[R296A/R360A] | ||
| ySKC02 | MatA, ade2-1, ura3-11, his3-11,15, leu2-3,12, can-100, trp1-1 | This study |
| mcm7Δ::hisG/pGEMCDC47 (ARS/CEN URA+ MCM7) mcm6::LEU2-PMCM5-mcm6[R296A/R360A] | ||
| ySKC03 | MatA, ade2-1, ura3-11, his3-11,15, leu2-3,12, can-100, trp1-1 | This study |
| mcm7Δ::hisG/pGEMCDC47 (ARS/CEN URA+ MCM7) mcm4::NatMX4-PMCM5-mcm4[R334A/K398A] |
Yeast plasmids used in this study
| Plasmids | Description | Source |
|---|---|---|
| pSKM002 | pRS307 (GAL1,10-MCM4, MCM5) | This study |
| pSKM003 | pRS404 (GAL1,10-MCM6, MCM7) | This study |
| pSKM004 | pRS403 (GAL1,10-MCM2, Flag-MCM3) | This study |
| pSKM007 | pRS305 (GAL1,10-Cdt1-Strep, GAL4) | This study |
| pSKM008 | pRS307 (GAL1,10-mcm4[R334A/K398A], MCM5) | This study |
| pSKM009 | pRS404 (GAL1,10-mcm6[R296A/R360A], MCM7) | This study |
| pSKM010 | pRS404 (GAL1,10-MCM6, mcm7[R247A/K314A]) | This study |
| pSKM011 | pRS404 (GAL1,10-mcm6[R296A/R360A], mcm7[R247A/K314A]) | This study |
| pSKS001 | pRS305 (GAL1,10-Cdc45-V5, Sld3-S) | This study |
| pSKS002 | pRS307 (GAL1,10-Dpb11-VSVG, Sld2-HSV) | This study |
| pSKS003 | pRS306 (Gal1,10-Cdc28, Clb5) | This study |
| pSKS004 | pRS403 (Gal1,10-Sld7) | This study |
| pSKC001 | pRS414 (PMCM5-mcm4[R334A/K398A]) | This study |
| pSKC002 | pRS414 (PMCM5- mcm6[R296A/R360A]) | This study |
| pSKC003 | pRS414 (PMCM5-mcm7[R247A/K314A]) | This study |
| pSKC004 | pRS414 (PMCM4-NatMX4-PMCM5- mcm4[R334A/K398A]) | This study |
| pSKC005 | pRS414 (PMCM6-LEU2-PMCM5- mcm6[R296A/R360A]) | This study |





